From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents
Abstract
:1. Introduction
2. Current Control Measures and Alternative Approaches: A One Health Perspective
Alternatives to Antibiotics
3. Phages in Agriculture and Food Industry
3.1. Phages in Primary Food Production
3.2. Other Applications of Phages in Livestock and Food Production
3.2.1. Phages as Growth Promoters
3.2.2. Phage Therapy in Crop Production
3.2.3. Phages as Biopreservatives and Biosanitisers
4. Streptococcus suis and Its Phages: A Case Study
4.1. Phages of S. suis
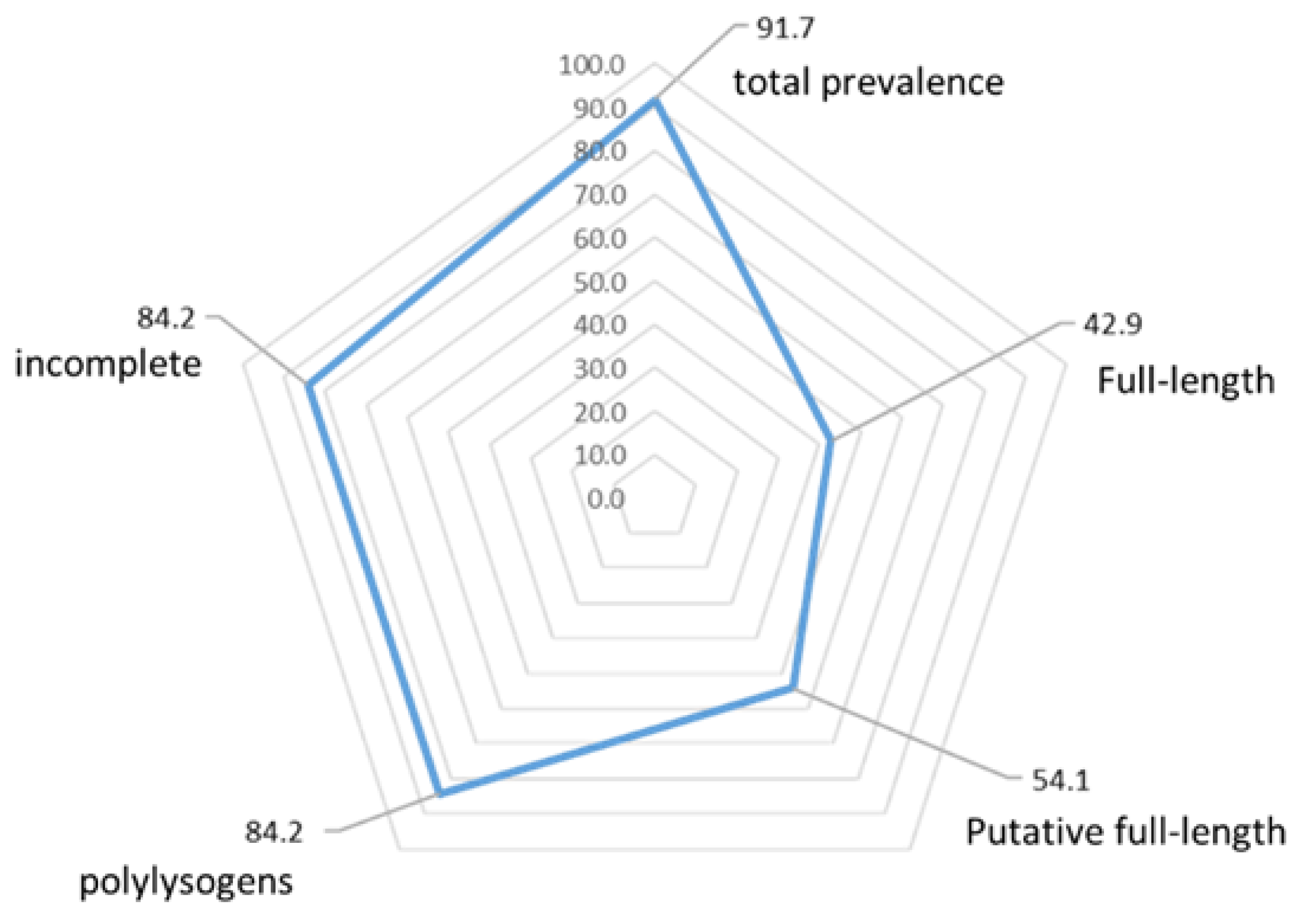
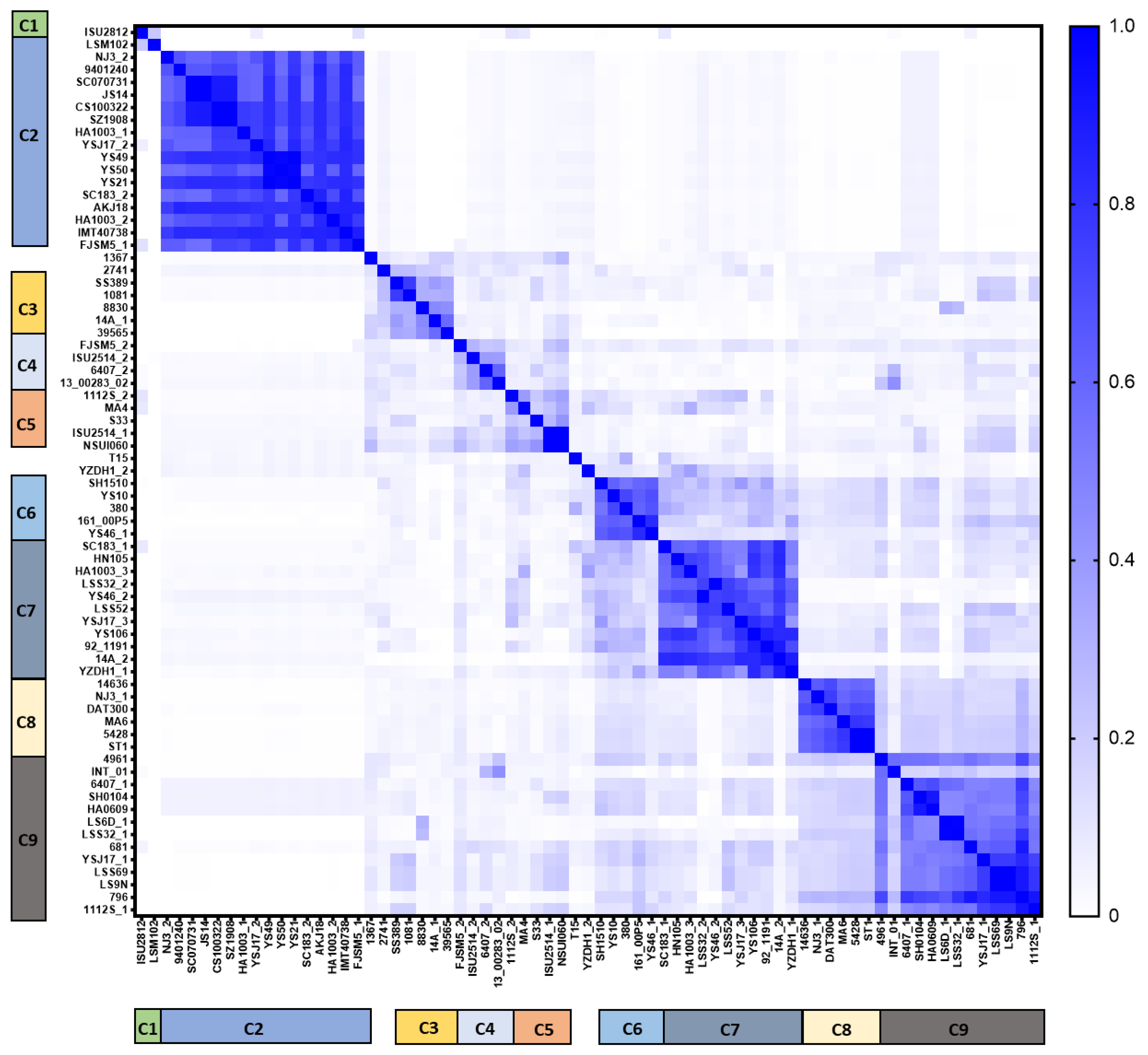
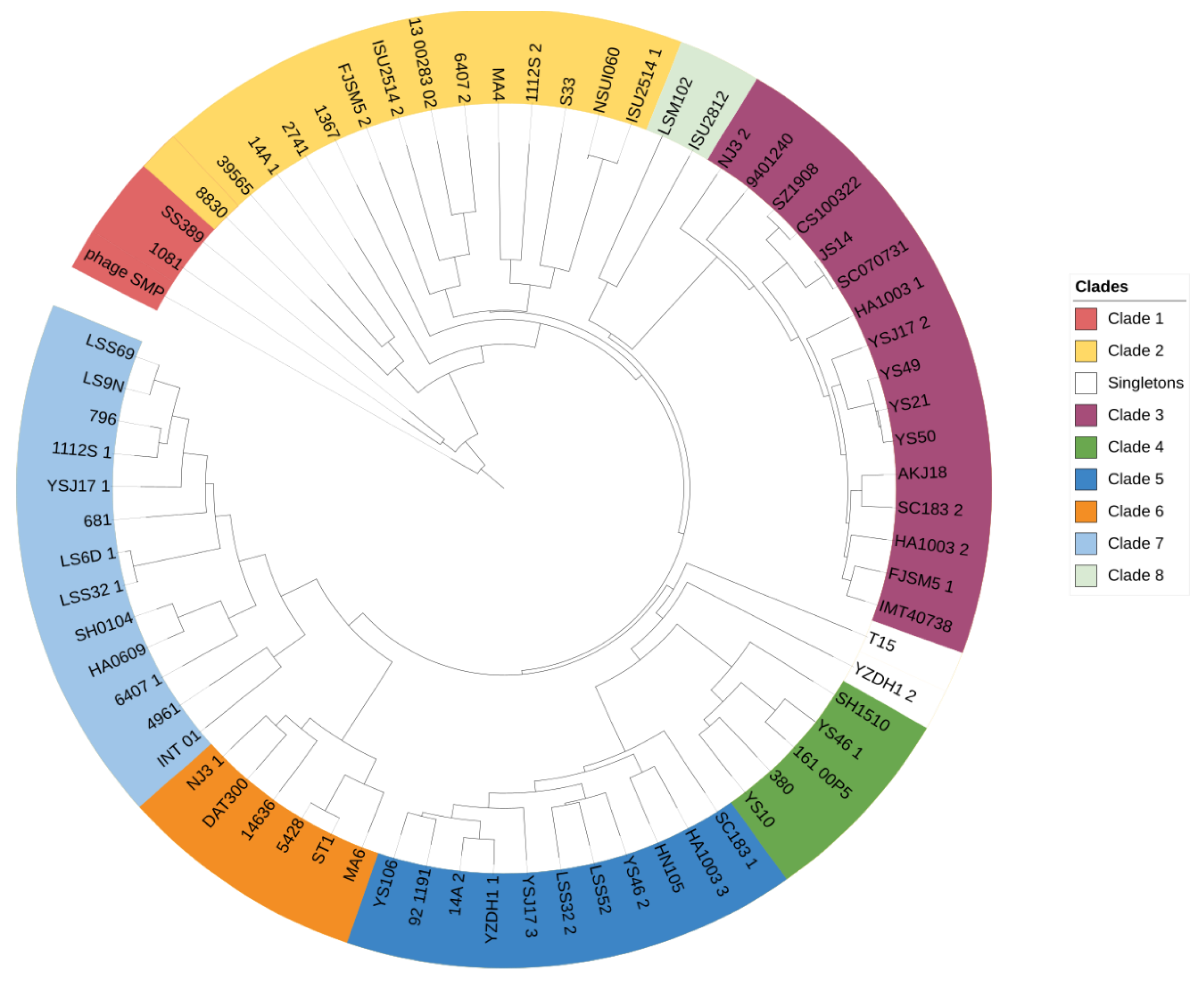
4.2. High Prophage Prevalence and Diversity in S. suis Genomes
4.3. Protein-Encoded S. suis Prophages
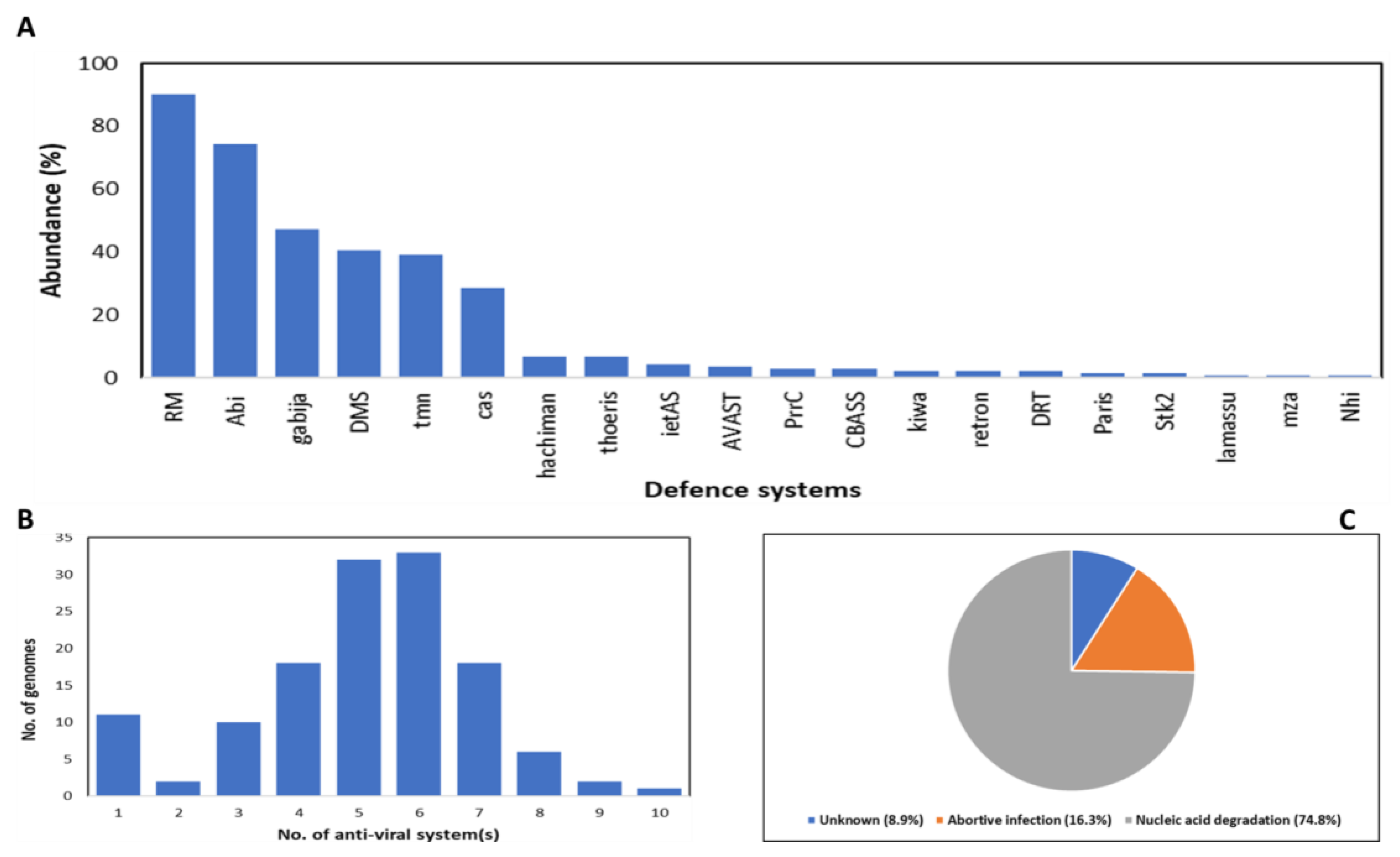
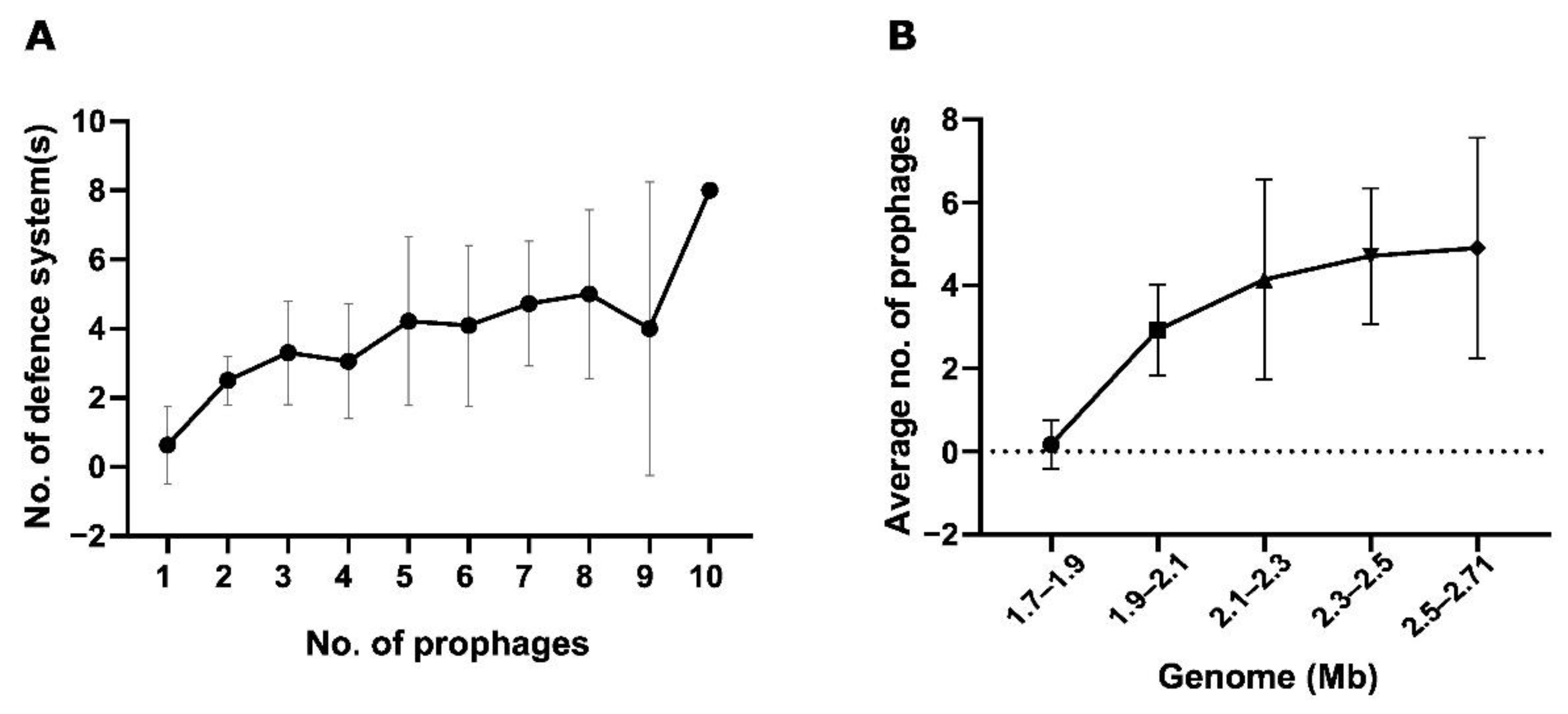
4.4. Relationship between Bacterial Genome Size, Number of Anti-Viral Defence, and Prevalence of Prophages
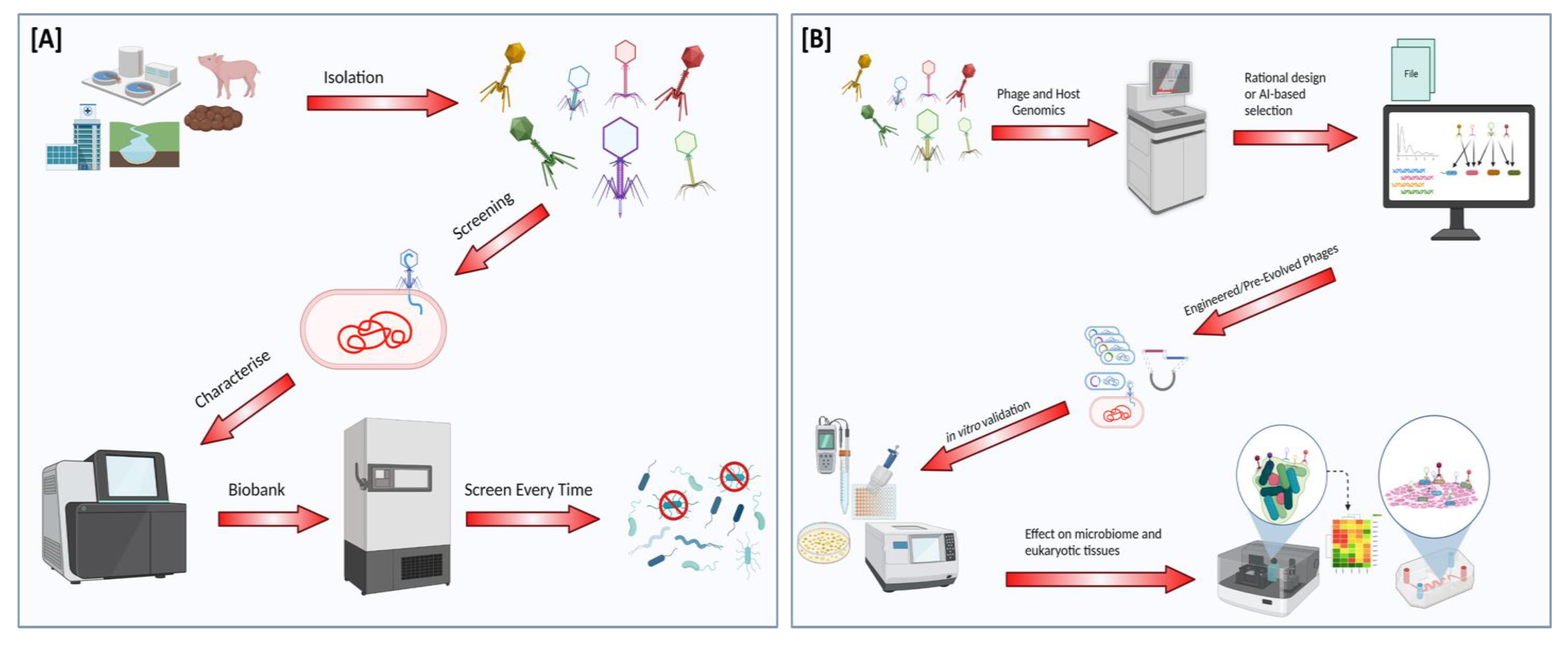
5. Implications for the Development of Therapeutics
Future Perspectives
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abubakar, M.; Iqbal, A.; Manzoor, S.; Arshed, M.J. Introductory Chapter: Livestock Health and Farming - Regional to Global Perspectives. In Livestock Health and Farming.; Abubakar, M., Ed.; IntechOpen: London, UK, 2020. [Google Scholar]
- Animal Production. Available online: http://www.fao.org/animal-production/en (accessed on 27 July 2022).
- Dewi, G.; Kollanoor Johny, A. Lactobacillus in Food Animal Production—A Forerunner for Clean Label Prospects in Animal-Derived Products. Front. Sustain. Food Syst. 2022, 6, 831195. [Google Scholar] [CrossRef]
- Barratt, A.S.; Rich, K.M.; Eze, J.I.; Porphyre, T.; Gunn, G.J.; Stott, A.W. Framework for Estimating Indirect Costs in Animal Health Using Time Series Analysis. Front. Vet. Sci. 2019, 6, 190. [Google Scholar] [CrossRef]
- World Bank. People, Pathogens, and Our Planet: Volume One—Towards a One Health Approach for Controlling Zoonotic Diseases; World Bank: Washington, DC, USA, 2010. [Google Scholar]
- Garner, M.G.; Whan, I.F.; Gard, G.P.; Phillips, D. The expected economic impact of selected exotic diseases on the pig industry of Australia. Rev. Sci. Tech. Int. Off. Epizoot. 2001, 20, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Gyles, C. One Medicine, One Health, One World. Can. Vet. J. 2016, 57, 345–346. [Google Scholar] [PubMed]
- Zinsstag, J.; Schelling, E.; Waltner-Toews, D.; Tanner, M. From “one medicine” to “one health” and systemic approaches to health and well-being. Prev. Vet. Med. 2011, 101, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140231. [Google Scholar] [CrossRef]
- One Health. Available online: https://www.who.int/news-room/questions-and-answers/item/one-health (accessed on 13 April 2022).
- One Health Basics|One Health|CDC. Available online: https://www.cdc.gov/onehealth/basics/index.html (accessed on 13 April 2022).
- Relun, A.; Charrier, F.; Trabucco, B.; Maestrini, O.; Molia, S.; Chavernac, D.; Grosbois, V.; Casabianca, F.; Etter, E.; Jori, F. Multivariate analysis of traditional pig management practices and their potential impact on the spread of infectious diseases in Corsica. Prev. Vet. Med. 2015, 121, 246–256. [Google Scholar] [CrossRef]
- Health-Management Interaction: Pigs—Management and Nutrition. Available online: https://www.msdvetmanual.com/management-and-nutrition/health-management-interaction-pigs/health-management-interaction-pigs (accessed on 13 April 2022).
- Disease Prevention—Teagasc|Agriculture and Food Development Authority. Available online: https://www.teagasc.ie/animals/amr/disease-prevention/ (accessed on 13 April 2022).
- Understanding and Managing Strep suis in Swine: The Essentials. 2020. Available online: https://ew-nutrition.com/strep-suis-essentials/ (accessed on 13 April 2022).
- Helke, K.L.; Ezell, P.C.; Duran-Struuck, R.; Swindle, M.M. Chapter 16—Biology and Diseases of Swine. In Laboratory Animal Medicine, 3rd ed.; Fox, J.G., Anderson, L.C., Otto, G.M., Pritchett-Corning, K.R., Whary, M.T., Eds.; American College of Laboratory Animal Medicine: Chester, NH, USA; Academic Press: Boston, MA, USA, 2015; pp. 695–769. ISBN 978-0-12-409527-4. [Google Scholar]
- Rieckmann, K.; Pendzialek, S.-M.; Vahlenkamp, T.; Baums, C.G. A critical review speculating on the protective efficacies of autogenous Streptococcus suis bacterins as used in Europe. Porc. Health Manag. 2020, 6, 12. [Google Scholar] [CrossRef]
- Baums, C.G.; Brüggemann, C.; Kock, C.; Beineke, A.; Waldmann, K.-H.; Valentin-Weigand, P. Immunogenicity of an Autogenous Streptococcus suis Bacterin in Preparturient Sows and Their Piglets in Relation to Protection after Weaning. Clin. Vaccine Immunol. CVI 2010, 17, 1589–1597. [Google Scholar] [CrossRef]
- Baums, C.G.; Kock, C.; Beineke, A.; Bennecke, K.; Goethe, R.; Schröder, C.; Waldmann, K.-H.; Valentin-Weigand, P. Streptococcus suis Bacterin and Subunit Vaccine Immunogenicities and Protective Efficacies against Serotypes 2 and 9. Clin. Vaccine Immunol. CVI 2009, 16, 200–208. [Google Scholar] [CrossRef] [Green Version]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; de Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Maruggi, G.; Chiarot, E.; Giovani, C.; Buccato, S.; Bonacci, S.; Frigimelica, E.; Margarit, I.; Geall, A.; Bensi, G.; Maione, D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine 2017, 35, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Van Rennings, L.; von Münchhausen, C.; Ottilie, H.; Hartmann, M.; Merle, R.; Honscha, W.; Käsbohrer, A.; Kreienbrock, L. Cross-Sectional Study on Antibiotic Usage in Pigs in Germany. PLoS ONE 2015, 10, e0119114. [Google Scholar] [CrossRef]
- Effects of Antibiotics on Animal Feed—Presentation. Available online: https://www1.udel.edu/chem/C465/senior/fall97/feed/present.html (accessed on 5 July 2022).
- Dall, C.; Cruz, C.R.; Van Beusekom, M.; Schnirring, L.; Soucheray, S.; Swain, M.W.; Vestin, N.; Wappes, J. FDA Reports Another Rise in Antibiotic Sales for Livestock. Available online: https://www.cidrap.umn.edu/news-perspective/2020/12/fda-reports-another-rise-antibiotic-sales-livestock (accessed on 5 July 2022).
- Gaio, D.; DeMaere, M.Z.; Anantanawat, K.; Eamens, G.J.; Zingali, T.; Falconer, L.; Chapman, T.A.; Djordjevic, S.; Darling, A.E. Phylogenetic diversity analysis of shotgun metagenomic reads describes gut microbiome development and treatment effects in the post-weaned pig. PLoS ONE 2022, 17, e0270372. [Google Scholar] [CrossRef]
- Antimicrobial resistance: A top ten global public health threat. EClinicalMedicine 2021, 41, 101221. [CrossRef]
- Ten Health Issues WHO Will Tackle This Year. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 14 April 2022).
- Wee, B.A.; Muloi, D.M.; Bunnik, B.A.D. van Quantifying the transmission of antimicrobial resistance at the human and livestock interface with genomics. Clin. Microbiol. Infect. 2020, 26, 1612–1616. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Zhu, L.; Wang, J. Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils. Sci. Total Environ. 2019, 654, 906–913. [Google Scholar] [CrossRef]
- Peng, J.-J.; Balasubramanian, B.; Ming, Y.-Y.; Niu, J.-L.; Yi, C.-M.; Ma, Y.; Liu, W.-C. Identification of antimicrobial resistance genes and drug resistance analysis of Escherichia coli in the animal farm environment. J. Infect. Public Health 2021, 14, 1788–1795. [Google Scholar] [CrossRef] [PubMed]
- Young, C.; Kharmacharya, D.; Bista, M.; Sharma, A.; Goldstein, T.; Anthony, S.J.; Smith, W.; Mazet, J.; Johnson, C. Sharing of antimicrobial resistance genes among animals, humans, and the environment in Nepal: A one health case study. Int. J. Infect. Dis. 2019, 79, 20. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Checcucci, A.; Trevisi, P.; Luise, D.; Modesto, M.; Blasioli, S.; Braschi, I.; Mattarelli, P. Exploring the Animal Waste Resistome: The Spread of Antimicrobial Resistance Genes Through the Use of Livestock Manure. Front. Microbiol. 2020, 11, 1416. [Google Scholar] [CrossRef]
- Food and Drug Administration. 2019 Summary Report On Antimicrobials Sold or Distributed for Use in Food-Producing Animals. AMR Insights 2020. Available online: https://www.fda.gov/media/144427/download (accessed on 14 April 2022).
- Phillips, I. Withdrawal of growth-promoting antibiotics in Europe and its effects in relation to human health. Int. J. Antimicrob. Agents 2007, 30, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Fifth OIE Annual Report on Antimicrobial Agents Intended for Use in Animals. WOAH—World Organisation for Animal Health. Available online: https://www.oie.int/en/document/fifth-oie-annual-report-on-antimicrobial-agents-intended-for-use-in-animals/ (accessed on 14 April 2022).
- Jackman, J.A.; Boyd, R.D.; Elrod, C.C. Medium-chain fatty acids and monoglycerides as feed additives for pig production: Towards gut health improvement and feed pathogen mitigation. J. Anim. Sci. Biotechnol. 2020, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, J.T.; Thomson, K.A.; Woodworth, J.C.; Dritz, S.S.; Tokach, M.D.; DeRouchey, J.M.; Goodband, R.D.; Jones, C.K.; Cochrane, R.A.; Niederwerder, M.C.; et al. Effect of dietary medium-chain fatty acids on nursery pig growth performance, fecal microbial composition, and mitigation properties against porcine epidemic diarrhea virus following storage. J. Anim. Sci. 2020, 98, skz358. [Google Scholar] [CrossRef]
- Correa-Fiz, F.; Neila-Ibáñez, C.; López-Soria, S.; Napp, S.; Martinez, B.; Sobrevia, L.; Tibble, S.; Aragon, V.; Migura-Garcia, L. Feed additives for the control of post-weaning Streptococcus suis disease and the effect on the faecal and nasal microbiota. Sci. Rep. 2020, 10, 20354. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa Bacteriocins: Diversity and New Developments. Int. J. Mol. Sci. 2012, 13, 16668–16707. [Google Scholar] [CrossRef]
- Sun, Y.; Veseli, I.A.; Vaillancourt, K.; Frenette, M.; Grenier, D.; Pombert, J.-F. The bacteriocin from the prophylactic candidate Streptococcus suis 90-1330 is widely distributed across S. suis isolates and appears encoded in an integrative and conjugative element. PLoS ONE 2019, 14, e0216002. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Gil, J.F.; Mesa, V.; Estrada-Ortiz, N.; Lopez-Obando, M.; Gómez, A.; Plácido, J. Viruses in Extreme Environments, Current Overview, and Biotechnological Potential. Viruses 2021, 13, 81. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage Therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Venturini, C.; Petrovic Fabijan, A.; Fajardo Lubian, A.; Barbirz, S.; Iredell, J. Biological foundations of successful bacteriophage therapy. EMBO Mol. Med. 2022, 14, e12435. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Ross, R.P.; Coffey, A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 2009, 33, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- Hoyle, N.; Zhvaniya, P.; Balarjishvili, N.; Bolkvadze, D.; Nadareishvili, L.; Nizharadze, D.; Wittmann, J.; Rohde, C.; Kutateladze, M. Phage therapy against Achromobacter xylosoxidans lung infection in a patient with cystic fibrosis: A case report. Res. Microbiol. 2018, 169, 540–542. [Google Scholar] [CrossRef]
- Corbellino, M.; Kieffer, N.; Kutateladze, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Tsertsvadze, G.; Rimoldi, S.G.; Nizharadze, D.; Hoyle, N.; et al. Eradication of a Multidrug-Resistant, Carbapenemase-Producing Klebsiella pneumoniae Isolate Following Oral and Intra-rectal Therapy With a Custom Made, Lytic Bacteriophage Preparation. Clin. Infect. Dis. 2020, 70, 1998–2001. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Little, J.S.; Dedrick, R.M.; Freeman, K.G.; Cristinziano, M.; Smith, B.E.; Benson, C.A.; Jhaveri, T.A.; Baden, L.R.; Solomon, D.A.; Hatfull, G.F. Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nat. Commun. 2022, 13, 2313. [Google Scholar] [CrossRef]
- Rubalskii, E.; Ruemke, S.; Salmoukas, C.; Boyle, E.C.; Warnecke, G.; Tudorache, I.; Shrestha, M.; Schmitto, J.D.; Martens, A.; Rojas, S.V.; et al. Bacteriophage Therapy for Critical Infections Related to Cardiothoracic Surgery. Antibiotics 2020, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Law, N.; Logan, C.; Yung, G.; Furr, C.-L.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Nick, J.A.; Dedrick, R.M.; Gray, A.L.; Vladar, E.K.; Smith, B.E.; Freeman, K.G.; Malcolm, K.C.; Epperson, L.E.; Hasan, N.A.; Hendrix, J.; et al. Host and pathogen response to bacteriophage engineered against Mycobacterium abscessus lung infection. Cell 2022, 185, 1860–1874.e2. [Google Scholar] [CrossRef] [PubMed]
- Román, S.; Sánchez-Siles, L.M.; Siegrist, M. The importance of food naturalness for consumers: Results of a systematic review. Trends Food Sci. Technol. 2017, 67, 44–57. [Google Scholar] [CrossRef]
- Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms 2020, 8, 1584. [Google Scholar] [CrossRef] [PubMed]
- Thanki, A.M.; Mignard, G.; Atterbury, R.J.; Barrow, P.; Millard, A.D.; Clokie, M.R.J. Prophylactic Delivery of a Bacteriophage Cocktail in Feed Significantly Reduces Salmonella Colonization in Pigs. Microbiol. Spectr. 2022, 10, e00422-22. [Google Scholar] [CrossRef]
- Wall, S.K.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Phage Therapy To Reduce Preprocessing Salmonella Infections in Market-Weight Swine. Appl. Environ. Microbiol. 2010, 76, 48–53. [Google Scholar] [CrossRef]
- Gebru, E.; Lee, J.S.; Son, J.C.; Yang, S.Y.; Shin, S.A.; Kim, B.; Kim, M.K.; Park, S.C. Effect of probiotic-, bacteriophage-, or organic acid-supplemented feeds or fermented soybean meal on the growth performance, acute-phase response, and bacterial shedding of grower pigs challenged with Salmonella enterica serotype Typhimurium1. J. Anim. Sci. 2010, 88, 3880–3886. [Google Scholar] [CrossRef]
- Mosimann, S.; Desiree, K.; Ebner, P. Efficacy of phage therapy in poultry: A systematic review and meta-analysis. Poult. Sci. 2021, 100, 101472. [Google Scholar] [CrossRef]
- Bumunang, E.W.; McAllister, T.A.; Stanford, K.; Anany, H.; Niu, Y.D.; Ateba, C.N. Characterization of Non-O157 STEC Infecting Bacteriophages Isolated from Cattle Faeces in North-West South Africa. Microorganisms 2019, 7, 615. [Google Scholar] [CrossRef] [PubMed]
- Montso, P.K.; Mnisi, C.M.; Ateba, C.N.; Mlambo, V. An Assessment of the Viability of Lytic Phages and Their Potency against Multidrug Resistant Escherichia coli O177 Strains under Simulated Rumen Fermentation Conditions. Antibiotics 2021, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Dini, C.; De Urraza, P.J. Isolation and selection of coliphages as potential biocontrol agents of enterohemorrhagic and Shiga toxin-producing E. coli (EHEC and STEC) in cattle. J. Appl. Microbiol. 2010, 109, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Guabiraba, R.; Schouler, C. Avian colibacillosis: Still many black holes. FEMS Microbiol. Lett. 2015, 362, fnv118. [Google Scholar] [CrossRef] [PubMed]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian Pathogenic Escherichia coli (APEC): An Overview of Virulence and Pathogenesis Factors, Zoonotic Potential, and Control Strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef]
- Kunert Filho, H.; Brito, K.; Cavalli, L.; Brito, B. Avian Pathogenic Escherichia coli (APEC)—An Update on the Control; Formatex Research Center: Badajoz, Spain, 2015; pp. 598–618. ISBN 978-84-942134-7-2. [Google Scholar]
- Nordin, N.; Sani, N.I.M.; Kadir, A.A.; Shaari, R.; Mohamed, M.; Reduan, M.F.H.; Nordin, M.L. Infectious bronchitis associated with Escherichia coli infection in commercial broiler chickens: A case report. J. Adv. Vet. Anim. Res. 2021, 8, 101–104. [Google Scholar] [CrossRef]
- Tawakol, M.M.; Nabil, N.M.; Samy, A. Evaluation of bacteriophage efficacy in reducing the impact of single and mixed infections with Escherichia coli and infectious bronchitis in chickens. Infect. Ecol. Epidemiol. 2019, 9, 1686822. [Google Scholar] [CrossRef]
- Eid, S.; Tolba, H.M.N.; Hamed, R.I.; Al-Atfeehy, N.M. Bacteriophage therapy as an alternative biocontrol against emerging multidrug resistant E. coli in broilers. Saudi J. Biol. Sci. 2022, 29, 3380–3389. [Google Scholar] [CrossRef]
- Naghizadeh, M.; Karimi Torshizi, M.A.; Rahimi, S.; Dalgaard, T.S. Synergistic effect of phage therapy using a cocktail rather than a single phage in the control of severe colibacillosis in quails. Poult. Sci. 2019, 98, 653–663. [Google Scholar] [CrossRef]
- Campylobacter. Available online: https://www.who.int/news-room/fact-sheets/detail/campylobacter (accessed on 6 June 2022).
- Richards, P.J.; Connerton, P.L.; Connerton, I.F. Phage Biocontrol of Campylobacter jejuni in Chickens Does Not Produce Collateral Effects on the Gut Microbiota. Front. Microbiol. 2019, 10, 476. [Google Scholar] [CrossRef]
- Ushanov, L.; Lasareishvili, B.; Janashia, I.; Zautner, A.E. Application of Campylobacter jejuni Phages: Challenges and Perspectives. Anim. Open Access J. MDPI 2020, 10, 279. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Jäckel, C.; Alter, T.; Janzcyk, P.; Stingl, K.; Knüver, M.T.; Hertwig, S. Reduction of Campylobacter jejuni in Broiler Chicken by Successive Application of Group II and Group III Phages. PLoS ONE 2014, 9, e114785. [Google Scholar] [CrossRef]
- Steffan, S.M.; Shakeri, G.; Kehrenberg, C.; Peh, E.; Rohde, M.; Plötz, M.; Kittler, S. Campylobacter Bacteriophage Cocktail Design Based on an Advanced Selection Scheme. Antibiotics 2022, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Kittler, S.; Fischer, S.; Abdulmawjood, A.; Glünder, G.; Klein, G. Effect of Bacteriophage Application on Campylobacter jejuni Loads in Commercial Broiler Flocks. Appl. Environ. Microbiol. 2013, 79, 7525–7533. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Wagenaar, J.A.; Bergen, M.A.P.V.; Mueller, M.A.; Wassenaar, T.M.; Carlton, R.M. Phage therapy reduces Campylobacter jejuni colonization in broilers. Vet. Microbiol. 2005, 109, 275–283. [Google Scholar] [CrossRef]
- Miller, R.W.; Skinner, J.; Sulakvelidze, A.; Mathis, G.F.; Hofacre, C.L. Bacteriophage Therapy for Control of Necrotic Enteritis of Broiler Chickens Experimentally Infected with Clostridium perfringens. Avian Dis. 2010, 54, 33–40. [Google Scholar] [CrossRef]
- Hosny, R.A.; Gaber, A.F.; Sorour, H.K. Bacteriophage mediated control of necrotic enteritis caused by C. perfringens in broiler chickens. Vet. Res. Commun. 2021, 45, 409–421. [Google Scholar] [CrossRef]
- The State of World Fisheries and Aquaculture 2020. FAO: Rome, Italy, 2020; Available online: https://www.fao.org/3/ca9229en/online/ca9229en.html (accessed on 31 May 2022)ISBN 978-92-5-132692-3.
- FAO. Food Outlook—Biannual Report on Global Food Markets: June 2020; Food Outlook; FAO: Rome, Italy, 2020; Available online: https://www.fao.org/documents/card/en/c/cb9427en/ (accessed on 31 May 2022)ISBN 978-92-5-132848-4.
- Culot, A.; Grosset, N.; Gautier, M. Overcoming the challenges of phage therapy for industrial aquaculture: A review. Aquaculture 2019, 513, 734423. [Google Scholar] [CrossRef]
- Xia, H.; Tang, Y.; Lu, F.; Luo, Y.; Yang, P.; Wang, W.; Jiang, J.; Li, N.; Han, Q.; Liu, F.; et al. The effect of Aeromonas hydrophila infection on the non-specific immunity of blunt snout bream (Megalobrama amblycephala). Cent. Eur. J. Immunol. 2017, 42, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Dien, L.T.; Ky, L.B.; Huy, B.T.; Mursalim, M.F.; Kayansamruaj, P.; Senapin, S.; Rodkhum, C.; Dong, H.T. Characterization and protective effects of lytic bacteriophage pAh6.2TG against a pathogenic multidrug-resistant Aeromonas hydrophila in Nile tilapia (Oreochromis niloticus). Transbound. Emerg. Dis. 2021, 69, e435–e450. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Leptihn, S.; Ramesh, N. Pharmacological and Immunological Aspects of Phage Therapy. Infect. Microbes Dis. 2019, 1, 34–42. [Google Scholar] [CrossRef]
- Górski, A.; Dąbrowska, K.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Łusiak-Szelachowska, M.; Jończyk-Matysiak, E.; Borysowski, J. Phages and immunomodulation. Future Microbiol. 2017, 12, 905–914. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity Studies of Proteins Forming the T4 Phage Head Surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Cong, C.; Wang, L.; Li, X.; Li, J.; Yang, H.; Li, S.; Xu, Y. Protective effectiveness of feeding phage cocktails in controlling Vibrio harveyi infection of turbot Scophthalmus maximus. Aquaculture 2021, 535, 736390. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Therapy: Various Perspectives on How to Improve the Art. In Host-Pathogen Interactions: Methods and Protocols; Methods in Molecular Biology; Medina, C., López-Baena, F.J., Eds.; Springer: New York, NY, USA, 2018; pp. 113–127. ISBN 978-1-4939-7604-1. [Google Scholar]
- Loh, B.; Gondil, V.S.; Manohar, P.; Khan, F.M.; Yang, H.; Leptihn, S. Encapsulation and Delivery of Therapeutic Phages. Appl. Environ. Microbiol. 2021, 87, e01979-20. [Google Scholar] [CrossRef]
- Lorenzo-Rebenaque, L.; Malik, D.J.; Catalá-Gregori, P.; Marin, C.; Sevilla-Navarro, S. In Vitro and In Vivo Gastrointestinal Survival of Non-Encapsulated and Microencapsulated Salmonella Bacteriophages: Implications for Bacteriophage Therapy in Poultry. Pharmaceuticals 2021, 14, 434. [Google Scholar] [CrossRef]
- Rotman, S.G.; Sumrall, E.; Ziadlou, R.; Grijpma, D.W.; Richards, R.G.; Eglin, D.; Moriarty, T.F. Local Bacteriophage Delivery for Treatment and Prevention of Bacterial Infections. Front. Microbiol. 2020, 11, 538060. [Google Scholar] [CrossRef]
- Ramasamy, P. Phage Therapy for Control of Bacterial Diseases. In Crustacea; Diarte-Plata, G., Escamilla-Montes, R., Eds.; InTech: London, UK, 2019; pp. 1–31. [Google Scholar]
- Pereira, C.; Duarte, J.; Costa, P.; Braz, M.; Almeida, A. Bacteriophages in the Control of Aeromonas sp. in Aquaculture Systems: An Integrative View. Antibiotics 2022, 11, 163. [Google Scholar] [CrossRef]
- Harshitha, N.; Rajasekhar, A.; Saurabh, S.; Sonalkar, R.; Tejashwini, M.; Mitra, S.D. Bacteriophages: Potential Biocontrol Agents and Treatment Options for Bacterial Pathogens. Clin. Microbiol. Newsl. 2022, 44, 41–50. [Google Scholar] [CrossRef]
- Upadhaya, S.D.; Ahn, J.M.; Cho, J.H.; Kim, J.Y.; Kang, D.K.; Kim, S.W.; Kim, H.B.; Kim, I.H. Bacteriophage cocktail supplementation improves growth performance, gut microbiome and production traits in broiler chickens. J. Anim. Sci. Biotechnol. 2021, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wang, Z.; Zou, T.; Chen, J.; Li, G.; Zheng, L.; Li, S.; You, J. Bacteriophage as an Alternative to Antibiotics Promotes Growth Performance by Regulating Intestinal Inflammation, Intestinal Barrier Function and Gut Microbiota in Weaned Piglets. Front. Vet. Sci. 2021, 8, 623899. [Google Scholar] [CrossRef] [PubMed]
- Rabiey, M.; Roy, S.R.; Holtappels, D.; Franceschetti, L.; Quilty, B.J.; Creeth, R.; Sundin, G.W.; Wagemans, J.; Lavigne, R.; Jackson, R.W. Phage biocontrol to combat Pseudomonas syringae pathogens causing disease in cherry. Microb. Biotechnol. 2020, 13, 1428–1445. [Google Scholar] [CrossRef] [PubMed]
- Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Jacobs-Sera, D.; Hatfull, G.F.; Hansen, L.H. Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses 2018, 10, 621. [Google Scholar] [CrossRef]
- Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Hansen, L.H. A novel six-phage cocktail reduces Pectobacterium atrosepticum soft rot infection in potato tubers under simulated storage conditions. FEMS Microbiol. Lett. 2019, 366, fnz101. [Google Scholar] [CrossRef]
- FAO. News Article: New Standards to Curb the Global Spread of Plant Pests and Diseases. Available online: https://www.fao.org/news/story/en/item/1187738/icode/ (accessed on 3 June 2022).
- WHO’s First Ever Global Estimates of Foodborne Diseases Find Children under 5 Account for Almost One Third of Deaths. Available online: https://www.who.int/news/item/03-12-2015-who-s-first-ever-global-estimates-of-foodborne-diseases-find-children-under-5-account-for-almost-one-third-of-deaths (accessed on 25 July 2022).
- Gómez, I.; Janardhanan, R.; Ibañez, F.C.; Beriain, M.J. The Effects of Processing and Preservation Technologies on Meat Quality: Sensory and Nutritional Aspects. Foods 2020, 9, 1416. [Google Scholar] [CrossRef]
- Amit, S.K.; Uddin, M.M.; Rahman, R.; Islam, S.M.R.; Khan, M.S. A review on mechanisms and commercial aspects of food preservation and processing. Agric. Food Secur. 2017, 6, 51. [Google Scholar] [CrossRef]
- Ziech, R.E.; Perin, A.P.; Lampugnani, C.; Sereno, M.J.; Viana, C.; Soares, V.M.; Pereira, J.G.; Pinto, J.P.d.A.N.; Bersot, L.d.S. Biofilm-producing ability and tolerance to industrial sanitizers in Salmonella spp. isolated from Brazilian poultry processing plants. LWT Food Sci. Technol. 2016, 68, 85–90. [Google Scholar] [CrossRef]
- Fink, R.; Oder, M.; Stražar, E.; Filip, S. Efficacy of cleaning methods for the removal of Bacillus cereus biofilm from polyurethane conveyor belts in bakeries. Food Control 2017, 80, 267–272. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, X.; Guo, D.; Shi, C.; Zhang, C.; Peng, X.; Yang, H.; Xia, X. Disinfectant Resistance Profiles and Biofilm Formation Capacity of Escherichia coli Isolated from Retail Chicken. Microb. Drug Resist. 2019, 25, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, C.; Duarte, A.C.; Escobedo, S.; Fernández, L.; Rodríguez, A.; García, P.; Martínez, B. Combined use of bacteriocins and bacteriophages as food biopreservatives. A review. Int. J. Food Microbiol. 2022, 368, 109611. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Vivas, J.; Elexpuru-Zabaleta, M.; Samano, M.L.; Barrera, A.P.; Forbes-Hernández, T.Y.; Giampieri, F.; Battino, M. Phages and Enzybiotics in Food Biopreservation. Molecules 2021, 26, 5138. [Google Scholar] [CrossRef] [PubMed]
- Byun, K.-H.; Han, S.H.; Choi, M.W.; Park, S.H.; Ha, S.-D. Isolation, characterization, and application of bacteriophages to reduce and inhibit Listeria monocytogenes in celery and enoki mushroom. Food Control 2022, 135, 108826. [Google Scholar] [CrossRef]
- Zhou, C.; Zhu, M.; Wang, Y.; Yang, Z.; Ye, M.; Wu, L.; Bao, H.; Pang, M.; Zhou, Y.; Wang, R.; et al. Broad host range phage vB-LmoM-SH3-3 reduces the risk of Listeria contamination in two types of ready-to-eat food. Food Control 2020, 108, 106830. [Google Scholar] [CrossRef]
- Esmael, A.; Azab, E.; Gobouri, A.A.; Nasr-Eldin, M.A.; Moustafa, M.M.A.; Mohamed, S.A.; Badr, O.A.M.; Abdelatty, A.M. Isolation and Characterization of Two Lytic Bacteriophages Infecting a Multi-Drug Resistant Salmonella Typhimurium and Their Efficacy to Combat Salmonellosis in Ready-to-Use Foods. Microorganisms 2021, 9, 423. [Google Scholar] [CrossRef]
- Guo, Y.; Li, J.; Islam, M.S.; Yan, T.; Zhou, Y.; Liang, L.; Connerton, I.F.; Deng, K.; Li, J. Application of a novel phage vB_SalS-LPSTLL for the biological control of Salmonella in foods. Food Res. Int. 2021, 147, 110492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-Y.; Sun, S.-F.; Zhang, Y.-S.; Hu, Q.; Zheng, X.-F.; Yang, Z.-Q.; Jiao, X.-A. Isolation and Characterization of a Virulent Bacteriophage for Controlling Salmonella Enteritidis Growth in Ready-to-Eat Mixed-Ingredient Salads. J. Food Prot. 2021, 84, 1629–1639. [Google Scholar] [CrossRef]
- Dewanggana, M.N.; Evangeline, C.; Ketty, M.D.; Waturangi, D.E.; Yogiara; Magdalena, S. Isolation, characterization, molecular analysis and application of bacteriophage DW-EC to control Enterotoxigenic Escherichia coli on various foods. Sci. Rep. 2022, 12, 495. [Google Scholar] [CrossRef]
- Liao, Y.-T.; Zhang, Y.; Salvador, A.; Harden, L.A.; Wu, V.C.H. Characterization of a T4-like Bacteriophage vB_EcoM-Sa45lw as a Potential Biocontrol Agent for Shiga Toxin-Producing Escherichia coli O45 Contaminated on Mung Bean Seeds. Microbiol. Spectr. 2022, 10, e02220-21. [Google Scholar] [CrossRef]
- Thung, T.Y.; Lee, E.; Mahyudin, N.A.; Wan Mohamed Radzi, C.W.J.; Mazlan, N.; Tan, C.W.; Radu, S. Partial characterization and in vitro evaluation of a lytic bacteriophage for biocontrol of Campylobacter jejuni in mutton and chicken meat. J. Food Saf. 2020, 40, e12770. [Google Scholar] [CrossRef]
- García, P.; Madera, C.; Martínez, B.; Rodríguez, A. Biocontrol of Staphylococcus aureus in curd manufacturing processes using bacteriophages. Int. Dairy J. 2007, 17, 1232–1239. [Google Scholar] [CrossRef]
- Shahin, K.; Bouzari, M. Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J. Food Sci. Technol. 2018, 55, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Ganegama Arachchi, G.J.; Cridge, A.G.; Dias-Wanigasekera, B.M.; Cruz, C.D.; McIntyre, L.; Liu, R.; Flint, S.H.; Mutukumira, A.N. Effectiveness of phages in the decontamination of Listeria monocytogenes adhered to clean stainless steel, stainless steel coated with fish protein, and as a biofilm. J. Ind. Microbiol. Biotechnol. 2013, 40, 1105–1116. [Google Scholar] [CrossRef]
- Wang, L.; Pang, X.; Zhao, J.; Jin, H.; Yang, X.; Fu, S.; Cheng, S.; Li, H.; Miao, C.; Man, C.; et al. Isolation and characteristics of new phage JK004 and application to control Cronobacter sakazakii on material surfaces and powdered infant formula. LWT 2022, 153, 112571. [Google Scholar] [CrossRef]
- Sadekuzzaman, M.; Mizan, M.F.R.; Yang, S.; Kim, H.-S.; Ha, S.-D. Application of bacteriophages for the inactivation of Salmonella spp. in biofilms. Food Sci. Technol. Int. 2018, 24, 424–433. [Google Scholar] [CrossRef]
- Wang, C.; Hang, H.; Zhou, S.; Niu, Y.D.; Du, H.; Stanford, K.; McAllister, T.A. Bacteriophage biocontrol of Shiga toxigenic Escherichia coli (STEC) O145 biofilms on stainless steel reduces the contamination of beef. Food Microbiol. 2020, 92, 103572. [Google Scholar] [CrossRef]
- Carvalho, C.; Costa, A.R.; Silva, F.; Oliveira, A. Bacteriophages and their derivatives for the treatment and control of food-producing animal infections. Crit. Rev. Microbiol. 2016, 43, 583–601. [Google Scholar] [CrossRef]
- Vötsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Watanabe, T.; Arai, S.; Kim, H.; Tohya, M.; Ishida-Kuroki, K.; Võ, T.H.; Nguyễn, T.P.B.; Nakagawa, I.; Osawa, R.; et al. Characterization of pig saliva as the major natural habitat of Streptococcus suis by analyzing oral, fecal, vaginal, and environmental microbiota. PLoS ONE 2019, 14, e0215983. [Google Scholar] [CrossRef]
- Gajdács, M.; Németh, A.; Knausz, M.; Barrak, I.; Stájer, A.; Mestyán, G.; Melegh, S.; Nyul, A.; Tóth, Á.; Ágoston, Z.; et al. Streptococcus suis: An Underestimated Emerging Pathogen in Hungary? Microorganisms 2020, 8, 1292. [Google Scholar] [CrossRef] [PubMed]
- Boonyagars, L.; Chongtrakool, P.; Watcharananan, S.P. Meningitis and spondylodiscitis due to Streptococcus suis. J. Infect. Dis. Antimicrob. Agents 2010, 27, 129–133. [Google Scholar]
- Estrada, A.A.; Gottschalk, M.; Rossow, S.; Rendahl, A.; Gebhart, C.; Marthaler, D.G. Serotype and Genotype (Multilocus Sequence Type) of Streptococcus suis Isolates from the United States Serve as Predictors of Pathotype. J. Clin. Microbiol. 2019, 57, e00377-19. [Google Scholar] [CrossRef] [PubMed]
- Tien, L.H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA–DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef]
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Segura, M.; Xu, J. Streptococcus suis infections in humans: The Chinese experience and the situation in North America. Anim. Health Res. Rev. 2007, 8, 29–45. [Google Scholar] [CrossRef]
- Li, P.; Liu, J.; Zhu, L.; Qi, C.; Bei, W.; Cai, X.; Sun, Y.; Feng, S. VirA: A virulence-related gene of Streptococcus suis serotype 2. Microb. Pathog. 2010, 49, 305–310. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Haas, B.; Bonifait, L.; Vaillancourt, K.; Charette, S.J.; Gottschalk, M.; Grenier, D. Characterization of DNase activity and gene in Streptococcus suis and evidence for a role as virulence factor. BMC Res. Notes 2014, 7, 424. [Google Scholar] [CrossRef]
- Gill, C.O.; Youssef, M.K. Microbiological Safety of Meat|Emerging Pathogens. In Encyclopedia of Meat Sciences, 2nd ed.; Dikeman, M., Devine, C., Eds.; Academic Press: Oxford, UK, 2014; pp. 340–344. ISBN 978-0-12-384734-8. [Google Scholar]
- Hlebowicz, M.; Jakubowski, P.; Smiatacz, T. Streptococcus suis Meningitis: Epidemiology, Clinical Presentation and Treatment. Vector-Borne Zoonotic Dis. 2019, 19, 557–562. [Google Scholar] [CrossRef]
- Maes, D.; Piñeiro, C.; Haesebrouck, F.; Boyen, F.; Rubio, P.; Manzanilla, E.G. 7—Control and prevention of bacterial diseases in swine. In Advancements and Technologies in Pig and Poultry Bacterial Disease Control; Foster, N., Kyriazakis, I., Barrow, P., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 171–198. ISBN 978-0-12-818030-3. [Google Scholar]
- Van der Ark, K.C.H.; Schultsz, C. Report on Retrospective Data on Incidence of S. suis Infection in Humans in the Partner Countries. Available online: https://ec.europa.eu/research/participants/documents/downloadPublic?documentIds=080166e5cfc2ea97&appId=PPGMS (accessed on 12 April 2022).
- Obradovic, M.R.; Segura, M.; Segalés, J.; Gottschalk, M. Review of the speculative role of co-infections in Streptococcus suis-associated diseases in pigs. Vet. Res. 2021, 52, 49. [Google Scholar] [CrossRef] [PubMed]
- Neila-Ibáñez, C.; Casal, J.; Hennig-Pauka, I.; Stockhofe-Zurwieden, N.; Gottschalk, M.; Migura-García, L.; Pailler-García, L.; Napp, S. Stochastic Assessment of the Economic Impact of Streptococcus suis-Associated Disease in German, Dutch and Spanish Swine Farms. Front. Vet. Sci. 2021, 8, 676002. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.; IJpelaar, J. Updated Estimates of the Costs Associated with Thirty Four Endemic Livestock Diseases in Great Britain: A Note. J. Agric. Econ. 2005, 56, 135–144. [Google Scholar] [CrossRef]
- Huong, V.T.L.; Turner, H.C.; Kinh, N.V.; Thai, P.Q.; Hoa, N.T.; Horby, P.; van Doorn, H.R.; Wertheim, H.F.L. Burden of disease and economic impact of human Streptococcus suis infection in Viet Nam. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Michaud, S.; Duperval, R.; Higgins, R. Streptococcus suis meningitis: First case reported in Quebec. Can. J. Infect. Dis. 1996, 7, 329–331. [Google Scholar]
- Arends, J.P.; Zanen, H.C. Meningitis Caused by Streptococcus suis in Humans. Rev. Infect. Dis. 1988, 10, 131–137. [Google Scholar] [CrossRef]
- Dong, X.; Chao, Y.; Zhou, Y.; Zhou, R.; Zhang, W.; Fischetti, V.A.; Wang, X.; Feng, Y.; Li, J. The global emergence of a novel Streptococcus suis clade associated with human infections. EMBO Mol. Med. 2021, 13, e13810. [Google Scholar] [CrossRef]
- Weinert, L.A.; Chaudhuri, R.R.; Wang, J.; Peters, S.E.; Corander, J.; Jombart, T.; Baig, A.; Howell, K.J.; Vehkala, M.; Välimäki, N.; et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015, 6, 6740. [Google Scholar] [CrossRef]
- Huong, V.T.L.; Ha, N.; Huy, N.T.; Horby, P.; Nghia, H.D.T.; Thiem, V.D.; Zhu, X.; Hoa, N.T.; Hien, T.T.; Zamora, J.; et al. Epidemiology, Clinical Manifestations, and Outcomes of Streptococcus suis Infection in Humans. Emerg. Infect. Dis. 2014, 20, 1105–1114. [Google Scholar] [CrossRef]
- Rayanakorn, A.; Goh, B.-H.; Lee, L.-H.; Khan, T.M.; Saokaew, S. Risk factors for Streptococcus suis infection: A systematic review and meta-analysis. Sci. Rep. 2018, 8, 13358. [Google Scholar] [CrossRef]
- Hughes, J.M.; Wilson, M.E.; Wertheim, H.F.L.; Nghia, H.D.T.; Taylor, W.; Schultsz, C. Streptococcus suis: An Emerging Human Pathogen. Clin. Infect. Dis. 2009, 48, 617–625. [Google Scholar] [CrossRef]
- Wangkaew, S.; Chaiwarith, R.; Tharavichitkul, P.; Supparatpinyo, K. Streptococcus suis infection: A series of 41 cases from Chiang Mai University Hospital. J. Infect. 2006, 52, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Manzin, A.; Palmieri, C.; Serra, C.; Saddi, B.; Princivalli, M.S.; Loi, G.; Angioni, G.; Tiddia, F.; Varaldo, P.E.; Facinelli, B. Streptococcus suis Meningitis without History of Animal Contact, Italy. Emerg. Infect. Dis. 2008, 14, 1946–1948. [Google Scholar] [CrossRef]
- Okwumabua, O.; Peterson, H.; Hsu, H.-M.; Bochsler, P.; Behr, M. Isolation and partial characterization of Streptococcus suis from clinical cases in cattle. J. Vet. Diagn. Investig. 2017, 29, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Muckle, A.; López, A.; Gottschalk, M.; López-Méndez, C.; Giles, J.; Lund, L.; Saab, M. Isolation of Streptococcus suis from 2 lambs with a history of lameness. Can. Vet. J. 2014, 55, 946–949. [Google Scholar] [PubMed]
- Muckle, A.; Giles, J.; Lund, L.; Stewart, T.; Gottschalk, M. Isolation of Streptococcus suis from the urine of a clinically ill dog. Can. Vet. J. 2010, 51, 773–774. [Google Scholar] [PubMed]
- Sánchez del Rey, V.; Fernández-Garayzábal, J.F.; Briones, V.; Iriso, A.; Domínguez, L.; Gottschalk, M.; Vela, A.I. Genetic analysis of Streptococcus suis isolates from wild rabbits. Vet. Microbiol. 2013, 165, 483–486. [Google Scholar] [CrossRef]
- Ma, Y.L.; Lu, C.P. Isolation and identification of a bacteriophage capable of infecting Streptococcus suis type 2 strains. Vet. Microbiol. 2008, 132, 340–347. [Google Scholar] [CrossRef]
- Laddika, L.; Malik, S.; Chauhan, R.; Qureshi, S.; Sahoo, M.; Patel, S.; Tiwari, A. Isolation and Partial Characterisation of Streptococcus suis Bacteriophage. Int J Curr Microbiol App Sci 2021, 10, 1300–1306. [Google Scholar]
- Harel, J.; Martinez, G.; Nassar, A.; Dezfulian, H.; Labrie, S.J.; Brousseau, R.; Moineau, S.; Gottschalk, M. Identification of an Inducible Bacteriophage in a Virulent Strain of Streptococcus suis Serotype 2. Infect. Immun. 2003, 71, 6104–6108. [Google Scholar] [CrossRef]
- Ma, Y.; Lu, C.; Fan, H. Isolation and identification of the temperate bacteriophage from isolated strains of Streptococcus suis serotype 2. Acta Microbiol. Sin. 2008, 48, 508–513. [Google Scholar]
- Tang, F.; Bossers, A.; Harders, F.; Lu, C.; Smith, H. Comparative genomic analysis of twelve Streptococcus suis (pro)phages. Genomics 2013, 101, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ma, J.; Shang, K.; Hu, X.; Liang, Y.; Li, D.; Wu, Z.; Dai, L.; Chen, L.; Wang, L. Evolution and Diversity of the Antimicrobial Resistance Associated Mobilome in Streptococcus suis: A Probable Mobile Genetic Elements Reservoir for Other Streptococci. Front. Cell. Infect. Microbiol. 2016, 6, 118. [Google Scholar] [CrossRef]
- Tang, F.; Bossers, A.; Harders, F.; Lu, C.; Smith, H. Complete Genome Sequence of the Streptococcus suis Temperate Bacteriophage ϕNJ2. Genome Announc. 2013, 1, e00008-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, L.J.; Meaden, S.; Mestre, M.R.; Palmer, C.; Toro, N.; Fineran, P.C.; Jackson, S.A. PADLOC: A web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res. 2022, 50, W541–W550. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef]
- Willemse, N.; Schultsz, C. Distribution of Type I Restriction–Modification Systems in Streptococcus suis: An Outlook. Pathogens 2016, 5, 62. [Google Scholar] [CrossRef]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011, 45, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.C.; Walker, A.R.; Maricic, N.; Chakraborty, B.; Underhill, S.A.M.; Burne, R.A. Repurposing the Streptococcus mutans CRISPR-Cas9 System to Understand Essential Gene Function. PLOS Pathog. 2020, 16, e1008344. [Google Scholar] [CrossRef] [PubMed]
- Brueggemann, A.B.; Harrold, C.L.; Rezaei Javan, R.; van Tonder, A.J.; McDonnell, A.J.; Edwards, B.A. Pneumococcal prophages are diverse, but not without structure or history. Sci. Rep. 2017, 7, 42976. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Fan, X.; Long, Q.; Deng, W.; Song, J.; Huang, E. Comparative analysis of prophages in Streptococcus mutans genomes. PeerJ 2017, 5, e4057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavrich, T.N.; Hatfull, G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017, 2, 17112. [Google Scholar] [CrossRef]
- Dupuis, M.-È.; Villion, M.; Magadán, A.H.; Moineau, S. CRISPR-Cas and restriction–modification systems are compatible and increase phage resistance. Nat. Commun. 2013, 4, 2087. [Google Scholar] [CrossRef]
- Modell, J.W.; Jiang, W.; Marraffini, L.A. CRISPR–Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 2017, 544, 101–104. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Rocha, E.P.C.; Touchon, M. The Adaptation of Temperate Bacteriophages to Their Host Genomes. Mol. Biol. Evol. 2013, 30, 737–751. [Google Scholar] [CrossRef]
- Sutcliffe, S.G.; Reyes, A.; Maurice, C.F. Bacteriophages Playing Nice: Lysogenic bacteriophage replication stable in the human gut microbiota. bioRxiv 2022. [Google Scholar]
- Mavrich, T.N.; Hatfull, G.F. Evolution of Superinfection Immunity in Cluster A Mycobacteriophages. mBio 2019, 10, e00971-19. [Google Scholar] [CrossRef]
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2019, 27, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Martel, B.; Moineau, S. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014, 42, 9504–9513. [Google Scholar] [CrossRef] [PubMed]
- Pryor, J.M.; Potapov, V.; Bilotti, K.; Pokhrel, N.; Lohman, G.J.S. Rapid 40 kb Genome Construction from 52 Parts through Data-optimized Assembly Design. ACS Synth. Biol. 2022, 11, 2036–2042. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Avrani, S.; Barrick, J.E.; Petrie, K.L.; Meyer, J.R. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2104592118. [Google Scholar] [CrossRef]
- Linden, S.B.; Alreja, A.B.; Nelson, D.C. Application of bacteriophage-derived endolysins to combat streptococcal disease: Current state and perspectives. Curr. Opin. Biotechnol. 2021, 68, 213–220. [Google Scholar] [CrossRef]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-Based Antibacterials as Therapeutics. In Therapeutic Enzymes: Function and Clinical Implications; Advances in Experimental Medicine and Biology; Labrou, N., Ed.; Springer: Singapore, 2019; pp. 233–253. ISBN 9789811377099. [Google Scholar]
- Wang, Z.; Ma, J.; Wang, J.; Yang, D.; Kong, L.; Fu, Q.; Cheng, Y.; Wang, H.; Yan, Y.; Sun, J. Application of the Phage Lysin Ply5218 in the Treatment of Streptococcus suis Infection in Piglets. Viruses 2019, 11, 715. [Google Scholar] [CrossRef]
- Hoopes, J.T.; Stark, C.J.; Kim, H.A.; Sussman, D.J.; Donovan, D.M.; Nelson, D.C. Use of a Bacteriophage Lysin, PlyC, as an Enzyme Disinfectant against Streptococcus equi. Appl. Environ. Microbiol. 2009, 75, 1388–1394. [Google Scholar] [CrossRef] [Green Version]

| Feature | Advantages | Limitations/Remarks |
|---|---|---|
| Specificity | Targets bacterial strains in a highly specific manner, thereby targeting only the intended bacteria and leaving bystander members of the resident microbiota unharmed | Strain specificity typically results in narrow target range compared to antibiotics. A mixture of several phages (cocktail) may be required for optimal bacteria clearance |
| Mode of action | Autodosing: ability to replicate at the site of infection and lyse bacterial cells | Temperate phages may integrate into bacterial chromosomes (prophages) and be passively replicated without resulting in bacterial lysis |
| Toxicity, safety profile and immunogenicity | Phages do not infect or have serious adverse effect on eukaryotic cells. Endotoxins can be easily removed during phage purification. | Efficacy of phages may be reduced through neutralisation by animal immune system |
| Efficacy against MDR | Effective against multidrug resistant bacteria | Some phages may encode antibiotic resistance to genes and toxins that confer extra fitness to bacteria |
| Resistance | Specificity of phages limits the widespread use of specific phages in different infections, thus reducing the chances of resistance development by bacteria | Bacteria encode anti-phage systems such as abortive infection, restriction-modification, gabija, CRISPR-Cas, DISARM, etc., that interfere with successful phage infection |
| Production | Natural; can be isolated from diverse clinical and environmental sources and characterised rapidly compared to antibiotic discovery and development | Difficulty in isolating good therapeutic phage candidates against specific species or strains such as S. suis Regulatory approval for the use of phages as therapeutics is onerous and has limited the commercialisation of phage products |
| Administration | Can be incorporated into feed or water and administered to animals | Challenges in formulation and stabilisation of phage preparation for therapy have been reported |
| Condition/Infection | Phage Intervention | Remarks | Reference |
|---|---|---|---|
| Cystic fibrosis/Achromobacter xylosoxidans | Phage cocktail (3 × 108 pfu/mL) for 20 d via inh, p.o. Treatment was repeated 4 times after initial PT at 1, 3, 6 and 12 mo. | Dyspnea resolved and cough reduced Increased lung function | [49] |
| Crohn’s disease/MDR Klebsiella pneumoniae | 3 week cycle single phage treatment 106 pfu/mL p.o., 106 pfu/mL rectal | 15 days after first PT treatment, no MDR K. pneumoniae was isolated from patient’s stools, rectal swabs, urine and the ureteral stents | [50] |
| Necrotising pancreatitis/systemic Acinetobacter baumannii | 3 different phage cocktails (1 × 109 pfu/mL i.c. for 18 weeks and 5 × 109 pfu/mL i.v. for 2 or 16 weeks) | Patient awoke from coma; mental health and renal function improvedPatient was discharged on day 245 | [51] |
| Aorto-cutaneous fistula/Pseudomonas aeruginosa | Single i.o. dose of 108 pfu/mL phage OMKO1 | Blood cultures tested negative for P. aeruginosa after for 4 weeks | [52] |
| Fracture-related infection (FRI)/Klebsiella pneumoniae | 100 mL of 108 pfu/mL on day 1 and 107 pfu/mL was instilled on surgical wound via catheter t.i.d. up to day 5 | Improved microbiological, radiological and blood parameters 3 months post-phage therapy FRI was controlled | [53] |
| Disseminated cutaneous/Mycobacterium chelonae infection | i.v. of 109 pfu/mL b.i.d. for > 6 months | Discharged on day 4 following no adverse effects and improved laboratory markers No evidence of granulomas 2 months after beginning of phage therapy | [54] |
| Chronic vascular graft infection/Staphylococcus aureus | local application of 20 mL 109 pfu/mL via drainage q.12 h for two days | Negative blood culture after last day of phage treatmentNo sign of graft infection | [55] |
| Sternal wound abscesses/P. aeruginosa | Single 4 ml 4 × 1010 pfu/mL i.o. | Wound was completely healed P. aeruginosa was undetectable post-phage therapy | [55] |
| Cystic fibrosis/P. aeruginosa | 8 weeks of 4 × 109 pfu/mL i.v., q.6 h | Resolution of renal function, white blood cell counts and fever No CF exacerbation or recurrence of P. aeruginosa 100 days post-PT | [56] |
| Cystic fibrosis/Mycobacterium abscessus | Single topical 109 pfu/mL cocktail on wound 109 pfu/mL i.v., q.12 h for 32 weeks | Negative serum and sputum cultures Positive skin nodule swabs up to 5 months post-PTNo sera phage neutralisation | [57] |
| Lung disease/M. abscessus | Up to 109 pfu/mL i.v., b.i.d. for > 6 months | M. abscessus cultures positive (6 of 7) through day 96 Most recent cultures (days 116–362) were negative (90%) Patient successfully underwent lung transplant post-PT | [58] |
| Food Matrix/Surface | Target Pathogen | Phage Treatment | Results/Log Unit Pathogen Reduction |
|---|---|---|---|
| Celery [116] Enoki mushrooms | L. monocytogenes | cocktail | Reduced initial 5.0 by 2.2 (celery) and 1.8 (mushroom) |
| Salmon meat [117] Orange juice | L. monocytogenes | SH3-3 phage | Target undetectable at 72 h compared to control (2.31) |
| Milk [118] Chicken | S. Typhimurium | cocktail | Milk: undetectable by 2 h (MOI 1000) or 12 h (MOI 100) at 25 °C Chicken: undetectable by 2 h (MOI 1000) or 6 h (MOI 100) at 25 °C |
| Milk [119] Apple juice | Salmonella | phage LPSTLL | Initial 3.0 reduced by 2.8 in milk and by 0.52 in apple juice |
| Chicken-lettuce salad [120] | S. Enteritidis | SapYZU01 | Initial 5.1 reduced by 3.4 |
| Meat and vegetables [121] | E. coli | Phage DW-EC | Initial 6.0 reduced by 43.38–87.89% on the foods |
| Mung beans [122] | E. coli | phage Sa45lw | Initial 4.8 reduced by 2.0 within 6 h |
| Chicken [123] Mutton | C. jejuni | phage CJ01 | Initial 4.0 reduced by 1.68 in chicken and 1.70 in mutton |
| Acid curd [124] Rennet curd | S. aureus | cocktail | undetectable by 4 h in acid curd or 1 h in rennet curd |
| Chicken breast [125] | Shigella Flexneri | phage SflS-ISF001 | Initial 4.0 reduced beyond 2.0 |
| Stainless steel [126] | L. monocytogenes | cocktail | Initial ~5.4 was undetectable by 75 min |
| Rubber [127] polyethylene Stainless steel (SS) | Cronobacter sakazakii | phage JK004 | inhibition rate for 6 h for rubber, polyethylene or SS was 99.95, 99.83, or 99.84%, respectively |
| Stainless steel [128] | S. Enteriditis and Typhimurium | phages BP1369 and BP1370 | Undetectable after 144 h at 10 °C |
| Stainless steel [129] | E. coli | phage AZO145A | Initially 4.8 reduced by 2.9 in 24 h |
| Product Name | Target Pathogen | Application | |
|---|---|---|---|
| Proteon Pharmaceuticals (Poland) | Bafasal® | Salmonella spp. | Feed or water additive |
| Bafador® | Aeromonas hydrophila and Pseudomonas fluorescens | Feed additive for aquaculture | |
| Intralytix (USA) | INT-401™ | Clostridium perfringens | In-feed or water additive |
| PLSV-1™ | Salmonella spp. | Animal health care | |
| Ecolicide PX™ | E. coli O157:H7 | Hides of live animals | |
| ListShield™ | L. monocytogenes | Food and surface decontamination | |
| ShigaShield™ | Shigella spp. | Decontamination of ready-to-eat (RTE) foods | |
| SalmoLyse® | Salmonella spp. | Decontamination of pet food | |
| ACD Pharma (Norway) | CUSTUS® YRS | Yersinia | Treatment of fish environment in aquaculture |
| Phagelux (China) | LUZON | Staphylococcus, E. coli, P. aeruginosa or Salmonella | Control of infections in pig farms |
| SHIJUNSHA | Staphylococcus, E. coli, P. aeruginosa or Salmonella | Control of infections in poultry farms | |
| OmniLytics Inc. (USA) | Agriphage™ | Xanthomonas campestris, Pseudomonas syringae | Infection control on pepper and tomato |
| CJ Cheiljedang Corp. (South Korea) | Biotector | Salmonella, C. perfringens and E. coli | Disease management on farms |
| Micreos Food Safety (The Netherlands) | PhageGuard Listex™ | L. monocytogenes | Decontamination of RTE and frozen foods |
| PhageGuard E | E. coli | Decontamination of food products | |
| PhageGuard S | Salmonella spp. | Decontamination of food products |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osei, E.K.; Mahony, J.; Kenny, J.G. From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents. Viruses 2022, 14, 1996. https://doi.org/10.3390/v14091996
Osei EK, Mahony J, Kenny JG. From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents. Viruses. 2022; 14(9):1996. https://doi.org/10.3390/v14091996
Chicago/Turabian StyleOsei, Emmanuel Kuffour, Jennifer Mahony, and John G. Kenny. 2022. "From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents" Viruses 14, no. 9: 1996. https://doi.org/10.3390/v14091996
APA StyleOsei, E. K., Mahony, J., & Kenny, J. G. (2022). From Farm to Fork: Streptococcus suis as a Model for the Development of Novel Phage-Based Biocontrol Agents. Viruses, 14(9), 1996. https://doi.org/10.3390/v14091996







