Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation, Purification, and Amplification of Bacteriophages
2.2. Microbiological Characterization of the Phage
2.2.1. Determination of the Host Range
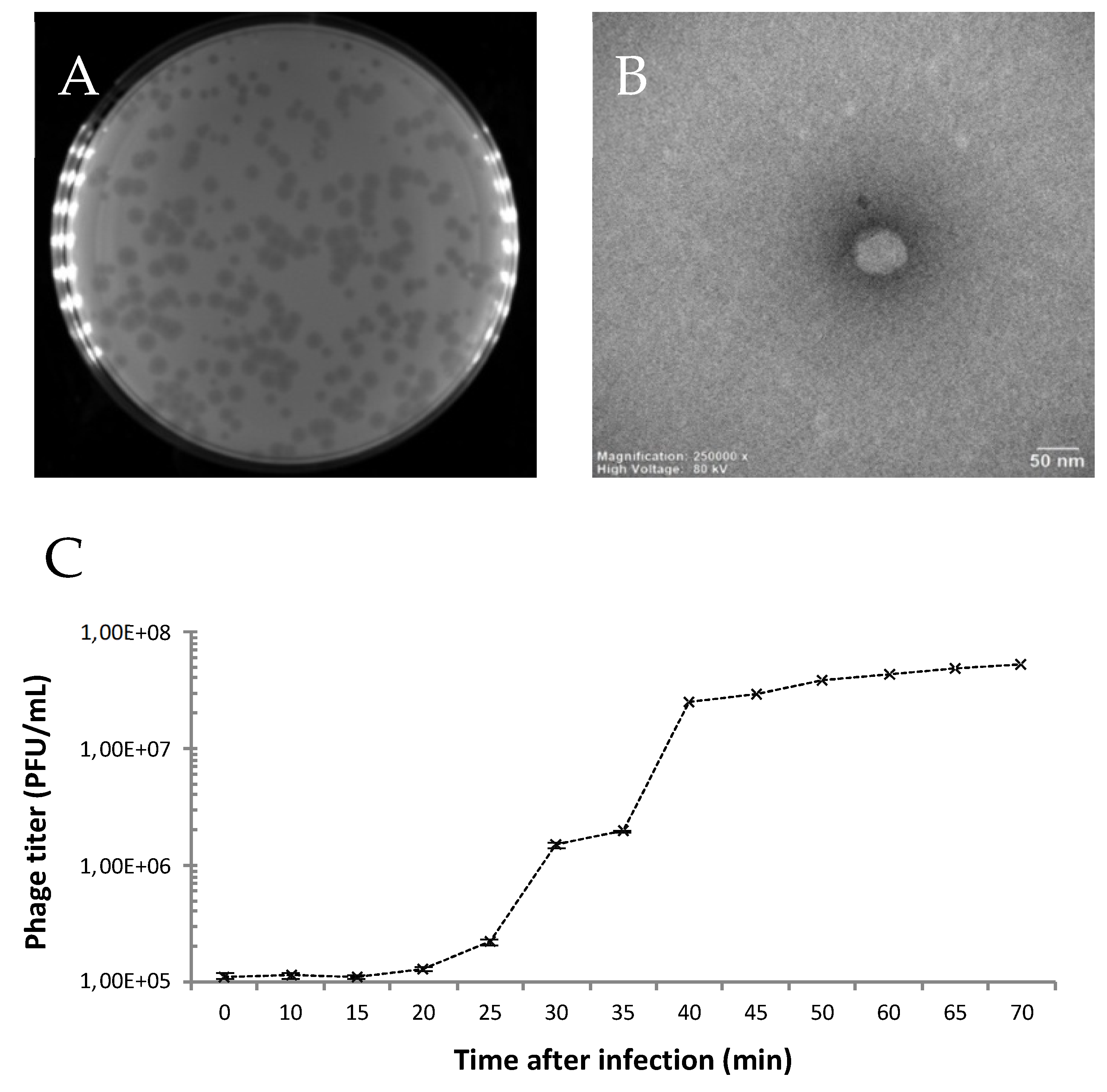
2.2.2. Electron Microscopy
2.2.3. One-Step Growth Curve
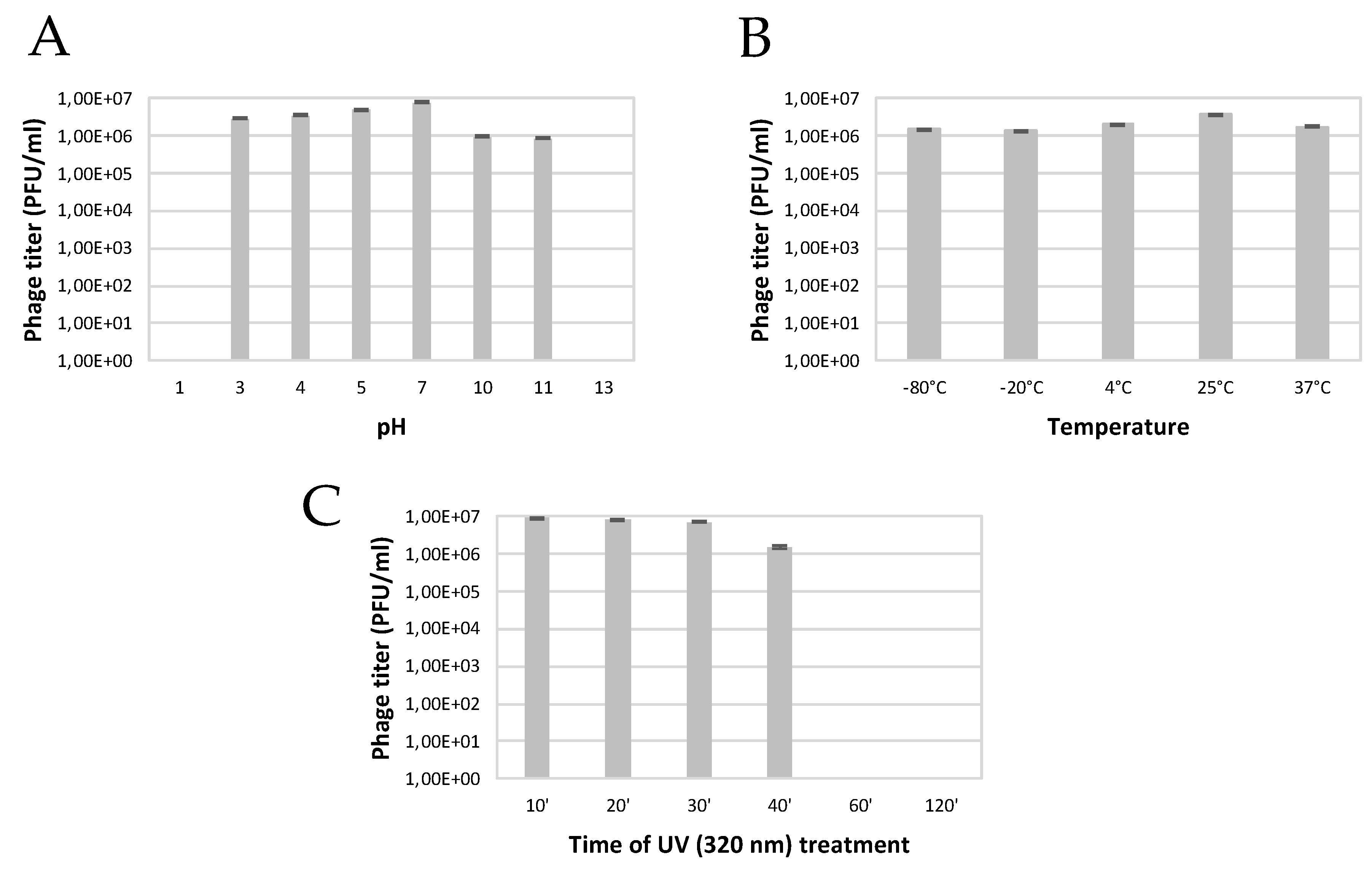
2.2.4. Influence of pH on Phage Viability
2.2.5. Effect of Temperature on Phage Viability
2.2.6. Effect of UV on Phage Viability
2.3. Molecular Characterization of the Phage
Genome Analysis
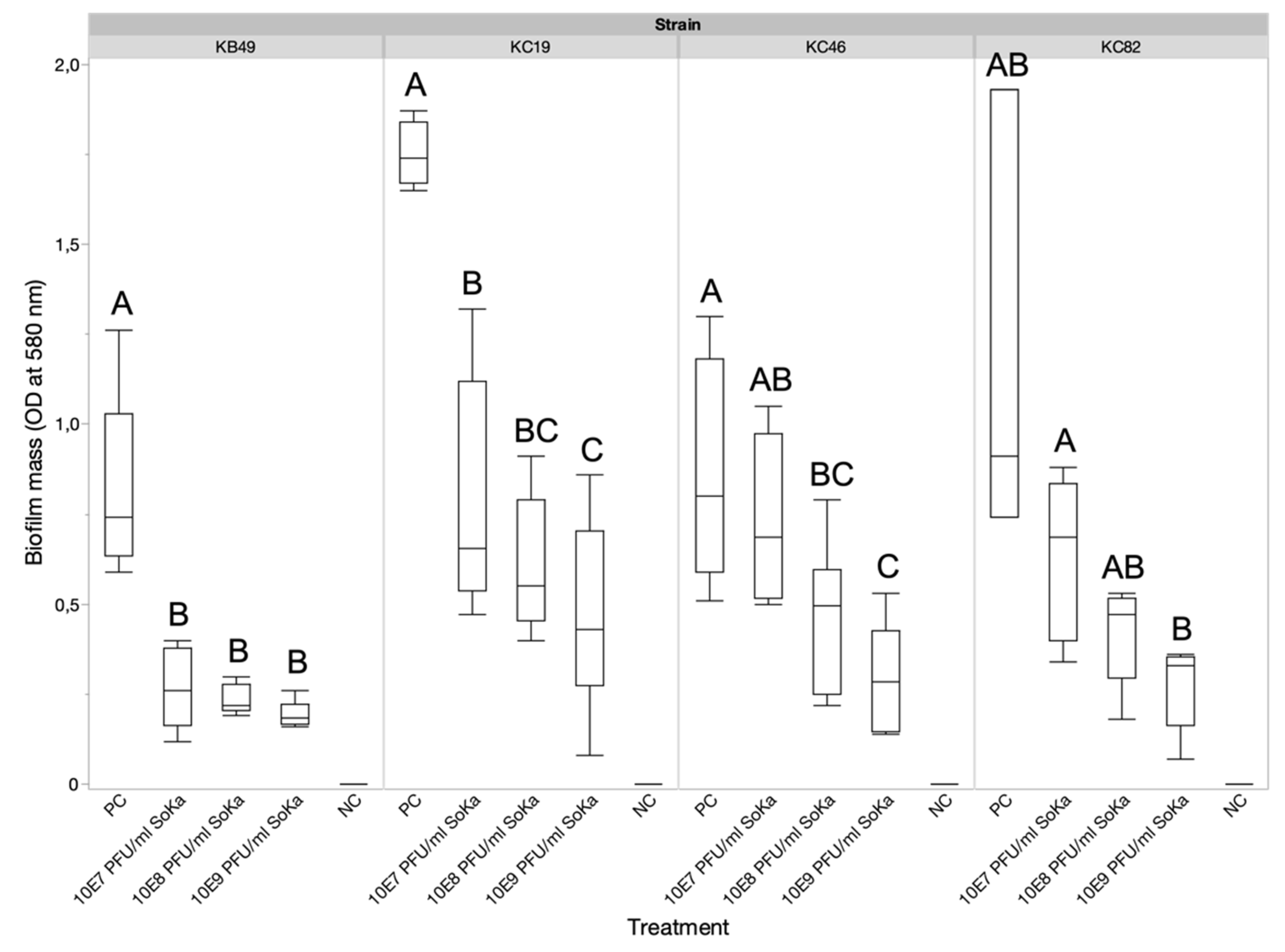
2.4. Effect of Bacteriophage on Biofilm Formation
2.4.1. Glass Slide Method
| Species and Pathovars | Strains | Plant Hosts | Infectivity | References |
|---|---|---|---|---|
| Pseudomonas syringae pv. syringae | E11 | Citrus | − | [23] |
| Pseudomonas syringae pv. syringae | EL1A | Citrus | − | [23] |
| Pseudomonas syringae pv. syringae | BE1 | Citrus | − | [23] |
| Pseudomonas syringae pv. syringae | BE3 | Citrus | − | [23] |
| Pseudomonas syringae pv. syringae | KC19 | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | KC46 | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | KC82 | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | TRR12 | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | TRR9 | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | E9A | Citrus | − | [23] |
| Pseudomonas syringae pv. syringae | E12A | Citrus | + | [23] |
| Pseudomonas syringae pv. syringae | IyGC | Citrus | + | [23] |
| Pseudomonas congelans | BE12A | Citrus | − | [23] |
| Pseudomonas nabeulensis | E10AB | Citrus | − | [25] |
| Pseudomonas kairouanensis | KC20 | Citrus | − | [25] |
| Pseudomonas savastanoi pv. savastanoi | 2C | Olive | − | unpublished |
| Pseudomonas savastanoi pv. savastanoi | 2E | Olive | − | unpublished |
| Agrobacterium rhizogenes | NCPPB 4042 | Cucumber | − | NCPPB |
| Agrobacterium rhizogenes | NCPPB 2659 | Cucumber | − | NCPPB |
| Agrobacterium tumefaciens | MAFF 210265 | Melon | − | MAFF |
| Dickeya dadantii | NCPPB 3537 | Potato | − | NCPPB |
| Dickeya chrysanthemi | NCPPB 402 | Chrysenthemum | − | NCPPB |
| Pseudomonas syringae pv. porri | CFBP 1770 | Leek | + | CFBP |
| Pseudomonas syringae pv. porri | GBBC 1269 | Leek | + | [39] |
| Pseudomonas syringae pv. porri | GBBC 3224 | Leek | + | [39] |
| Pseudomonas syringae pv. porri | Pspo 1277 | Leek | + | [39] |
| Pseudomonas syringae pv. porri | P55 | Leek | + | [40] |
2.4.2. Microplate Method
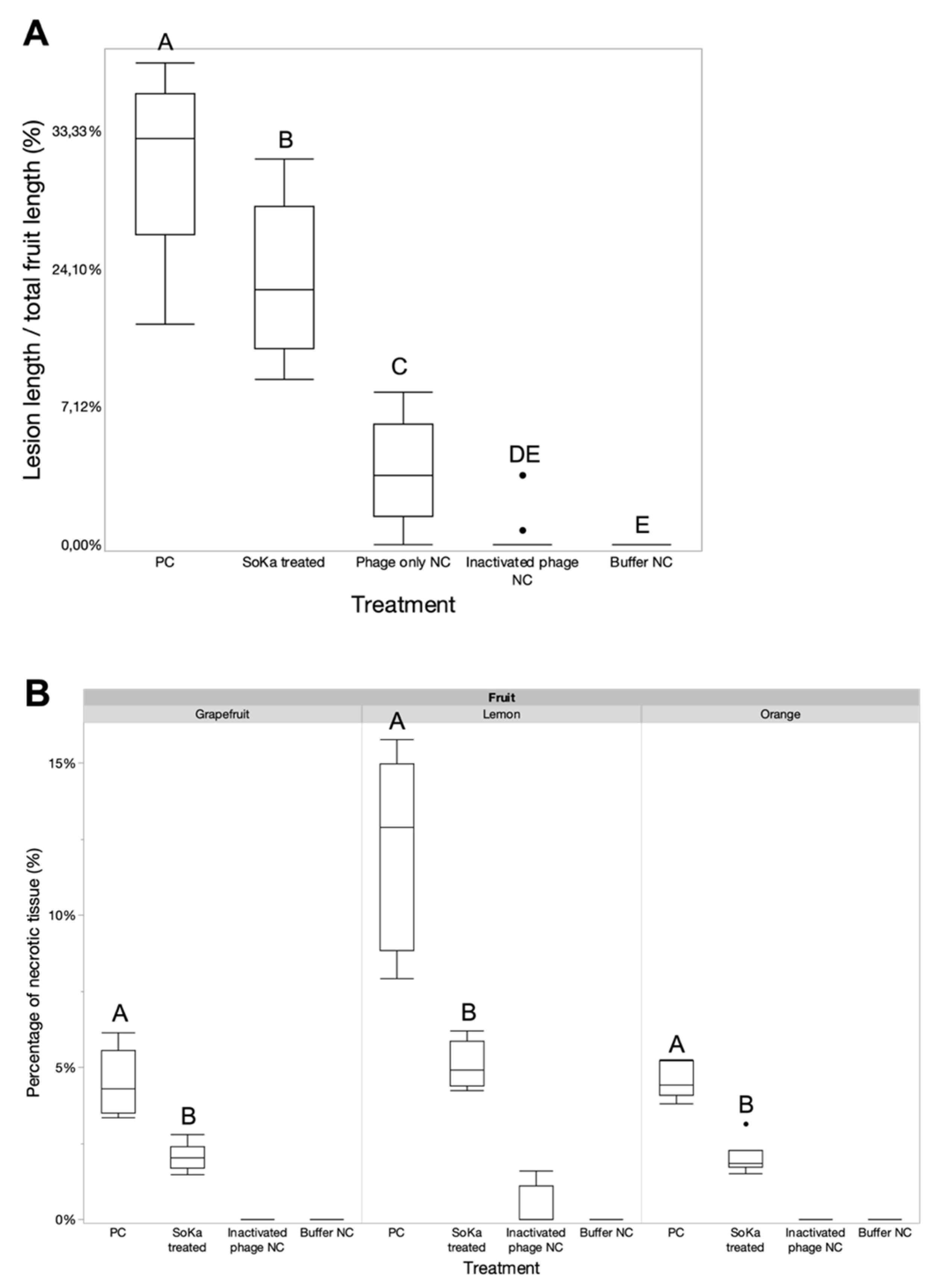
2.5. Bioassay on Detached Fruits
2.6. Statistical Data Analysis
3. Results
3.1. Isolation and Microbiological Characterization of the P. syringae Podovirus SoKa
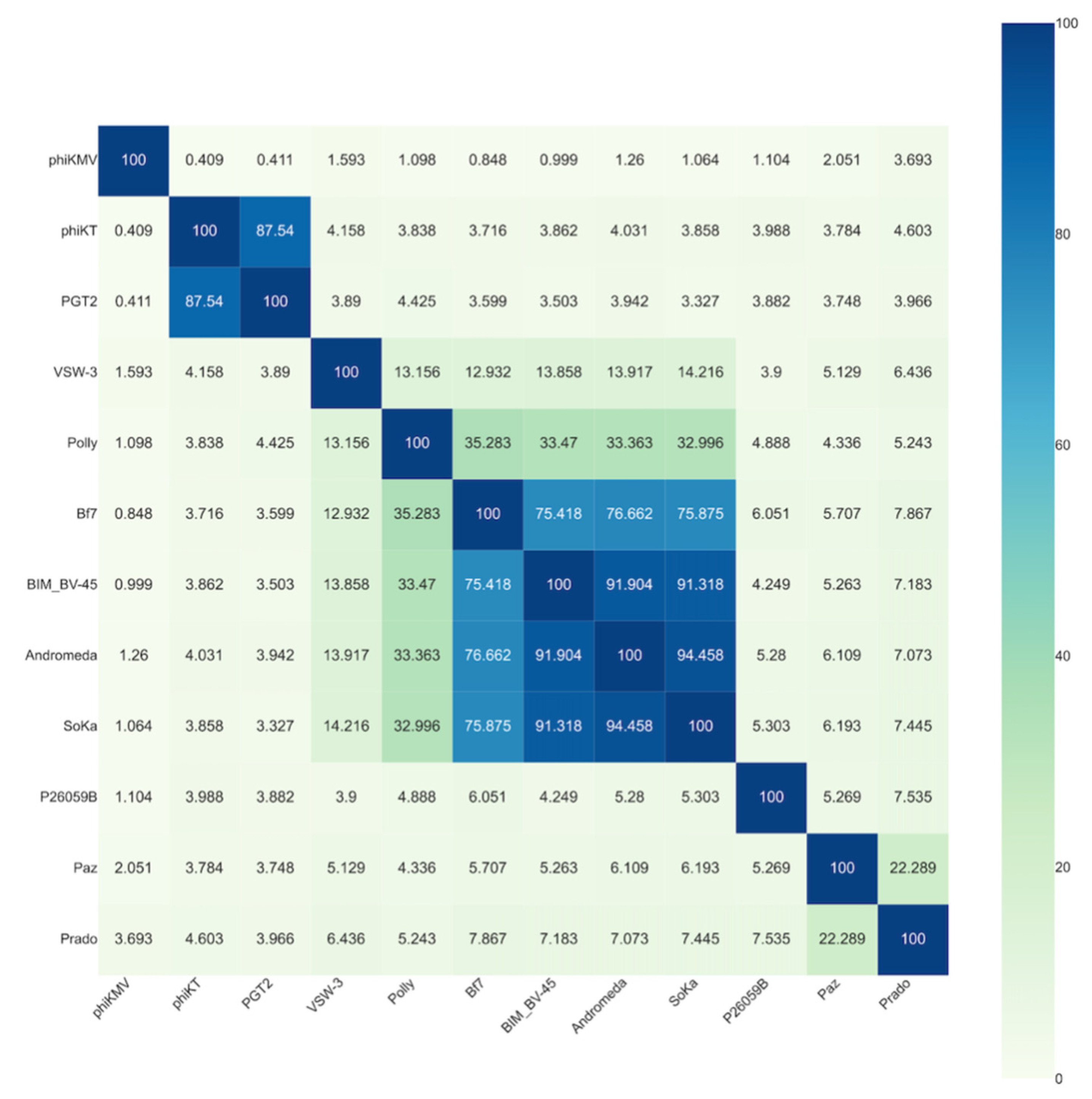
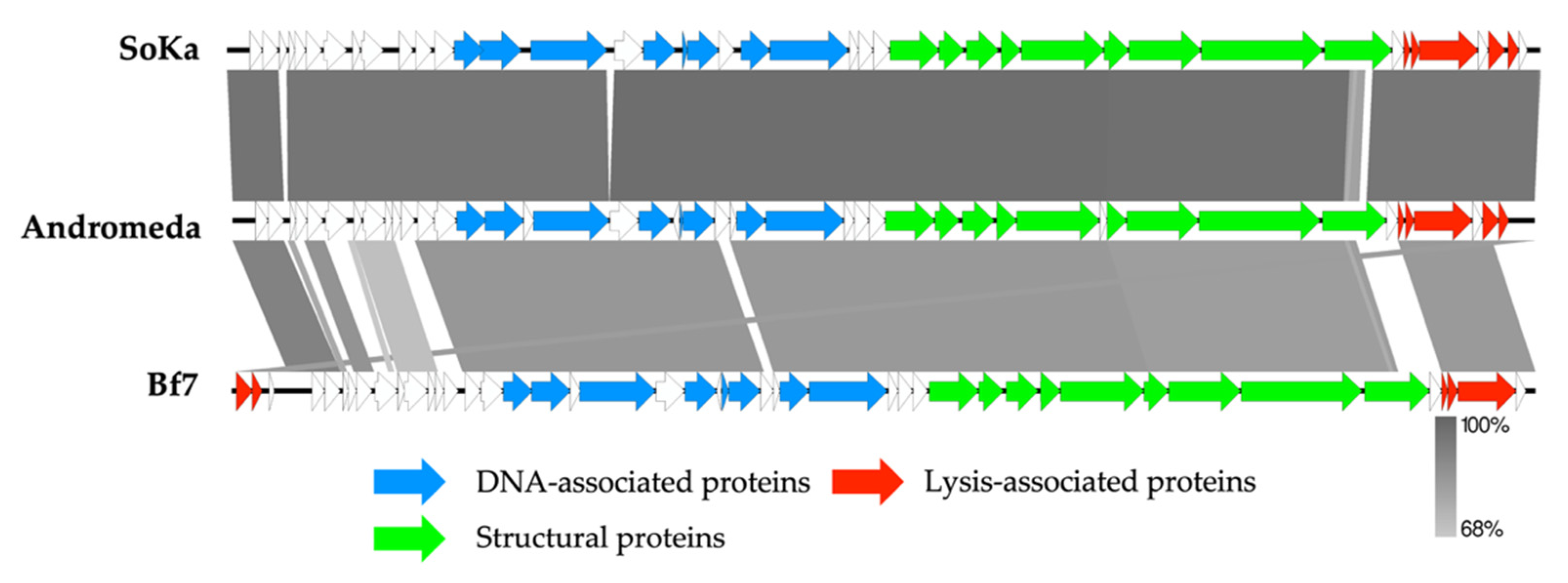
3.2. SoKa Is a Proposed New Species within the Bifseptvirus Genus of the Autographiviridae Family
3.3. SoKa Displays Significant Antibiofilm Activity
3.4. A Fruit Bioassay Shows the ex Planta Efficacy of SoKa
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it Takes to be a Pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, J.R.; Messéan, A.; Morris, C.E. Insights into Epidemiology and Control of Diseases of Annual Plants Caused by the Pseudomonas syringae Species Complex. J. Gen. Plant Pathol. 2015, 81, 331–350. [Google Scholar] [CrossRef]
- Kennelly, M.M.; Cazorla, F.M.; de Vicente, A.; Ramos, C.; Sundin, G.W. Pseudomonas syringae Diseases of Fruit Trees: Progress Toward Understanding and Control. Plant Dis. 2007, 91, 4–17. [Google Scholar] [CrossRef]
- Svircev, A.; Roach, D.; Castle, A. Framing the Future with Bacteriophages in Agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef]
- Holtappels, D.; Fortuna, K.; Lavigne, R.; Wagemans, J. The Future of Phage Biocontrol in Integrated Plant Protection for Sustainable Crop Production. Curr. Opin. Biotechnol. 2021, 68, 60–71. [Google Scholar] [CrossRef]
- Sieiro, C.; Areal-Hermida, L.; Pichardo-Gallardo, Á.; Almuiña-González, R.; de Miguel, T.; Sánchez, S.; Sánchez-Pérez, Á.; Villa, T.G. A Hundred Years of Bacteriophages: Can Phages Replace Antibiotics in Agriculture and Aquaculture? Antibiotics 2020, 9, 493. [Google Scholar] [CrossRef]
- Farooq, T.; Hussain, M.D.; Shakeel, M.T.; Tariqjaveed, M.; Aslam, M.N.; Naqvi, S.A.H.; Amjad, R.; Tang, Y.; She, X.; He, Z. Deploying Viruses against Phytobacteria: Potential Use of Phage Cocktails as a Multifaceted Approach to Combat Resistant Bacterial Plant Pathogens. Viruses 2022, 14, 171. [Google Scholar] [CrossRef]
- Jones, J.B.; Jackson, L.E.; Balogh, B.; Obradovic, A.; Iriarte, F.B.; Momol, M.T. Bacteriophages for Plant Disease Control. Annu Rev. Phytopathol. 2007, 45, 245–262. [Google Scholar] [CrossRef]
- Rombouts, S.; Volckaert, A.; Venneman, S.; Declercq, B.; Vandenheuvel, D.; Allonsius, C.N.; van Malderghem, C.; Jang, H.B.; Briers, Y.; Noben, J.P.; et al. Characterization of Novel Bacteriophages for Biocontrol of Bacterial Blight in Leek Caused by Pseudomonas syringae pv. porri. Front. Microbiol. 2016, 7, 279. [Google Scholar] [CrossRef]
- Born, Y.; Bosshard, L.; Duffy, B.; Loessner, M.J.; Fieseler, L. Protection of Erwinia amylovora Bacteriophage Y2 from UV-Induced Damage by Natural Compounds. Bacteriophage 2015, 5, e1074330. [Google Scholar] [CrossRef] [Green Version]
- Iriarte, F.B.; Obradović, A.; Wernsing, M.H.; Jackson, L.E.; Balogh, B.; Hong, J.A.; Momol, M.T.; Jones, J.B.; Vallad, G.E. Soil-Based Systemic Delivery and Phyllosphere in vivo Propagation of Bacteriophages: Two Possible Strategies for Improving Bacteriophage Persistence for Plant Disease Control. Bacteriophage 2012, 2, 215–224. [Google Scholar] [CrossRef]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic Discovery of Antiphage Defense Systems in the Microbial Pangenome. Science 2018, 359, 6379. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Sorek, R. Contemporary Phage Biology: From Classic Models to New Insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Tock, M.R.; Dryden, D.T.F. The Biology of Restriction and Anti-Restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; de Jager, V.; Siwinska, J.; Smolarska, A.; Ossowicki, A.; Narajczyk, M.; Lojkowska, E. Genomic, Proteomic and Morphological Characterization of Two Novel Broad Host Lytic Bacteriophages ΦPD10.3 and ΦPD23.1 Infecting Pectinolytic Pectobacterium spp. and Dickeya spp. PLoS ONE 2015, 10, e0119812. [Google Scholar] [CrossRef] [PubMed]
- Cemen, A.; Saygili, H.; Horuz, S.; Aysan, Y. Potential of Bacteriophages to Control Bacterial Speck of Tomato (Pseudomonas syringae pv. tomato). Fresenius Environ. Bull. 2018, 27, 9366–9373. [Google Scholar]
- Quiñones-Aguilar, E.E.; Reyes-Tena, A.; Hernández-Montiel, L.G.; Rincón Enríquez, G. Bacteriófagos En El Control Biológico de Pseudomonas syringae pv. phaseolicola Agente Causal Del Tizón de Halo Del Frijol. Ecosistemas Recur. Agropecu. 2018, 5, 191–202. [Google Scholar] [CrossRef]
- Martino, G.; Holtappels, D.; Vallino, M.; Chiapello, M.; Turina, M.; Lavigne, R.; Wagemans, J.; Ciuffo, M. Molecular Characterization and Taxonomic Assignment of Three Phage Isolates from a Collection Infecting Pseudomonas syringae pv. actinidiae and P. syringae pv. phaseolicola from Northern Italy. Viruses 2021, 13, 2083. [Google Scholar] [CrossRef]
- Yin, Y.; Ni, P.; Deng, B.; Wang, S.; Xu, W.; Wang, D. Isolation and Characterisation of Phages against Pseudomonas syringae pv. actinidiae. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2019, 69, 199–208. [Google Scholar] [CrossRef]
- Pinheiro, L.A.M.; Pereira, C.; Esther Barreal, M.; Pablo Gallego, P.; Balcao, V.M.; Almeida, A. Use of Phage Phi 6 to Inactivate Pseudomonas syringae pv. actinidiae in Kiwifruit Plants: In vitro and ex vivo Experiments. Appl. Microbiol. Biotechnol. 2020, 104, 1319–1330. [Google Scholar] [CrossRef]
- James, S.L.; Rabiey, M.; Neuman, B.W.; Percival, G.; Jackson, R.W. Isolation, Characterisation and Experimental Evolution of Phage That Infect the Horse Chestnut Tree Pathogen, Pseudomonas syringae pv. aesculi. Curr. Microbiol. 2020, 77, 1438–1447. [Google Scholar] [CrossRef] [Green Version]
- Rabiey, M.; Roy, S.R.; Holtappels, D.; Franceschetti, L.; Quilty, B.J.; Creeth, R.; Sundin, G.W.; Wagemans, J.; Lavigne, R.; Jackson, R.W. Phage Biocontrol to Combat Pseudomonas syringae Pathogens Causing Disease in Cherry. Microb. Biotechnol. 2020, 13, 1428–1445. [Google Scholar] [CrossRef]
- Oueslati, M.; Mulet, M.; Zouaoui, M.; Chandeysson, C.; Lalucat, J.; Hajlaoui, M.R.; Berge, O.; García-Valdés, E.; Sadfi-Zouaoui, N. Diversity of Pathogenic Pseudomonas Isolated from Citrus in Tunisia. AMB Express. 2020, 10, 198. [Google Scholar] [CrossRef]
- Berge, O.; Monteil, C.L.; Bartoli, C.; Chandeysson, C.; Guilbaud, C.; Sands, D.C.; Morris, C.E. A User’s Guide to a Database of the Diversity of Pseudomonas syringae and Its Application to Classifying Strains in This Phylogenetic Complex. PLoS ONE 2014, 9, e105547. [Google Scholar] [CrossRef]
- Oueslati, M.; Mulet, M.; Gomila, M.; Berge, O.; Hajlaoui, M.R.; Lalucat, J.; Sadfi-Zouaoui, N.; García-Valdés, E. New Species of Pathogenic Pseudomonas Isolated from Citrus in Tunisia: Proposal of Pseudomonas kairouanensis sp. nov. and Pseudomonas nabeulensis sp. nov. Syst. Appl. Microbiol. 2019, 42, 348–359. [Google Scholar] [CrossRef]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Adriaenssens, E.M.; van Vaerenbergh, J.; Vandenheuvel, D.; Dunon, V.; Ceyssens, P.-J.; de Proft, M.; Kropinski, A.M.; Noben, J.-P.; Maes, M.; Lavigne, R. T4-Related Bacteriophage LIMEstone Isolates for the Control of Soft Rot on Potato Caused by ‘Dickeya solani’. PLoS ONE 2012, 7, e33227. [Google Scholar] [CrossRef]
- Makalatia, K.; Kakabadze, E.; Wagemans, J.; Grdzelishvili, N.; Bakuradze, N.; Natroshvili, G.; Macharashvili, N.; Sedrakyan, A.; Arakelova, K.; Ktsoyan, Z.; et al. Characterization of Salmonella Isolates from Various Geographical Regions of the Caucasus and Their Susceptibility to Bacteriophages. Viruses 2020, 12, 1418. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The Viral Proteomic Tree Server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI-Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing 1. bioRXiv 2020, 198606. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antibiotic Resistance Genes. 2016. Available online: https://github.com/tseemann/abricate (accessed on 1 July 2022).
- Merrill, B.D.; Ward, A.T.; Grose, J.H.; Hope, S. Software-Based Analysis of Bacteriophage Genomes, Physical Ends, and Packaging Strategies. BMC Genom. 2016, 17, 679. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Shafique, M.; Alvi, I.A.; Abbas, Z.; Ur Rehman, S. Assessment of Biofilm Removal Capacity of a Broad Host Range Bacteriophage JHP against Pseudomonas aeruginosa. APMIS 2017, 125, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, S.; van Vaerenbergh, J.; Volckaert, A.; Baeyen, S.; de Langhe, T.; Declercq, B.; Lavigne, R.; Maes, M. Isolation and Characterization of Pseudomonas syringae pv. porri from Leek in Flanders. Eur. J. Plant Pathol. 2016, 144, 185–198. [Google Scholar] [CrossRef]
- van Overbeek, L.S.; Nijhuis, E.H.M.; Koenraadt, H.; Visser, J.; van Kruistum, G. The Role of Crop Waste and Soil in Pseudomonas syringae pathovar porri Infection of Leek (Allium porrum). Appl. Soil Ecol. 2010, 46, 457–463. [Google Scholar] [CrossRef]
- Magill, D.J.; Kulakov, L.A.; Skvortsov, T.A. Genomic Hypervariability of Phage Andromeda Is Unique among Known DsDNA Viruses. bioRxiv 2020, 619015. [Google Scholar] [CrossRef]
- Sajben-Nagy, E.; Maróti, G.; Kredics, L.; Horváth, B.; Párducz, A.; Vágvölgyi, C.; Manczinger, L. Isolation of New Pseudomonas tolaasii Bacteriophages and Genomic Investigation of the Lytic Phage BF7. FEMS Microbiol. Lett. 2012, 332, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Husseini, A.; Akköprü, A. The Possible Mechanisms of Copper Resistance in the Pathogen Pseudomonas syringae Pathovars in Stone Fruit Trees. Phytoparasitica 2020, 48, 705–718. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Iriarte, F.B.; Balogh, B.; Momol, M.T.; Smith, L.M.; Wilson, M.; Jones, J.B. Factors Affecting Survival of Bacteriophage on Tomato Leaf Surfaces. Appl. Environ. Microbiol. 2007, 73, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage Φ6 for Biocontrol of Pseudomonas syringae pv. syringae: An in vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef]
- Jones, J.B.; Svircev, A.M.; Obradović, A.Ž. Crop Use of Bacteriophages. In Bacteriophages; Harper, D., Abedon, S., Burrowes, B., McConville, M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 839–856. [Google Scholar]
- Holtappels, D.; Fortuna, K.J.; Moons, L.; Broeckaert, N.; Bäcker, L.E.; Venneman, S.; Rombouts, S.; Lippens, L.; Baeyen, S.; Pollet, S.; et al. The Potential of Bacteriophages to Control Xanthomonas campestris pv. campestris at Different Stages of Disease Development. Microb. Biotechnol. 2022, 15, 1762–1782. [Google Scholar] [CrossRef]
- Balogh, B.; Nga, N.T.T.; Jones, J.B. Relative Level of Bacteriophage Multiplication in vitro or in Phyllosphere May Not Predict in planta Efficacy for Controlling Bacterial Leaf Spot on Tomato Caused by Xanthomonas perforans. Front. Microbiol. 2018, 9, 2176. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oueslati, M.; Holtappels, D.; Fortuna, K.; Hajlaoui, M.R.; Lavigne, R.; Sadfi-Zouaoui, N.; Wagemans, J. Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia. Viruses 2022, 14, 1949. https://doi.org/10.3390/v14091949
Oueslati M, Holtappels D, Fortuna K, Hajlaoui MR, Lavigne R, Sadfi-Zouaoui N, Wagemans J. Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia. Viruses. 2022; 14(9):1949. https://doi.org/10.3390/v14091949
Chicago/Turabian StyleOueslati, Maroua, Dominique Holtappels, Kiandro Fortuna, Mohamed Rabeh Hajlaoui, Rob Lavigne, Najla Sadfi-Zouaoui, and Jeroen Wagemans. 2022. "Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia" Viruses 14, no. 9: 1949. https://doi.org/10.3390/v14091949
APA StyleOueslati, M., Holtappels, D., Fortuna, K., Hajlaoui, M. R., Lavigne, R., Sadfi-Zouaoui, N., & Wagemans, J. (2022). Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia. Viruses, 14(9), 1949. https://doi.org/10.3390/v14091949








