Viruses Ubiquity and Diversity in Atacama Desert Endolithic Communities
Abstract
:1. Introduction
2. Materials and Methods
3. Results
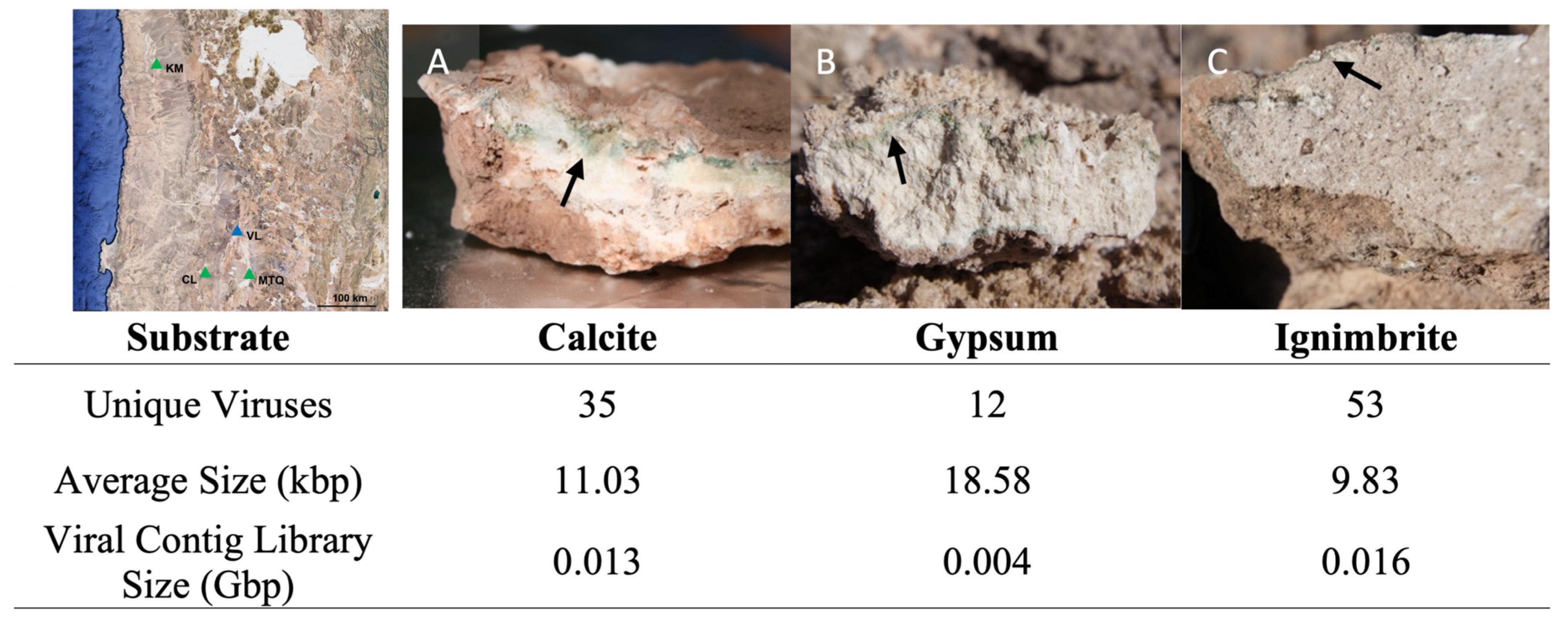
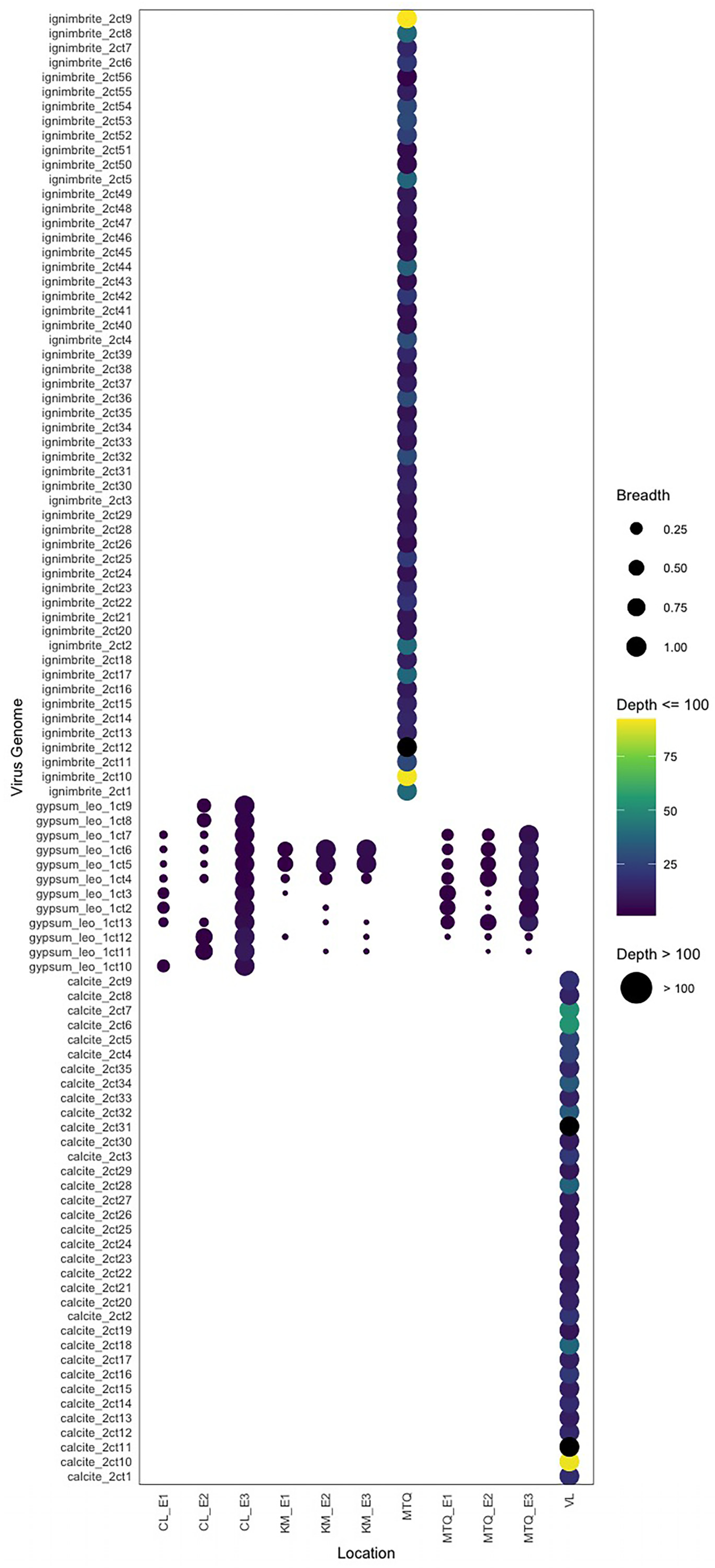
3.1. Endolithic Metagenomes from the Atacama Desert Harbor Diverse Viruses
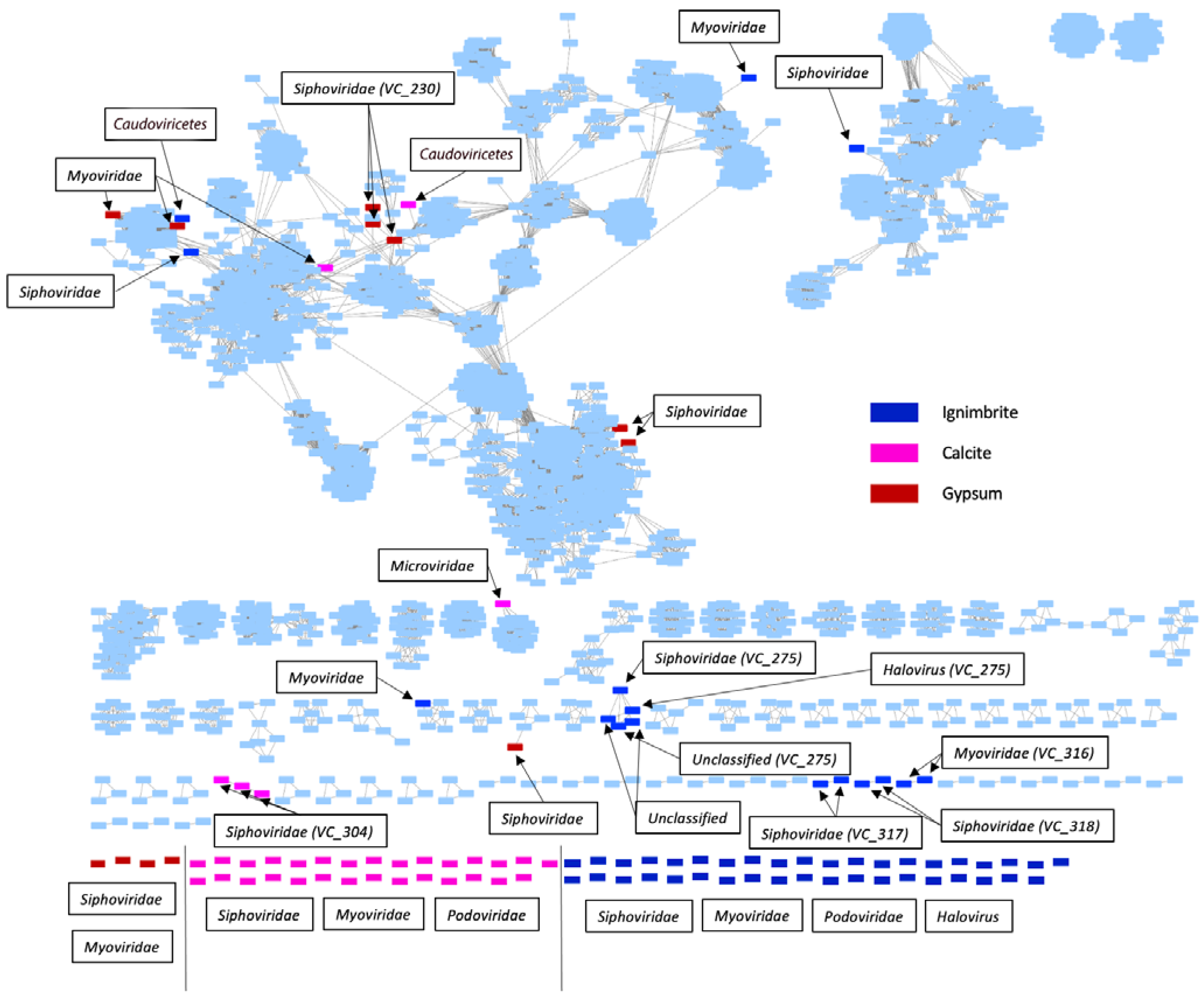
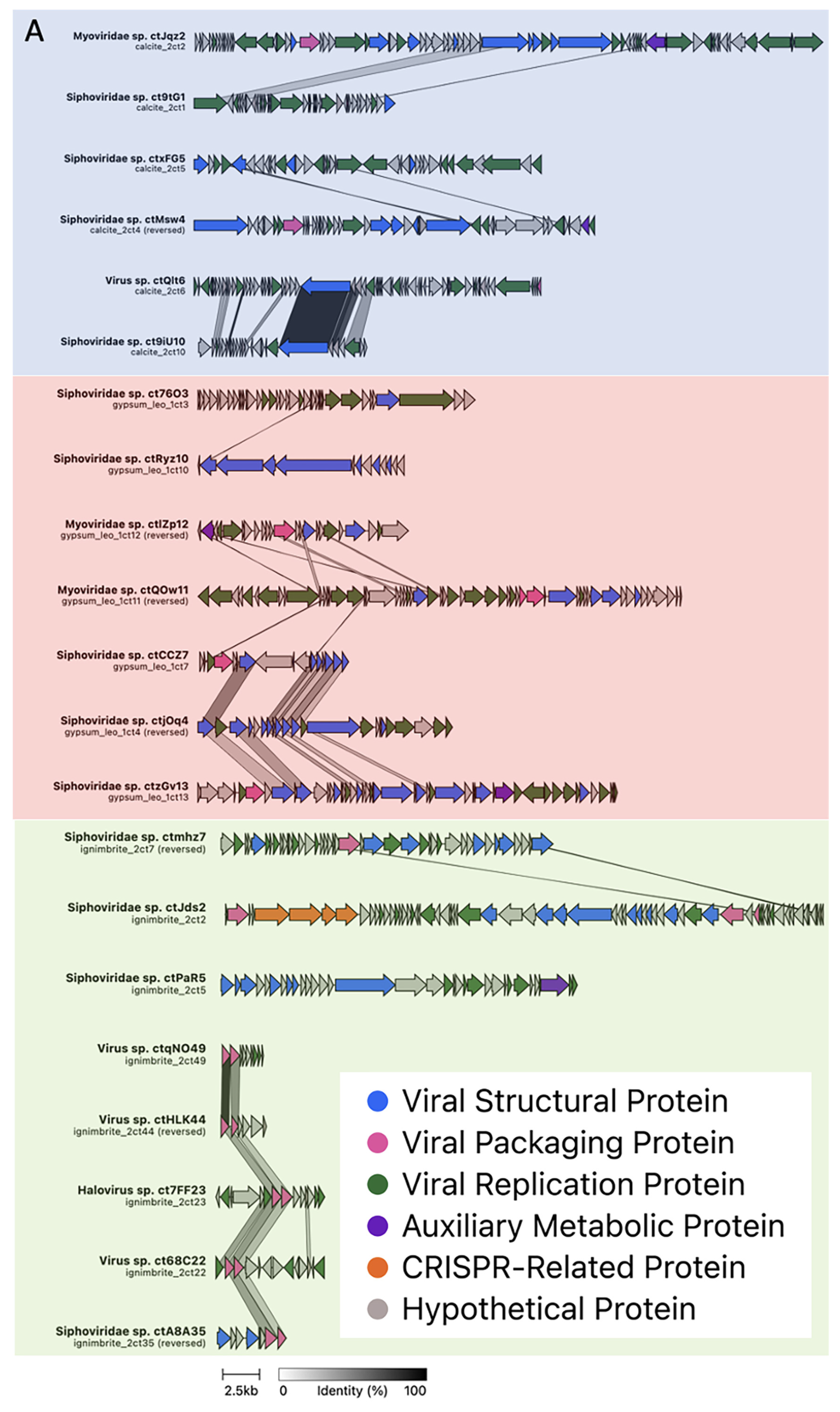
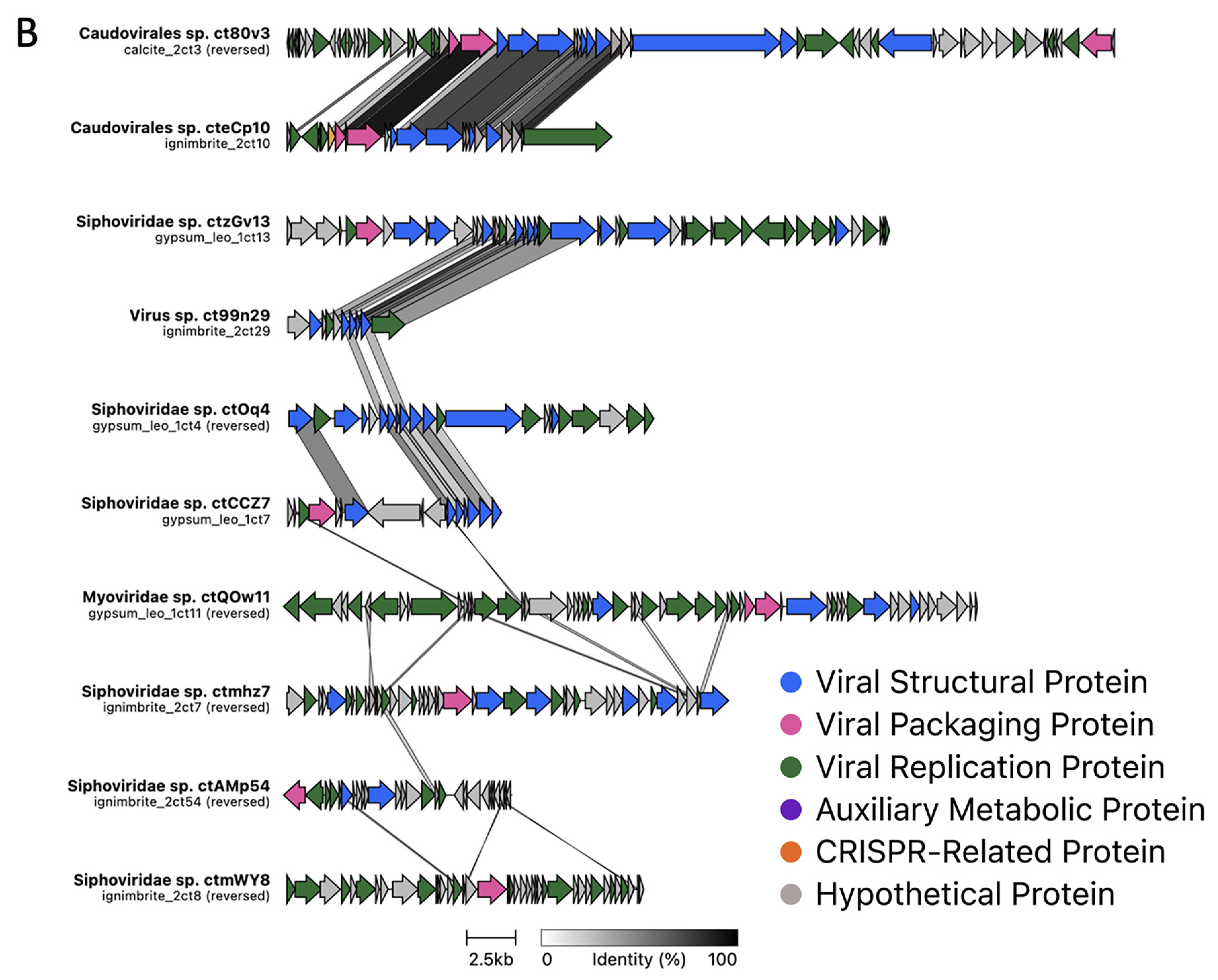
3.2. Viral Genome Organization Is Highly Diverse across Endoliths
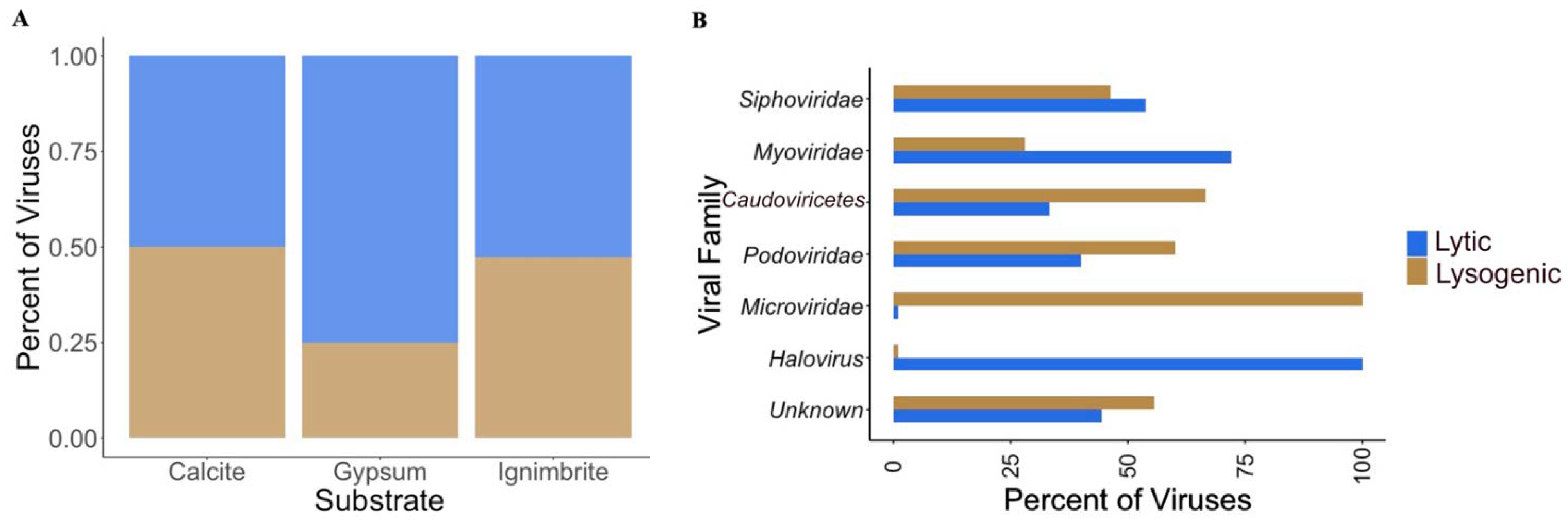
3.3. Viral Infective Strategies Vary by Viral Family
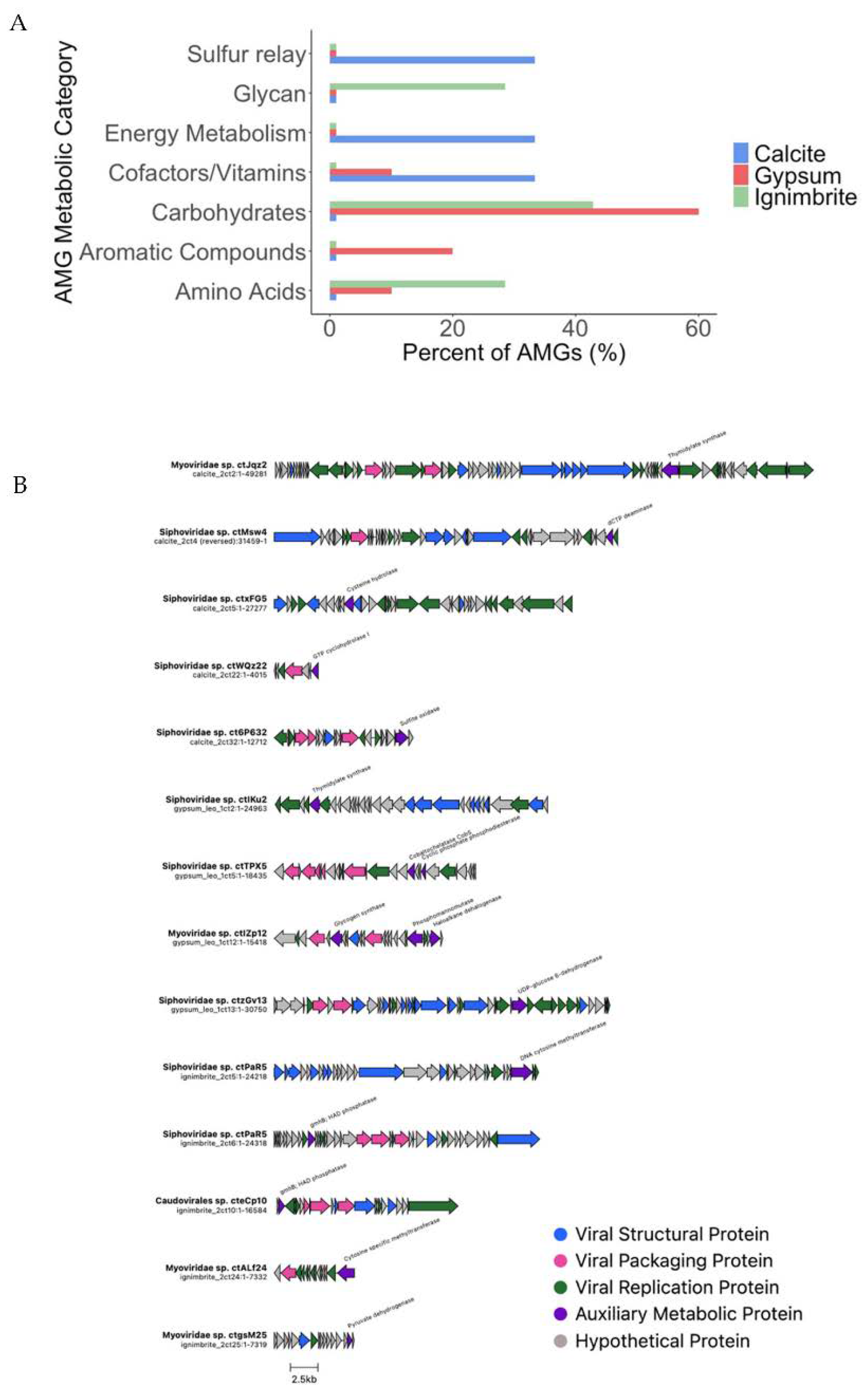
3.4. Endolithic Viruses Contain a Diverse Array of Auxiliary Metabolic Genes
3.5. Viral–Host Relationship Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suttle, C. The viriosphere: The greatest biological diversity on Earth and driver of global processes. Environ Microbiol. 2005, 7, 481–482. [Google Scholar] [CrossRef] [PubMed]
- Sabet, S. Halophilic viruses. In Advances in Understanding the Biology of Halophilic Microorganisms; Vreeland, R.H., Ed.; Springer: Heidelberg, Germany; London, UK; New York, NY, USA, 2012; pp. 81–116. [Google Scholar]
- Tarafder, A.K.; von Kugelgen, A.; Mellul, A.J.; Schulze, U.; Aarts, D.; Bharat, T.A.M. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 4724–4731. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.J.; Lee, S.H.; Cho, J.C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Zayed, A.A.; Wainaina, J.M.; Dominguez-Huerta, G.; Pelletier, E.; Guo, J.; Mohssen, M.; Tian, F.; Pratama, A.A.; Bolduc, B.; Zablocki, O.; et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 2022, 376, 156–162. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Heyerhoff, B.; Engelen, B.; Bunse, C. Auxiliary Metabolic Gene Functions in Pelagic and Benthic Viruses of the Baltic Sea. Front. Microbiol. 2022, 13, 863620. [Google Scholar] [CrossRef]
- Hwang, Y.; Rahlff, J.; Schulze-Makuch, D.; Schloter, M.; Probst, A.J. Diverse Viruses Carrying Genes for Microbial Extremotolerance in the Atacama Desert Hyperarid Soil. mSystems 2021, 6, e00385-21. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s virome. Nature 2016, 536, 425–430. [Google Scholar] [CrossRef]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef]
- Davidson, A.R.; Lu, W.T.; Stanley, S.Y.; Wang, J.; Mejdani, M.; Trost, C.N.; Hicks, B.T.; Lee, J.; Sontheimer, E.J. Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annu. Rev. Biochem. 2020, 89, 309–332. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, K.; Duhaime, M.B.; Breier, J.A.; Wendt, K.A.; Toner, B.M.; Dick, G.J. Sulfur oxidation genes in diverse deep-sea viruses. Science 2014, 344, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed]
- Kepner, R.L., Jr.; Wharton, R.A., Jr.; Suttle, C.A. Viruses in Antarctic lakes. Limnol. Oceanogr. 1998, 43, 1754–1761. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Kramer, R.; Van Goethem, M.W.; Makhalanyane, T.P.; Hogg, I.; Cowan, D.A. Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome 2017, 5, 83. [Google Scholar] [CrossRef]
- Zablocki, O.; van Zyl, L.; Adriaenssens, E.M.; Rubagotti, E.; Tuffin, M.; Cary, S.C.; Cowan, D. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of antarctic soils. Appl. Environ. Microbiol. 2014, 80, 6888–6897. [Google Scholar] [CrossRef] [PubMed]
- Bezuidt, O.K.I.; Lebre, P.H.; Pierneef, R.; León-Sobrino, C.; Adriaenssens, E.M.; Cowan, D.A.; Van de Peer, Y.; Makhalanyane, T.P. Phages Actively Challenge Niche Communities in Antarctic Soils. mSystems 2020, 5, e00234-20. [Google Scholar] [CrossRef]
- Prigent, M.; Leroy, M.; Confalonieri, F.; Dutertre, M.; DuBow, M.S. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles 2005, 9, 289–296. [Google Scholar] [CrossRef]
- Scola, V.; Ramond, J.B.; Frossard, A.; Zablocki, O.; Adriaenssens, E.M.; Johnson, R.M.; Seely, M.; Cowan, D.A. Namib Desert Soil Microbial Community Diversity, Assembly, and Function Along a Natural Xeric Gradient. Microb. Ecol. 2018, 75, 193–203. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Van Zyl, L.J.; Cowan, D.A.; Trindade, M.I. Metaviromics of Namib Desert Salt Pans: A Novel Lineage of Haloarchaeal Salterproviruses and a Rich Source of ssDNA Viruses. Viruses 2016, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Zablocki, O.; Adriaenssens, E.M.; Frossard, A.; Seely, M.; Ramond, J.B.; Cowan, D. Metaviromes of Extracellular Soil Viruses along a Namib Desert Aridity Gradient. Genome Announc. 2017, 5, e01470-16. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Van Zyl, L.; De Maayer, P.; Rubagotti, E.; Rybicki, E.; Tuffin, M.; Cowan, D.A. Metagenomic analysis of the viral community in Namib Desert hypoliths. Environ. Microbiol. 2015, 17, 480–495. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Gelsinger, D.R.; Ma, B.; Wierzchos, J.; Ravel, J.; Ascaso, C.; Artieda, O.; Davila, A.; DiRuggiero, J. Functional analysis of the archaea, bacteria, and viruses from a halite endolithic microbial community. Environ. Microbiol. 2016, 18, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Uritskiy, G.; Tisza, M.J.; Gelsinger, D.R.; Munn, A.; Taylor, J.; DiRuggiero, J. Cellular life from the three domains and viruses are transcriptionally active in a hypersaline desert community. Environ. Microbiol. 2020, 23, 3401–3417. [Google Scholar] [CrossRef]
- Pointing, S.B.; Belnap, J. Microbial colonization and controls in dryland systems. Nat. Rev. Microbiol. 2012, 10, 551–562. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Robinson, C.K.; Barnum, T.; Fricke, W.F.; Davila, A.F.; Jedynak, B.; McKay, C.; DiRuggiero, J. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 2013, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A.; Hopkins, D.W.; Jones, B.E.; Maggs-Kolling, G.; Majewska, R.; Ramond, J.B. Microbiomics of Namib Desert habitats. Extremophiles 2020, 24, 17–29. [Google Scholar] [CrossRef]
- Qu, E.; Ertekin, E.; DiRuggiero, J. Biology of Desert Endolithic Habitats. In Microbiology of Hot Deserts; Cowan, D., Ramond, J.B., Eds.; Springer Nature: Berlin, Germany, 2022. [Google Scholar]
- Wierzchos, J.; Casero, M.C.; Artieda, O.; Ascaso, C. Endolithic microbial habitats as refuges for life in polyextreme environment of the Atacama Desert. Curr. Opin. Microbiol. 2018, 43, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.J.; Pace, N.R. Endolithic microbial ecosystems. Annu. Rev. Microbiol. 2007, 61, 331–347. [Google Scholar] [CrossRef]
- Uritskiy, G.; Getsin, S.; Munn, A.; Gomez-Silva, B.; Davila, A.; Glass, B.; Taylor, J.; DiRuggiero, J. Halophilic microbial community compositional shift after a rare rainfall in the Atacama Desert. ISME J. 2019, 13, 2737–2749. [Google Scholar] [CrossRef] [Green Version]
- DiRuggiero, J.; Wierzchos, J.; Robinson, C.K.; Souterre, T.; Ravel, R.; Artieda, O.; Souza-Egipsy, V.; Ascaso, C. Microbial Colonization of Chasmoendolithic Habitats in the Hyper-arid Zone of the Atacama Desert. Biogeosciences 2013, 10, 2439–2450. [Google Scholar] [CrossRef]
- Meslier, V.; Casero, M.C.; Daily, M.; Wierchos, J.; Ascaso, C.; Artieda, O.; McCullough, P.R.; DiRuggiero, J. Fundamental drivers for endolithic microbial community assemblies in the hyperarid Atacama Desert. Env. Microbiol. 2018, 20, 1765–1781. [Google Scholar] [CrossRef]
- Qu, E.B.; Omelon, C.R.; Oren, A.; Meslier, V.; Cowan, D.A.; Maggs-Kolling, G.; DiRuggiero, J. Trophic Selective Pressures Organize the Composition of Endolithic Microbial Communities From Global Deserts. Front. Microbiol. 2019, 10, 2952. [Google Scholar] [CrossRef] [PubMed]
- Wierzchos, J.; DiRuggiero, J.; Vítek, P.; Artieda, O.; Souza-Egipsy, V.; Skaloud, P.; Tisza, M.; Davila, A.F.; Vílchez, C.; Garbayo, I.; et al. Adaptation strategies of endolithic chlorophototrophs to survive the hyperarid and extreme solar radiation environment of the Atacama Desert. Front. Microbiol. 2015, 6, 934. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.A. Antiquity of aridity in the Chilean Atacama Desert. Geomorphology 2006, 73, 101–114. [Google Scholar] [CrossRef]
- Connon, S.A.; Lester, E.D.; Shafaat, H.S.; Obenhuber, D.C.; Ponce, A. Bacterial diversity in hyperarid Atacama Desert soils. J. Geophys. Res. 2007, 112, G04S17. [Google Scholar] [CrossRef]
- Houston, J. Variability of precipitation in the Atacama desert: Its causes and hydrological impact. Int. J. Climatol. 2006, 26, 2181–2198. [Google Scholar] [CrossRef]
- Ertekin, E.; Meslier, V.; Browning, A.; Treadgold, J.; DiRuggiero, J. Functional and taxonomic diversity is driven by substrate architecture in endolithic communities from extreme environments. Env. Microbial. 2021, 23, 3937–3956. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitua, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Camargo, A.P.; Schulz, F.; Eloe-Fadrosh, E.; Roux, S.; Kyrpides, N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef]
- Tisza, M.J.; Belford, A.K.; Dominguez-Huerta, G.; Bolduc, B.; Buck, C.B. Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus. Evol. 2021, 7, veaa100. [Google Scholar] [CrossRef] [PubMed]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e20. [Google Scholar] [CrossRef]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef]
- Shaffer, M.; Borton, M.A.; McGivern, B.B.; Zayed, A.A.; La Rosa, S.L.; Solden, L.M.; Liu, P.; Narrowe, A.B.; Rodriguez-Ramos, J.; Bolduc, B.; et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020, 48, 8883–8900. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Munoz, D.; Shah, S.A.; Sorensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Prestel, E.; Regeard, C.; Salamitou, S.; Neveu, J.; Dubow, M.S. The bacteria and bacteriophages from a Mesquite Flats site of the Death Valley desert. Antonie Van Leeuwenhoek 2013, 103, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
- Prestel, E.; Salamitou, S.; DuBow, M.S. An examination of the bacteriophages and bacteria of the Namib desert. J. Microbiol. 2008, 46, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Villamor, J.; Ramos-Barbero, M.D.; Gonzalez-Torres, P.; Gabaldon, T.; Rossello-Mora, R.; Meseguer, I.; Martinez-Garcia, M.; Santos, F.; Anton, J. Characterization of ecologically diverse viruses infecting co-occurring strains of cosmopolitan hyperhalophilic Bacteroidetes. ISME J. 2018, 12, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, B.; Bhattacharjee, A.S.; Coutinho, F.H.; Goel, R.K. Viruses and Their Interactions with Bacteria and Archaea of Hypersaline Great Salt Lake. Front. Microbiol. 2021, 12, 701414. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fernandez, C.A.; Wilburn, P.; Davila, A.; DiRuggiero, J. Adaptations of endolithic communities to abrupt environmental changes in a hyper-arid desert. Sci. Rept. 2022, in press.
- Robinson, C.K.; Wierzchos, J.; Black, C.; Crits-Christoph, A.; Ma, B.; Ravel, J.; Ascaso, C.; Artieda, O.; Valea, S.; Roldan, M.; et al. Microbial diversity and the presence of algae in halite endolithic communities are correlated to atmospheric moisture in the hyper-arid zone of the Atacama Desert. Environ. Microbiol. 2015, 17, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Uritskiy, G.; Munn, A.; Dailey, M.; Gelsinger, D.R.; Getsin, S.; Davila, A.; McCullough, P.R.; Taylor, J.; DiRuggiero, J. Environmental Factors Driving Spatial Heterogeneity in Desert Halophile Microbial Communities. Front. Microbiol. 2020, 11, 578669. [Google Scholar] [CrossRef]
- Thompson, L.R.; Zeng, Q.; Kelly, L.; Huang, K.H.; Singer, A.U.; Stubbe, J.; Chisholm, S.W. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, E757–E764. [Google Scholar] [CrossRef]
- Tiedje, J.; Cho, J.; Murray, A.; Treves, D.; Xia, B.; Zhou, J. Soil teeming with life: New frontiers in soil science. In Sustainable Management of Soil Organic Matter; Rees, R.M., Ball, B., Campbell, C., Watson, C.A., Eds.; CAB International: Wallingford, UK, 2001; pp. 393–412. [Google Scholar]
- Stovicek, A.; Kim, M.; Or, D.; Gillor, O. Microbial community response to hydration-desiccation cycles in desert soil. Sci. Rep. 2017, 7, 45735. [Google Scholar] [CrossRef] [Green Version]
| Virus-ID | tRNA Length (bp) | % GC | tRNA Identity | Putative Host (1) | Number of Predicted Hosts |
|---|---|---|---|---|---|
| Gypsum_1ct11 (Myoviridae sp. ctQ0w11) | 94 | 83.0 | Alanine | Bacteria | 1 |
| Calcite_2ct6 (virus sp. ctQIt6) | 76 | 47.4 | Valine | Bacteria Cyanobacteria | 3 1 |
| Calcite_2ct17 (Siphoviridae sp. Ct19o17) | 79 | 82.3 | Alanine | Bacteria | 2 |
| Calcite_2ct32 (Siphoviridae sp. Ct6P632) | 85 | 61.2 | Tyrosine | Bacteria | 1 |
| Ignimbrite_2ct7 (Siphoviridae sp. ctmhz7) | 77 | 70.1 | Methionine | Bacteria | 1 |
| Ignimbrite_2ct8 (Siphoviridae sp. ctmWY8) | 76 | 73.7 | Glycine | Bacteria | 1 |
| Virus | Viral CRISPR Protein | Putative Host (1) | % Identity | e-Value |
|---|---|---|---|---|
| Ignimbrite_2ct1 (Myoviridae sp. ctz491) | CRISPR-associated endonuclease Cas12f3 | Bacteria | 84 | 5 × 10−19 |
| Spacer 1 | Bacteria | 100 | 4 × 10−10 | |
| Spacer 2 | Bacteria | 100 | 4 × 10−10 | |
| Spacer 3 | Bacteria | 100 | 1 × 10−10 | |
| Spacer 4 | Bacteria | 100 | 5 × 10−8 | |
| Spacer 5 | Bacteria | 100 | 9 × 10−13 | |
| Ignimbrite_2ct2 (Siphoviridae sp. ctJds2) | CRISPR-associated protein Csx17 | Bacteria | 100 | 0.0 |
| Actinobacteria | 39 | 3 × 10−78 | ||
| Proteobacteria | 40 | 3 × 10−61 | ||
| CRISPR-associated endonuclease/helicase Cas3 | Bacteria | 100 | 0.0 | |
| Actinobacteria | 50 | 0.0 | ||
| CRISPR-associated protein Csb1 | Bacteria | 100 | 0.0 | |
| Actinobacteria | 53 | 7 × 10−105 | ||
| CRISPR-associated protein Csb2 | Bacteria | 100 | 0.0 | |
| Actinobacteria | 79 | 6 × 10−138 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busse, L.; Tisza, M.; DiRuggiero, J. Viruses Ubiquity and Diversity in Atacama Desert Endolithic Communities. Viruses 2022, 14, 1983. https://doi.org/10.3390/v14091983
Busse L, Tisza M, DiRuggiero J. Viruses Ubiquity and Diversity in Atacama Desert Endolithic Communities. Viruses. 2022; 14(9):1983. https://doi.org/10.3390/v14091983
Chicago/Turabian StyleBusse, Leora, Mike Tisza, and Jocelyne DiRuggiero. 2022. "Viruses Ubiquity and Diversity in Atacama Desert Endolithic Communities" Viruses 14, no. 9: 1983. https://doi.org/10.3390/v14091983
APA StyleBusse, L., Tisza, M., & DiRuggiero, J. (2022). Viruses Ubiquity and Diversity in Atacama Desert Endolithic Communities. Viruses, 14(9), 1983. https://doi.org/10.3390/v14091983







