Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains, Media, and Growth Conditions
2.2. RNA Modification
2.3. Isolation of Total RNA
2.4. Primer Extension and Data Processing
2.5. RNA Secondary Structure Modeling and Analysis
2.6. Signal-To-Background Ratio and Correlation Calculation
2.7. Comparison of SHAPE Reactivities and SHAPE-Directed Structural Models of Ty1 gRNA
2.8. Computing Regions of Large Absolute SHAPE Reactivities Changes
3. Results
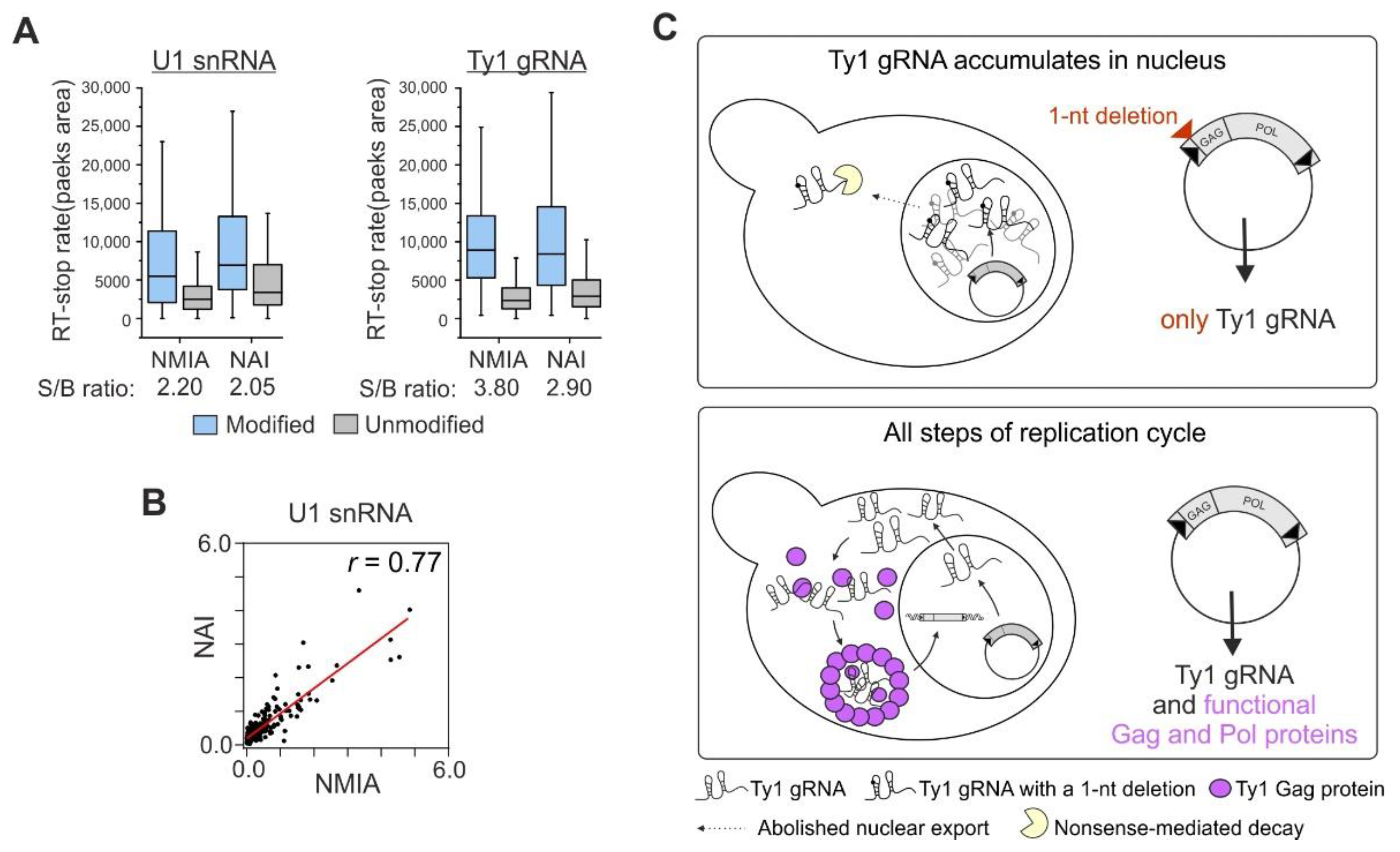
3.1. NMIA Successfully Modifies RNA in the Yeast Cell Nucleus
3.2. The Experimental Strategy Used to Explore Ty1 gRNA Structure in the Nucleus
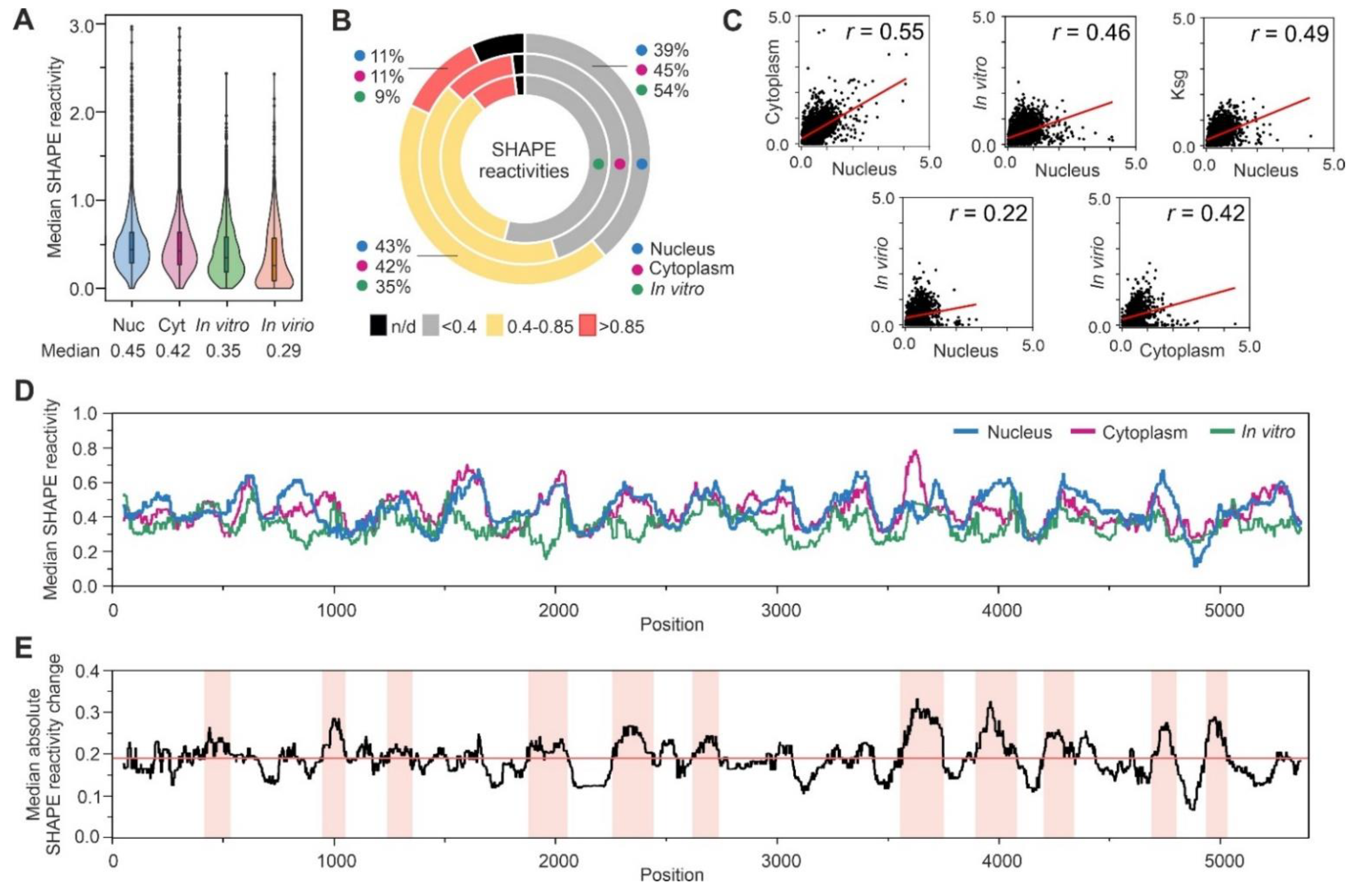
3.3. Analysis of SHAPE Structural Data for Nuclear Ty1 Grna
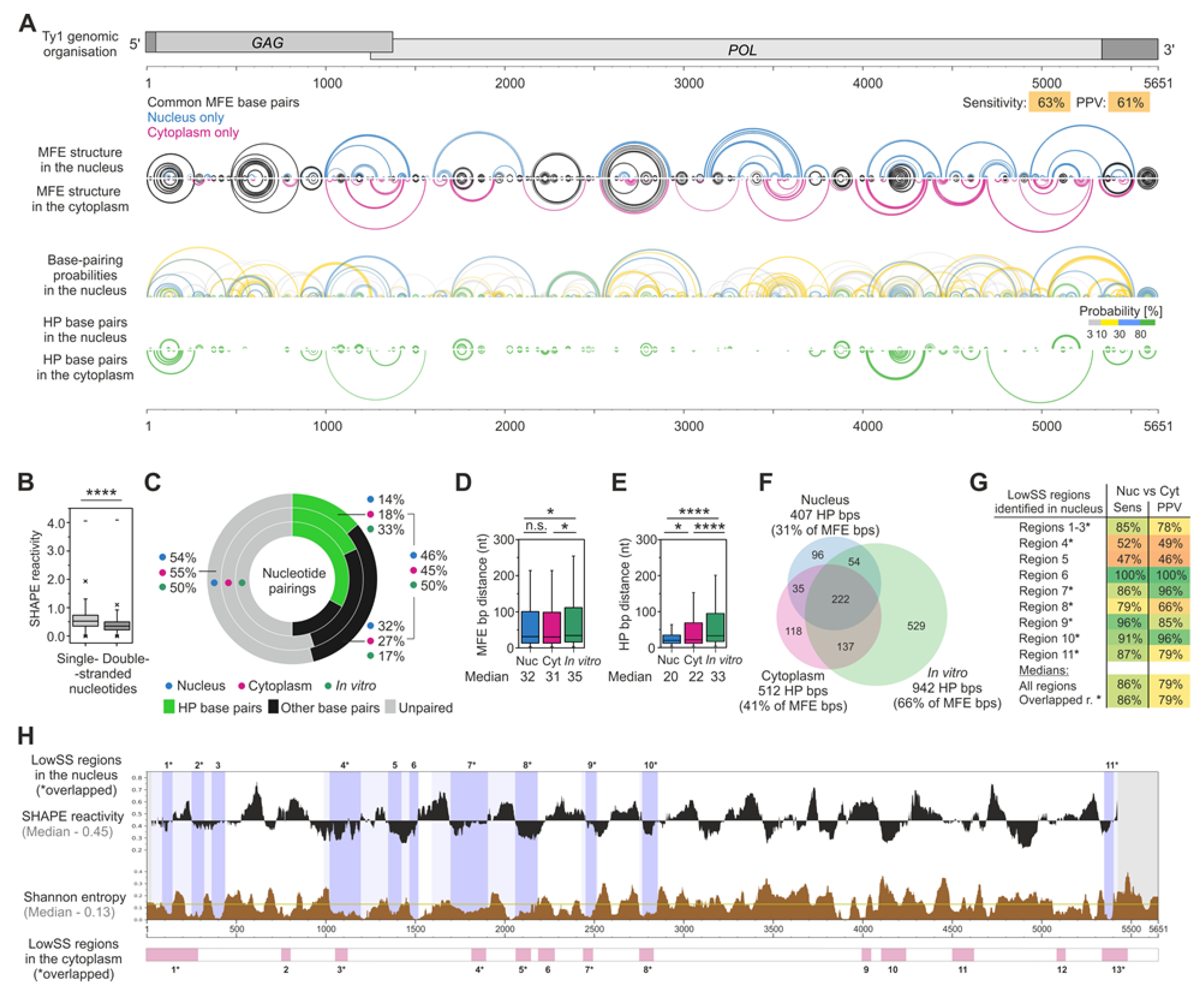
3.4. The MFE Structure Model of the Nuclear Ty1 RNA Genome
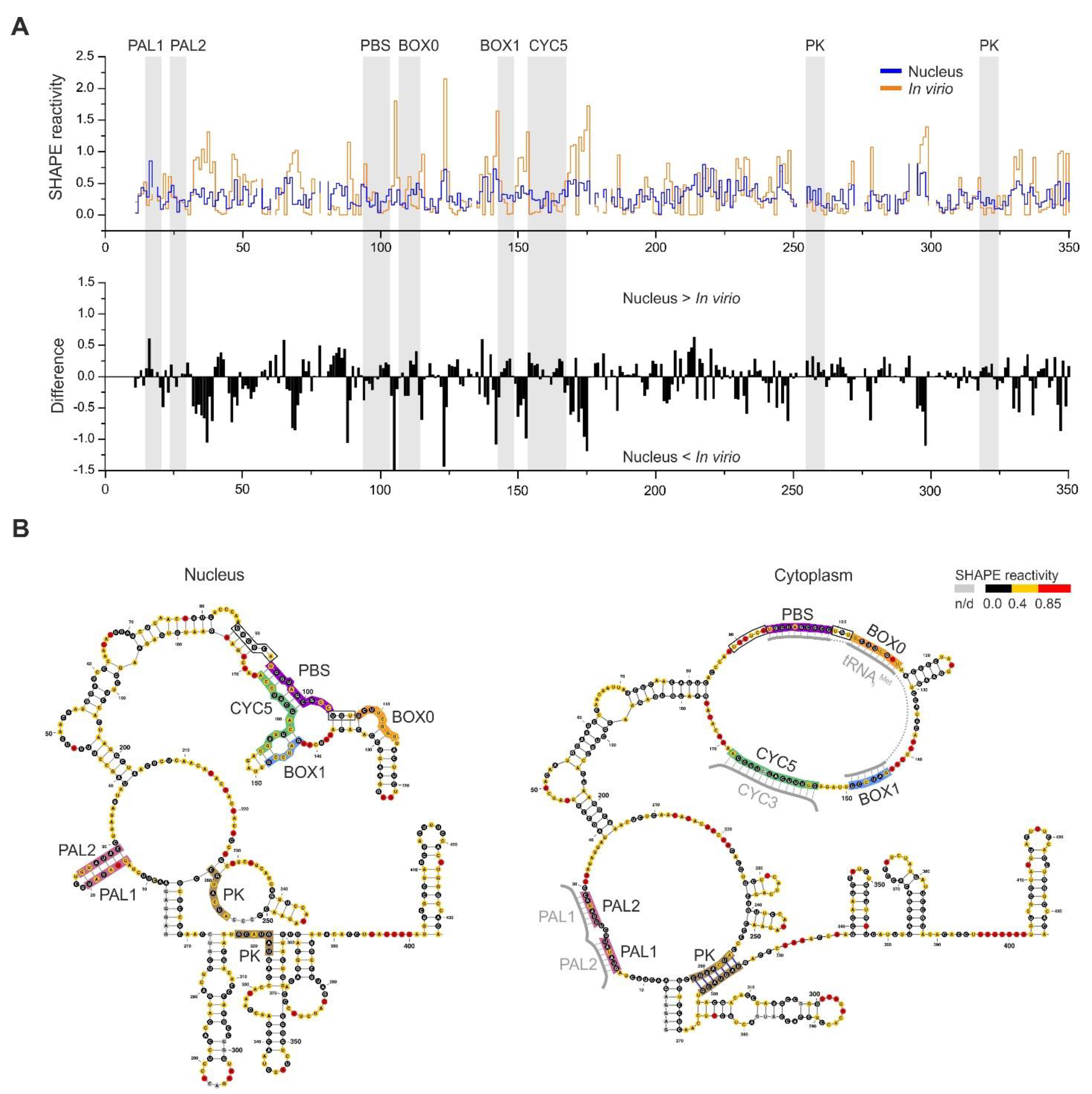
3.5. RNA–RNA Interactions Mediated by Gag In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eichler, E.E.; Sankoff, D. Structural dynamics of eukaryotic chromosome evolution. Science 2003, 301, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Blomberg, J.; Coffin, J.M.; Dasgupta, I.; Fan, H.; Geering, A.D.; Gifford, R.; Harrach, B.; Hull, R.; Johnson, W.; et al. Ortervirales: New Virus Order Unifying Five Families of Reverse-Transcribing Viruses. J. Virol. 2018, 92, e00515-18. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Bleykasten-Grosshans, C.; Fabrizio, R.; Friedrich, A.; Schacherer, J. Species-Wide Transposable Element Repertoires Retrace the Evolutionary History of the Saccharomyces cerevisiae Host. Mol. Biol. Evol. 2021, 38, 4334–4345. [Google Scholar] [CrossRef]
- Czaja, W.; Bensasson, D.; Ahn, H.W.; Garfinkel, D.J.; Bergman, C.M. Evolution of Ty1 copy number control in yeast by horizontal transfer and recombination. PLoS Genet. 2020, 16, e1008632. [Google Scholar] [CrossRef]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty elements transpose through an RNA intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef]
- Mellor, J.; Fulton, S.M.; Dobson, M.J.; Wilson, W.; Kingsman, S.M.; Kingsman, A.J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature 1985, 313, 243–246. [Google Scholar] [CrossRef]
- Elder, R.T.; Loh, E.Y.; Davis, R.W. Rna from the Yeast Transposable Element Ty1 Has Both Ends in the Direct Repeats, a Structure Similar to Retrovirus Rna. Proc. Natl. Acad. Sci. USA 1983, 80, 2432–2436. [Google Scholar] [CrossRef]
- Curcio, M.J.; Lutz, S.; Lesage, P. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 2015, 3, 1–35. [Google Scholar]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef]
- Pachulska-Wieczorek, K.; Le Grice, S.F.J.; Purzycka, K.J. Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3. Viruses-Basel. 2016, 8, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malagon, F.; Jensen, T.H. T-body formation precedes virus-like particle maturation in S-cerevisiae. RNA Biol. 2011, 8, 184–189. [Google Scholar] [CrossRef]
- Checkley, M.A.; Mitchell, J.A.; Eizenstat, L.D.; Lockett, S.J.; Garfinkel, D.J. Ty1 gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 2013, 14, 57–69. [Google Scholar] [CrossRef]
- Nishida, Y.; Pachulska-Wieczorek, K.; Blaszczyk, L.; Saha, A.; Gumna, J.; Garfinkel, D.J.; Purzycka, K.J. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res. 2015, 43, 7414–7431. [Google Scholar] [CrossRef] [PubMed]
- Gumna, J.; Purzycka, K.J.; Ahn, H.W.; Garfinkel, D.J.; Pachulska-Wieczorek, K. Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA. RNA Biol. 2019, 16, 1749–1763. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, G.; Bampi, C.; Wilhelm, M.; Wilhelm, F.X.; Darlix, J.L. A 5′—3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002, 21, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, G.; Ficheux, D.; Darlix, J.L. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000, 275, 19210–19217. [Google Scholar] [CrossRef]
- Malagon, F.; Jensen, T.H. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 2008, 28, 6022–6032. [Google Scholar] [CrossRef]
- Nonet, M.; Scafe, C.; Sexton, J.; Young, R. Eukaryotic Rna-Polymerase Conditional Mutant That Rapidly Ceases Messenger-Rna Synthesis. Mol. Cell. Biol. 1987, 7, 1602–1611. [Google Scholar] [CrossRef]
- Curcio, M.J.; Hedge, A.M.; Boeke, J.D.; Garfinkel, D.J. Ty Rna Levels Determine the Spectrum of Retrotransposition Events That Activate Gene-Expression in Saccharomyces-Cerevisiae. Mol. Gen. Genet. 1990, 220, 213–221. [Google Scholar] [CrossRef]
- Friant, S.; Heyman, T.; Bystrom, A.S.; Wilhelm, M.; Wilhelm, F.X. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol. 1998, 18, 799–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamache, E.R.; Doh, J.H.; Ritz, J.; Laederach, A.; Bellaousov, S.; Mathews, D.H.; Curcio, M.J. Structure-Function Model for Kissing Loop Interactions That Initiate Dimerization of Ty1 RNA. Viruses 2017, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Boeke, J.D. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: Analysis of mini-Ty1 elements. Mol. Cell. Biol. 1990, 10, 2695–2702. [Google Scholar] [CrossRef]
- Andrzejewska, A.; Zawadzka, M.; Gumna, J.; Garfinkel, D.J.; Pachulska-Wieczorek, K. In vivo structure of the Ty1 retrotransposon RNA genome. Nucleic Acids Res. 2021, 49, 2878–2893. [Google Scholar] [CrossRef]
- Garfinkel, D.J.; Nyswaner, K.; Wang, J.; Cho, J.Y. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 2003, 165, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.P.; Liti, G.; Stefanisko, K.M.; Nyswaner, K.M.; Chang, C.; Louis, E.J.; Garfinkel, D.J. Analysis of a Ty1-less variant of Saccharomyces paradoxus: The gain and loss of Ty1 elements. Yeast 2004, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, D.J.; Mastrangelo, M.F.; Sanders, N.J.; Shafer, B.K.; Strathern, J.N. Transposon tagging using Ty elements in yeast. Genetics 1988, 120, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Winston, F.; Dollard, C.; Ricupero-Hovasse, S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 1995, 11, 53–55. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Vasa, S.M.; Guex, N.; Wilkinson, K.A.; Weeks, K.M.; Giddings, M.C. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 2008, 14, 1979–1990. [Google Scholar] [CrossRef]
- Gumna, J.; Zok, T.; Figurski, K.; Pachulska-Wieczorek, K.; Szachniuk, M. RNAthor—fast, accurate normalization, visualization and statistical analysis of RNA probing data resolved by capillary electrophoresis. PLoS ONE 2020, 15, e0239287. [Google Scholar]
- Smola, M.J.; Rice, G.M.; Busan, S.; Siegfried, N.A.; Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015, 10, 1643–1669. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Hajdin, C.E.; Bellaousov, S.; Huggins, W.; Leonard, C.W.; Mathews, D.H.; Weeks, K.M. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA 2013, 110, 5498–5503. [Google Scholar] [CrossRef]
- Purzycka, K.J.; Legiewicz, M.; Matsuda, E.; Eizentstat, L.D.; Lusvarghi, S.; Saha, A.; Le Grice, S.F.J.; Garfinkel, D.J. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 2013, 41, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M. SHAPE Directed Discovery of New Functions in Large RNAs. Acc. Chem. Res. 2021, 54, 2502–2517. [Google Scholar] [CrossRef]
- Marinus, T.; Fessler, A.B.; Ogle, C.A.; Incarnato, D. A novel SHAPE reagent enables the analysis of RNA structure in living cells with unprecedented accuracy. Nucleic Acids Res. 2021, 49, e34. [Google Scholar] [CrossRef]
- Busan, S.; Weidmann, C.A.; Sengupta, A.; Weeks, K.M. Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry 2019, 58, 2655–2664. [Google Scholar] [CrossRef]
- Lee, B.; Flynn, R.A.; Kadina, A.; Guo, J.K.; Kool, E.T.; Chang, H.Y. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 2017, 23, 169–174. [Google Scholar] [CrossRef]
- Huang, Q.; Purzycka, K.J.; Lusvarghi, S.; Li, D.H.; LeGrice, S.F.J.; Boeke, J.D. Retrotransposon Ty1 RNA contains a 5′-terminal long-range pseudoknot required for efficient reverse transcription. RNA 2013, 19, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, L.; Biesiada, M.; Saha, A.; Garfinkel, D.J.; Purzycka, K.J. Structure of Ty1 Internally Initiated RNA Influences Restriction Factor Expression. Viruses 2017, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; McQueary, H.; Hall, D.W.; Philippsen, P.; Garfinkel, D.J.; Bergman, C.M. Genome Assembly of the Ty1-Less Saccharomyces paradoxus Strain DG1768. Microbiol. Resour. Announc. 2022, 11, e0086821. [Google Scholar] [CrossRef] [PubMed]
- Schuwirth, B.S.; Day, J.M.; Hua, C.W.; Janssen, G.R.; Dahlberg, A.E.; Cate, J.H.; Vila-Sanjurjo, A. Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 2006, 13, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fazal, F.M.; Li, P.; Broughton, J.P.; Lee, B.; Tang, L.; Huang, W.Z.; Kool, E.T.; Chang, H.Y.; Zhang, Q.F.C. RNA structure maps across mammalian cellular compartments. Nat. Struct. Mol. Biol. 2019, 26, 322–330. [Google Scholar]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.E.; Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [Google Scholar] [CrossRef]
- Chapman, K.B.; Bystrom, A.S.; Boeke, J.D. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 1992, 89, 3236–3240. [Google Scholar] [CrossRef]
- Keeney, J.B.; Chapman, K.B.; Lauermann, V.; Voytas, D.F.; Astrom, S.U.; von Pawel-Rammingen, U.; Bystrom, A.; Boeke, J.D. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol. 1995, 15, 217–226. [Google Scholar] [CrossRef]
- Kawakami, K.; Pande, S.; Faiola, B.; Moore, D.P.; Boeke, J.D.; Farabaugh, P.J.; Strathern, J.N.; Nakamura, Y.; Garfinkel, D.J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 1993, 135, 309–320. [Google Scholar] [CrossRef]
- Huber, R.G.; Lim, X.N.; Ng, W.C.; Sim, A.Y.L.; Poh, H.X.; Shen, Y.; Lim, S.Y.; Sundstrom, K.B.; Sun, X.Y.; Aw, J.G.; et al. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Na.t Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Simon, L.M.; Morandi, E.; Luganini, A.; Gribaudo, G.; Martinez-Sobrido, L.; Turner, D.H.; Oliviero, S.; Incarnato, D. In vivo analysis of influenza A mRNA secondary structures identifies critical regulatory motifs. Nucleic Acids Res. 2019, 47, 7003–7017. [Google Scholar] [CrossRef] [PubMed]
- Manfredonia, I.; Nithin, C.; Ponce-Salvatierra, A.; Ghosh, P.; Wirecki, T.K.; Marinus, T.; Ogando, N.S.; Snijder, E.J.; van Hemert, M.J.; Bujnicki, J.M.; et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020, 48, 12436–12452. [Google Scholar] [CrossRef] [PubMed]
- Huston, N.C.; Wan, H.; Strine, M.S.; Tavares, R.D.A.; Wilen, C.B.; Pyle, A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell. 2021, 81, 584–598.e5. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Adams, R.L.; Lindenbach, B.D.; Pyle, A.M. The In Vivo and In Vitro Architecture of the Hepatitis C Virus RNA Genome Uncovers Functional RNA Secondary and Tertiary Structures. J. Virol. 2022, 96, e0194621. [Google Scholar] [CrossRef] [PubMed]
- Herschlag, D.; Bonilla, S.; Bisaria, N. The Story of RNA Folding, as Told in Epochs. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Morandi, E.; Anselmi, F.; Simon, L.M.; Basile, G.; Oliviero, S. In vivo probing of nascent RNA structures reveals principles of cotranscriptional folding. Nucleic Acids Res. 2017, 45, 9716–9725. [Google Scholar] [CrossRef]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 3692–3697. [Google Scholar] [CrossRef]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513–11518. [Google Scholar] [CrossRef]
- Liu, Z.S.; Liu, Q.; Yang, X.F.; Zhang, Y.Y.; Norris, M.; Chen, X.X.; Cheema, J.; Zhang, H.K.; Ding, Y.L. In vivo nuclear RNA structurome reveals RNA-structure regulation of mRNA processing in plants. Genome Biol. 2021, 22, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Novoa, E.M.; Vejnar, C.E.; Yartseva, V.; Takacs, C.M.; Kellis, M.; Giraldez, A.J. Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nat. Struct. Mol. Biol. 2018, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zawadzka, M.; Andrzejewska-Romanowska, A.; Gumna, J.; Garfinkel, D.J.; Pachulska-Wieczorek, K. Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome. Viruses 2022, 14, 2007. https://doi.org/10.3390/v14092007
Zawadzka M, Andrzejewska-Romanowska A, Gumna J, Garfinkel DJ, Pachulska-Wieczorek K. Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome. Viruses. 2022; 14(9):2007. https://doi.org/10.3390/v14092007
Chicago/Turabian StyleZawadzka, Małgorzata, Angelika Andrzejewska-Romanowska, Julita Gumna, David J. Garfinkel, and Katarzyna Pachulska-Wieczorek. 2022. "Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome" Viruses 14, no. 9: 2007. https://doi.org/10.3390/v14092007
APA StyleZawadzka, M., Andrzejewska-Romanowska, A., Gumna, J., Garfinkel, D. J., & Pachulska-Wieczorek, K. (2022). Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome. Viruses, 14(9), 2007. https://doi.org/10.3390/v14092007





