Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections
Abstract
:1. Introduction
2. Common Characteristics of the Viruses That Are Able to Induce ADE
3. Molecular Mechanisms Underlying the ADE of Viral Infections
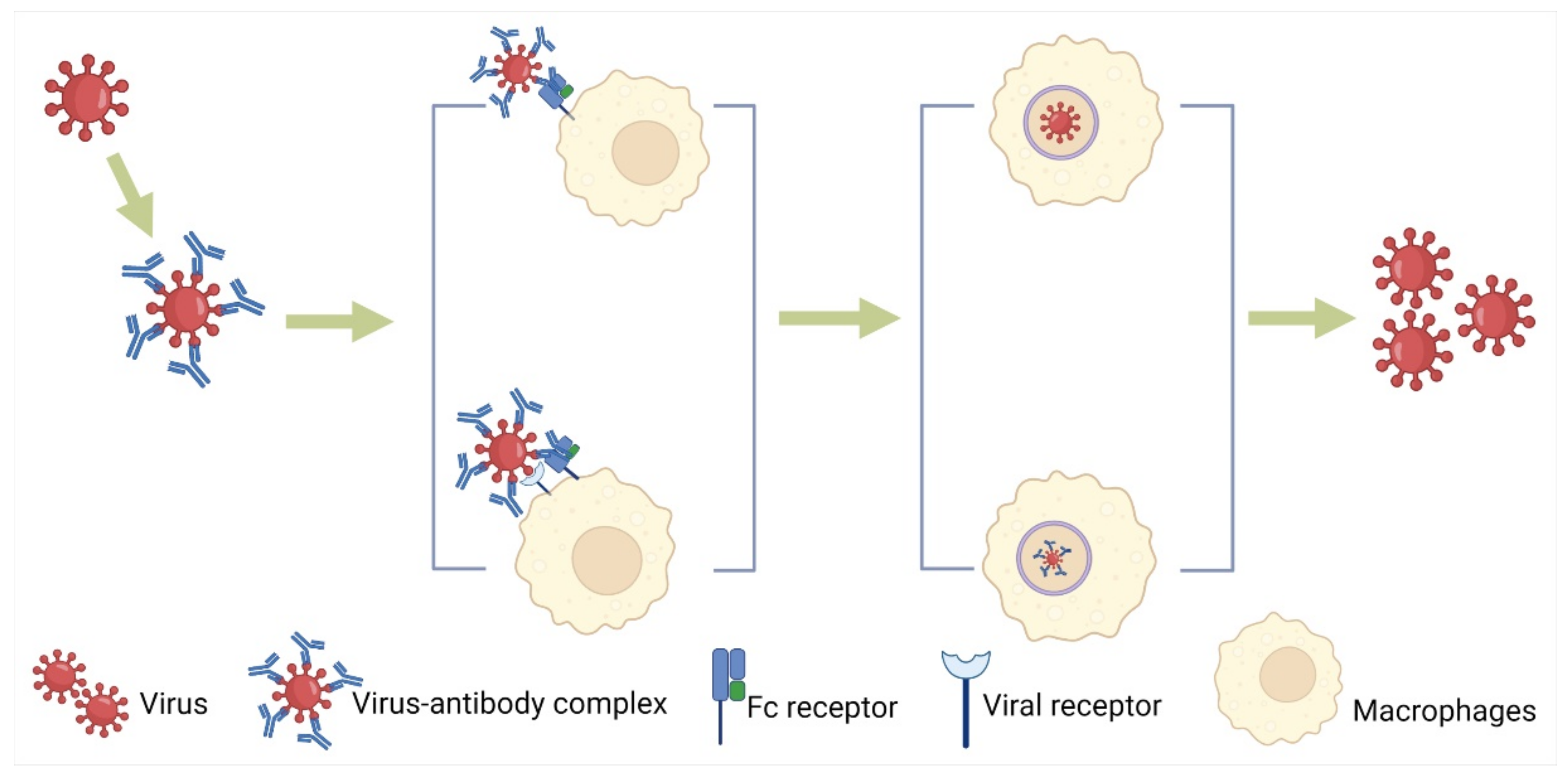
3.1. Facilitating Virus Entry into the Target Cells with or without Viral Receptors
3.2. Changing the Innate Immune Response of Host Cells
3.3. Changing the Transcriptional Levels of Host Molecules Supporting Viral Replication
4. Challenges and Prospects of Viral ADE
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rogers, T.F.; Goodwin, E.; Briney, B.; Sok, D.; Beutler, N.; Strubel, A.; Nedellec, R.; Le, K.; Brown, M.E.; Burton, D.R.; et al. Zika virus activates de novo and cross-reactive memory B cell responses in dengue-experienced donors. Sci. Immunol. 2017, 2, eaan6809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, M.J.; Broecker, F.; Freyn, A.W.; Choi, A.; Brown, J.A.; Fedorova, N.; Simon, V.; Lim, J.K.; Evans, M.J.; Garcia-Sastre, A.; et al. Human monoclonal antibodies potently neutralize Zika virus and select for escape mutations on the lateral ridge of the envelope protein. J. Virol. 2019, 93, e00405–e00419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderven, H.A.; Jegaskanda, S.; Wheatley, A.K.; Kent, S.J. Antibody-dependent cellular cytotoxicity and influenza virus. Curr. Opin. Virol. 2017, 22, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.W.; Yuan, S.; Poon, K.M.; Wen, L.; Yang, D.; Sun, Z.; Li, C.; Hu, M.; Shuai, H.; Zhou, J.; et al. Antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin head region of pandemic H1N1 influenza virus play detrimental roles in H1N1-infected mice. Front. Immunol. 2017, 8, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenzel, K.; Lehmann, J.; Kruger, D.H.; Martin-Parras, L.; Uharek, L.; Hofmann, J. Combination of immunoglobulins and natural killer cells in the context of CMV and EBV infection. Med. Microbiol. Immunol. 2014, 203, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Oliviero, B.; Mantovani, S.; Varchetta, S.; Mele, D.; Grossi, G.; Ludovisi, S.; Nuti, E.; Rossello, A.; Mondelli, M. Hepatitis C virus-induced NK cell activation causes metzincin-mediated CD16 cleavage and impaired antibody-dependent cytotoxicity. J. Hepatol. 2017, 66, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, M.L.; Qian, S.; Zhang, Z.N.; Fu, Y.J.; Xu, J.J.; Han, X.X.; Ding, H.B.; Dong, T.; Shang, H.; et al. The early antibody-dependent cell-mediated cytotoxicity response is associated with lower viral set point in individuals with primary HIV infection. Front. Immunol. 2018, 9, 2322. [Google Scholar] [CrossRef] [Green Version]
- Forthal, D.N.; Finzi, A. Antibody-dependent cellular cytotoxicity in HIV infection. AIDS 2018, 32, 2439–2451. [Google Scholar] [CrossRef]
- Holder, K.A.; Lajoie, J.; Grant, M.D. Natural killer cells adapt to cytomegalovirus along a functionally static phenotypic spectrum in human immunodeficiency virus infection. Front. Immunol. 2018, 9, 2494. [Google Scholar] [CrossRef]
- Sicca, F.; Neppelenbroek, S.; Huckriede, A. Effector mechanisms of influenza-specific antibodies: Neutralization and beyond. Expert Rev. Vaccines 2018, 17, 785–795. [Google Scholar] [CrossRef]
- Yu, W.H.; Cosgrove, C.; Berger, C.T.; Cheney, P.C.; Alter, G. ADCC-mediated CD56DIM NK cell responses are associated with early HBsAg clearance in acute HBV infection. Pathog. Immun. 2018, 3, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Jegaskanda, S.; Co, M.; Cruz, J.; Subbarao, K.; Ennis, F.A.; Terajima, M. Induction of H7N9-cross-reactive antibody-dependent cellular cytotoxicity antibodies by human seasonal influenza a viruses that are directed toward the nucleoprotein. J. Infect. Dis. 2017, 215, 818–823. [Google Scholar]
- Halstead, S.B.; Medicine, S. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977, 146, 201–217. [Google Scholar] [CrossRef]
- Halstead, S.B.; O’Rourke, E.J. Antibody-enhanced dengue virus infection in primate leukocytes. Nature 1977, 265, 739–741. [Google Scholar] [CrossRef]
- Halstead, S.B.; Medicine, S. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J. Exp. Med. 1977, 146, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B.; Mahalingam, S.; Marovich, M.A.; Ubol, S.; Mosser, D.M. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: Disease regulation by immune complexes. Lancet Infect. Dis. 2010, 10, 712–722. [Google Scholar] [CrossRef] [Green Version]
- Gollins, S.W.; Porterfield, J.S. Flavivirus infection enhancement in macrophages: An electron microscopic study of viral cellular entry. J. Gen. Virol. 1985, 66, 1969–1982. [Google Scholar] [CrossRef]
- Gollins, S.W.; Porterfield, J. Flavivirus infection enhancement in macrophages: Radioactive and biological studies on the effect of antibody on viral fate. J. Gen. Virol. 1984, 65, 1261–1272. [Google Scholar] [CrossRef]
- Linn, M.L.; Aaskov, J.G.; Suhrbier, A. Antibody-dependent enhancement and persistence in macrophages of an arbovirus associated with arthritis. J. Gen. Virol. 1996, 77, 407–411. [Google Scholar] [CrossRef]
- Takada, A.; Kawaoka, Y. Antibody-dependent enhancement of viral infection: Molecular mechanisms and in vivo implications. Rev. Med. Virol. 2003, 13, 387–398. [Google Scholar] [CrossRef]
- Kimman, T.G.; Cornelissen, L.A.; Moormann, R.J.; Rebel, J.M.; Stockhofe-Zurwieden, N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 2009, 27, 3704–3718. [Google Scholar] [CrossRef]
- Yoon, K.J.; Wu, L.L.; Zimmerman, J.J.; Hill, H.T.; Platt, K.B. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996, 9, 51–63. [Google Scholar] [CrossRef]
- Weiss, R.C.; Scott, F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: Comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 175–189. [Google Scholar] [CrossRef]
- Lager, K.M.; Mengeling, W.L.; Brockmeier, S.L. Evaluation of protective immunity in gilts inoculated with the NADC-8 isolate of porcine reproductive and respiratory syndrome virus (PRRSV) and challenge-exposed with an antigenically distinct PRRSV isolate. Am. J. Vet. Res. 1999, 60, 1022–1027. [Google Scholar]
- Balsitis, S.J.; Williams, K.L.; Lachica, R.; Flores, D.; Harris, E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 2010, 6, e1000790. [Google Scholar] [CrossRef] [Green Version]
- Mccann, P.A.; Amirfeyz, R.; Wakeley, C.; Bhatia, R. The volar anatomy of the distal radius--An MRI study of the FCR approach. Injury 2010, 41, 1012–1014. [Google Scholar] [CrossRef]
- Qiao, S.; Liu, Y.; Zhang, J.; Yang, S.; Wan, B.; Shi, P.; Zhang, H.; Guo, J.; Zhang, G. Genetic characterization and ligand specificity of the ovine Fc gamma receptor I (ovFc gamma RI). Vet. Immunol. Immunopathol. 2010, 137, 317–321. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Bruhns, P.; Horiuchi, K.; Ravetch, J.V. FcγRIV: A novel FcR with distinct IgG subclass specificity. Immunity 2005, 23, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Bournazos, S.; Wang, T.; Dahan, R.; Maamary, J.; Ravetch, J.V. Signaling by antibodies: Recent progress. Annu. Rev. Immunol. 2017, 35, 285–311. [Google Scholar] [CrossRef] [Green Version]
- Shi, P.; Zhang, L.; Wang, J.; Lu, D.; Li, Y.; Ren, J.; Shen, M.; Zhang, L.; Huang, J. Porcine FcεRI mediates porcine reproductive and respiratory syndrome virus multiplication and regulates the inflammatory reaction. Virol. Sin. 2018, 33, 249–260. [Google Scholar] [CrossRef]
- Cardosa, M.J.; Porterfield, J.S.; Gordon, S. Complement receptor mediates enhanced flavivirus replication in macrophages. J. Exp. Med. 1983, 158, 258–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polack, F.P.; Teng, M.N.; Collins, P.L.; Prince, G.A.; Exner, M.; Regele, H.; Lirman, D.D.; Rabold, R.; Hoffman, S.J.; Karp, C.L.; et al. A role for immune complexes in enhanced respiratory syncytial virus disease. J. Exp. Med. 2002, 196, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Gralinski, L.E.; Sheahan, T.P.; Morrison, T.E.; Menachery, V.D.; Jensen, K.; Leist, S.R.; Whitmore, A.; Heise, M.T.; Baric, R.S. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio 2018, 9, e01753-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef]
- Ubol, S.; Halstead, S.B. How innate immune mechanisms contribute to antibody-enhanced viral infections. Clin. Vaccine Immunol. 2010, 17, 1829–1835. [Google Scholar] [CrossRef] [Green Version]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [Green Version]
- Rodenhuis-Zybert, I.A.; van der Schaar, H.M.; da Silva Voorham, J.M.; van der Ende-Metselaar, H.; Lei, H.Y.; Wilschut, J.; Smit, J.M. Immature dengue virus: A veiled pathogen? PLoS Pathog. 2010, 6, e1000718. [Google Scholar] [CrossRef] [Green Version]
- Shi, P.; Su, Y.; Li, Y.; Zhang, L.; Lu, D.; Li, R.; Zhang, L.; Huang, J. The alternatively spliced porcine FcγRI regulated PRRSV-ADE infection and proinflammatory cytokine production. Dev. Comp. Immunol. 2019, 90, 186–198. [Google Scholar] [CrossRef]
- Flipse, J.; Wilschut, J.; Smit, J.M. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic 2013, 14, 25–35. [Google Scholar] [CrossRef]
- Ayala-Nunez, N.V.; Hoornweg, T.E.; van de Pol, D.P.; Sjollema, K.A.; Flipse, J.; van der Schaar, H.M.; Smit, J.M. How antibodies alter the cell entry pathway of dengue virus particles in macrophages. Sci. Rep. 2016, 6, 28768. [Google Scholar] [CrossRef]
- Bardina, S.V.; Bunduc, P.; Tripathi, S.; Duehr, J.; Frere, J.J.; Brown, J.A.; Nachbagauer, R.; Foster, G.A.; Krysztof, D.; Tortorella, D.; et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 2017, 356, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Lilienthal, G.M.; Rahmoller, J.; Petry, J.; Bartsch, Y.C.; Leliavski, A.; Ehlers, M. Potential of murine IgG1 and human IgG4 to inhibit the classical complement and Fc gamma receptor activation pathways. Front. Immunol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Oostindie, S.C.; Lazar, G.A.; Schuurman, J.; Parren, P.W. Avidity in antibody effector functions and biotherapeutic drug design. Nat. Rev. Drug Discov. 2022, 5, 1–21. [Google Scholar] [CrossRef]
- Cardosa, M.J.; Gordon, S.; Hirsch, S.; Springer, T.A.; Porterfield, J.S. Interaction of West Nile virus with primary murine macrophages: Role of cell activation and receptors for antibody and complement. J. Virol. 1986, 57, 952–959. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Tai, W.; Zhou, Y.; Jiang, S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev. Vaccines 2016, 15, 1123–1134. [Google Scholar] [CrossRef]
- Takano, T.; Hohdatsu, T.; Toda, A.; Tanabe, M.; Koyama, H. TNF-alpha, produced by feline infectious peritonitis virus (FIPV)-infected macrophages, upregulates expression of type II FIPV receptor feline aminopeptidase N in feline macrophages. Virology 2007, 364, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Thavorasak, T.; Chulanetra, M.; Glab-Ampai, K.; Teeranitayatarn, K.; Songserm, T.; Yodsheewan, R.; Sae-Lim, N.; Lekcharoensuk, P.; Sookrung, N.; Chaicumpa, W. Novel neutralizing epitope of PEDV S1 protein identified by IgM monoclonal antibody. Viruses 2022, 14, 125. [Google Scholar] [CrossRef]
- Park, J.E.; Jang, H.; Kim, J.H.; Hyun, B.H.; Shin, H.J. Immunization with porcine epidemic diarrhea virus harbouring Fc domain of IgG enhances antibody production in pigs. Vet. Q. 2020, 40, 183–189. [Google Scholar] [CrossRef]
- Ubol, S.; Phuklia, W.; Kalayanarooj, S.; Modhiran, N. Mechanisms of immune evasion induced by a complex of dengue virus and preexisting enhancing antibodies. J. Infect. Dis. 2010, 201, 923–935. [Google Scholar] [CrossRef] [Green Version]
- Dejnirattisai, W.; Wongwiwat, W.; Supasa, S.; Zhang, X.K.; Dai, X.H.; Rouvinsky, A.; Jumnainsong, A.; Edwards, C.; Quyen, N.; Duangchinda, T.; et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015, 16, 785. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, J.; Wang, D.; Li, N.; Qin, Y.; Du, D.; Yang, M.; Xia, P. Ligation of porcine Fc gamma receptor III inhibits levels of antiviral cytokine in response to PRRSV infection in vitro. Res. Vet. Sci. 2016, 105, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Janoff, E.N.; Wahl, S.M.; Thomas, K.; Smith, P.D. Modulation of human immunodeficiency virus type 1 infection of human monocytes by IgA. J. Infect. Dis. 1995, 172, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Winarski, K.L.; Tang, J.; Klenow, L.; Lee, J.; Coyle, E.M.; Manischewitz, J.; Turner, H.L.; Takeda, K.; Ward, A.B.; Golding, H.; et al. Antibody-dependent enhancement of influenza disease promoted by increase in hemagglutinin stem flexibility and virus fusion kinetics. Proc. Natl. Acad. Sci. USA 2019, 116, 15194–15199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Sreenivasan, C.; Uprety, T.; Gao, R.; Huang, C.; Lee, E.J.; Lawson, S.; Nelson, J.; Christopher-Hennings, J.; Kaushik, R.S.; et al. Piglet immunization with a spike subunit vaccine enhances disease by porcine epidemic diarrhea virus. NPJ Vaccines 2021, 6, 22. [Google Scholar] [CrossRef]
- Thavorasak, T.; Chulanetra, M.; Glab-Ampai, K.; Mahasongkram, K.; Sae-Lim, N.; Teeranitayatarn, K.; Songserm, T.; Yodsheewan, R.; Nilubol, D.; Chaicumpa, W.; et al. Enhancing epitope of PEDV spike protein. Front. Microbiol. 2022, 16, 1664. [Google Scholar] [CrossRef]
- Cui, G.H.; Si, L.L.; Wang, Y.; Zhou, J.M.; Yan, H.J.; Jiang, L.F. Antibody-dependent enhancement (ADE) of dengue virus: Identification of the key amino acid that is vital in DENV vaccine research. J. Gene Med. 2021, 23, e3297. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [Green Version]
- Tetro, J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020, 22, 72–73. [Google Scholar] [CrossRef]
- Ricke, D.O. Two different antibody-dependent enhancement (ADE) risks for SARS-CoV-2 antibodies. Front. Immunol. 2021, 12, 640093. [Google Scholar] [CrossRef]
- Pierson, T.C.; Xu, Q.; Nelson, S.; Oliphant, T.; Nybakken, G.E.; Fremont, D.H.; Diamond, M.S. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 2007, 1, 135–145. [Google Scholar] [CrossRef] [Green Version]
- De Wispelaere, M.; Ricklin, M.; Souque, P.; Frenkiel, M.P.; Paulous, S.; Garcia-Nicolas, O.; Summerfield, A.; Charneau, P.; Despres, P.A. Lentiviral vector expressing Japanese encephalitis virus-like particles elicits broad neutralizing antibody response in pigs. PLoS Negl. Trop. Dis. 2015, 9, e0004081. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.Y.Y.; Low, J.Z.H.; Gan, E.S.; Ong, E.Z.; Zhang, S.L.X.; Tan, H.C.; Chai, X.R.; Ghosh, S.; Ooi, E.E.; Chan, K.R. Antibody-dependent dengue virus entry modulates cell intrinsic responses for enhanced infection. mSphere 2019, 4, e00528-19. [Google Scholar] [CrossRef] [Green Version]
- Beltramello, M.; Williams, K.L.; Simmons, C.P.; Macagno, A.; Simonelli, L.; Quyen, N.T.; Sukupolvi-Petty, S.; Navarro-Sanchez, E.; Young, P.R.; de Silva, A.M.; et al. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010, 8, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Stiasny, K.; Kiermayr, S.; Holzmann, H.; Heinz, F.X. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 2006, 80, 9557–9568. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, P.; Sabeena, S.P.; Varma, M.; Arunkumar, G. Current understanding of the pathogenesis of dengue virus infection. Curr. Microbiol. 2021, 78, 17–32. [Google Scholar] [CrossRef]
- Brown, J.A.; Singh, G.; Acklin, J.A.; Lee, S.; Duehr, J.E.; Chokola, A.N.; Frere, J.J.; Hoffman, K.W.; Foster, G.A.; Krysztof, D.; et al. Dengue virus immunity increases Zika virus-induced damage during pregnancy. Immunity 2019, 50, 751–762. [Google Scholar] [CrossRef] [Green Version]
- Shukla, R.; Ramasamy, V.; Shanmugam, R.K.; Ahuja, R.; Khanna, N. Antibody-dependent enhancement: A challenge for developing a safe dengue vaccine. Front. Cell. Infect. Microbiol. 2020, 10, 572681. [Google Scholar] [CrossRef]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004, 427, 313–319. [Google Scholar] [CrossRef]
- Stiasny, K.; Bressanelli, S.; Lepault, J.; Rey, F.A.; Heinz, F.X. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J. Virol. 2004, 78, 3178–3183. [Google Scholar] [CrossRef] [Green Version]
- Slon-Campos, J.L.; Dejnirattisai, W.; Jagger, B.W.; Lopez-Camacho, C.; Wongwiwat, W.; Durnell, L.A.; Winkler, E.S.; Chen, R.E.; Reyes-Sandoval, A.; Rey, F.A.; et al. A protective Zika virus E-dimer-based subunit vaccine engineered to abrogate antibody-dependent enhancement of dengue infection. Nat. Immunol. 2019, 20, 1291–1298. [Google Scholar] [CrossRef]
- Castanha, P.; Nascimento, E.; Braga, C.; Cordeiro, M.T.; de Carvalho, O.V.; de Mendonca, L.R.; Azevedo, E.; Franca, R.; Dhalia, R.; Marques, E. Dengue virus-specific antibodies enhance Brazilian Zika virus infection. J. Infect. Dis. 2017, 215, 781–785. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.K.; Bhattacharjee, S. Dengue virus: Epidemiology, biology, and disease aetiology. Can. J. Microbiol. 2021, 67, 687–702. [Google Scholar] [CrossRef]
- Castro, A.; Carreño, J.M.; Duehr, J.; Krammer, F.; Kane, R.S. Refocusing the immune response to selected epitopes on a Zika virus protein antigen by nanopatterning. Adv. Healthc. Mater. 2021, 10, e2002140. [Google Scholar] [CrossRef]
- Lai, H.; Paul, A.M.; Sun, H.; He, J.; Yang, M.; Bai, F.; Chen, Q. A plant-produced vaccine protects mice against lethal West Nile virus infection without enhancing Zika or dengue virus infectivity. Vaccine 2018, 36, 1846–1852. [Google Scholar] [CrossRef]
- Blome, S.; Gabriel, C.; Beer, M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine 2014, 32, 3879–3882. [Google Scholar] [CrossRef]
- Sunwoo, S.Y.; Pérez-Núñez, D.; Morozov, I.; Sánchez, E.G.; Gaudreault, N.N.; Trujillo, J.D.; Mur, L.; Nogal, M.; Madden, D.; Urbaniak, K.; et al. DNA-protein vaccination strategy does not protect from challenge with African swine fever virus Armenia 2007 strain. Vaccines 2019, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Flipse, J.; Diosa-Toro, M.A.; Hoornweg, T.E.; van de Pol, D.; Urcuqui-Inchima, S.; Smit, J.M. Antibody-dependent enhancement of dengue virus infection in primary human macrophages; balancing higher fusion against antiviral responses. Sci. Rep. 2016, 6, 29201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The dark side of antibody dependent enhancement during dengue infection. Front. Cell. Infect. Microbiol. 2020, 10, 580096. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.R.; Ong, E.Z.; Tan, H.C.; Zhang, S.L.; Zhang, Q.; Tang, K.F.; Kaliaperumal, N.; Lim, A.P.; Hibberd, M.L.; Chan, S.H.; et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. USA 2014, 111, 2722–2727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, D.; Wang, R.; Qiao, S.; Wan, B.; Wang, Y.; Liu, M.; Shi, X.; Guo, J.; Zhang, G. Antibody-dependent enhancement of PRRSV infection down-modulates TNF-α and IFN-β transcription in macrophages. Vet. Immunol. Immunopathol. 2013, 156, 128–134. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Ooi, E.E.; Horstick, O.; Wills, B. Dengue. Lancet 2019, 393, 350–363. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Rupali, P. Estimating the dengue burden in India. Lancet Glob. Health 2019, 7, e988–e989. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.F.; Tseng, S.P.; Yen, C.H.; Yang, J.Y.; Tsao, C.H.; Shen, C.W.; Chen, K.H.; Liu, F.T.; Liu, W.T.; Chen, Y.M.A.; et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014, 451, 208–214. [Google Scholar] [CrossRef]
- Wan, Y.S.; Shang, J.; Sun, S.H.; Tai, W.B.; Chen, J.; Geng, Q.B.; He, L.; Chen, Y.H.; Wu, J.M.; Shi, Z.L.; et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020, 94, e02015–e02019. [Google Scholar] [CrossRef] [Green Version]
- Karthik, K.; Senthilkumar, T.; Udhayavel, S.; Raj, G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccin. Immunother. 2020, 16, 3055–3060. [Google Scholar] [CrossRef]
- Du, L.; Tai, W.; Yang, Y.; Zhao, G.; Zhu, Q.; Sun, S.; Liu, C.; Tao, X.; Tseng, C.K.; Perlman, S.; et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016, 7, 13473. [Google Scholar] [CrossRef]
- Hohdatsu, T.; Nakamura, M.; Ishizuka, Y.; Yamada, H.; Koyama, H. A study on the mechanism of antibody-dependent enhancement of feline infectious peritonitis virus infection in feline macrophages by monoclonal antibodies. Arch. Virol. 1991, 120, 207–217. [Google Scholar] [CrossRef]
- Takano, T.; Yamada, S.; Doki, T.; Hohdatsu, T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 2019, 81, 911–915. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Deng, T.; Zhang, Y.; Niu, W.; Nie, Q.; Yang, S.; Liu, P.; Pei, P.; Chen, L.; Li, H. ACE2 can act as the secondary receptor in the FcγR-dependent ADE of SARS-CoV-2. IScience 2022, 25, 103720. [Google Scholar] [CrossRef]
- Liu, Y.; Soh, W.T.; Kishikawa, J.I.; Hirose, M.; Nakayama, E.E.; Li, S.; Sasai, M.; Suzuki, T.; Tada, A.; Arakawa, A.; et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell 2021, 184, 3452–3466. [Google Scholar] [CrossRef]
- Halstead, S.B.; Katzelnick, L. COVID-19 vaccines: Should we fear ADE? J. Infect. Dis. 2020, 222, 1946–1950. [Google Scholar] [CrossRef]
- Cloutier, M.; Nandi, M.; Ihsan, A.U.; Chamard, H.A.; Ilangumaran, S.; Ramanathan, S. ADE and hyperinflammation in SARS-CoV2 infection-comparison with dengue hemorrhagic fever and feline infectious peritonitis. Cytokine 2020, 136, 155256. [Google Scholar] [CrossRef]
- Taylor, A.; Foo, S.S.; Bruzzone, R.; Dinh, L.V.; King, N.J.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef] [Green Version]
- Tamura, M.; Webster, R.G.; Ennis, F.A. Neutralization and infection-enhancement epitopes of influenza A virus hemagglutinin. J. Immunol. 1993, 151, 1731–1738. [Google Scholar]
- Maemura, T.; Kuroda, M.; Armbrust, T.; Yamayoshi, S.; Halfmann, P.J.; Kawaoka, Y. Antibody-dependent enhancement of SARS-CoV-2 infection is mediated by the IgG receptors FcγRIIA and FcγRIIIA but does not contribute to aberrant cytokine production by macrophages. mBio 2021, 12, e0198721. [Google Scholar] [CrossRef]
- Wan, B.; Chen, X.; Li, Y.; Pang, M.; Chen, H.; Nie, X.; Pan, Y.; Qiao, S.; Bao, D. Porcine FcγRIIb mediated PRRSV ADE infection through inhibiting IFN-β by cytoplasmic inhibitory signal transduction. Int. J. Biol. Macromol. 2019, 138, 198–206. [Google Scholar] [CrossRef]
- Ruggeri, J.; Ferlazzo, G.; Boniotti, M.B.; Capucci, L.; Guarneri, F.; Barbieri, I.; Alborali, G.L.; Amadori, M. Characterization of the IgA response to PRRS virus in pig oral fluids. PLoS ONE 2020, 15, e0229065. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, W.; Sun, Y.; Kong, L.; Xu, P.; Xia, P.; Zhang, G. Antibody-mediated porcine reproductive and respiratory syndrome virus infection downregulates the production of interferon-α and tumor necrosis factor-α in porcine alveolar macrophages via Fc gamma receptor I and III. Viruses 2020, 12, 187. [Google Scholar] [CrossRef] [Green Version]
- Chareonsirisuthigul, T.; Kalayanarooj, S.; Ubol, S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 2007, 88, 365–375. [Google Scholar] [CrossRef]
- Valliyott, L.; Dungdung, R.; Pilankatta, R. Semi-quantification of antibody-dependent enhancement (ADE) in the uptake of adenovirus serotype 5 into THP-1 cells. Anal. Biochem. 2020, 15, 591. [Google Scholar] [CrossRef] [PubMed]
- Ubol, S.; Masrinoul, P.; Chaijaruwanich, J.; Kalayanarooj, S.; Charoensirisuthikul, T.; Kasisith, J. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J. Infect. Dis. 2008, 197, 1459–1467. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, L.; Jin, P.; Li, N.; Meng, Y.; Wang, C.; Xu, M.; Zhang, Y.; Bian, J.; Deng, X. Methane alleviates carbon tetrachloride induced liver injury in mice: Anti-inflammatory action demonstrated by increased PI3K/Akt/GSK-3β-mediated IL-10 expression. J. Mol. Histol. 2017, 48, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Ruckwardt, T.J.; Morabito, K.M.; Graham, B.S. Immunological lessons from respiratory syncytial virus vaccine development. Immunity 2019, 51, 429–442. [Google Scholar] [CrossRef]
- Sanchez Vargas, L.A.; Adam, A.; Masterson, M.; Smith, M.; Lyski, Z.L.; Dowd, K.A.; Pierson, T.C.; Messer, W.B.; Currier, J.R.; Mathew, A. Non-structural protein 1-specific antibodies directed against Zika virus in humans mediate antibody-dependent cellular cytotoxicity. Immunology 2021, 164, 386–397. [Google Scholar] [CrossRef]
- Scribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: Myths and facts. Virus Res. 2013, 173, 101–109. [Google Scholar] [CrossRef]
- Sánchez-Zuno, G.A.; Matuz-Flores, M.G.; González-Estevez, G.; Nicoletti, F.; Turrubiates-Hernández, F.J.; Mangano, K.; Muñoz-Valle, J.F. A review: Antibody-dependent enhancement in COVID-19: The not so friendly side of antibodies. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211050199. [Google Scholar] [CrossRef]
- Yip, M.S.; Leung, N.H.; Cheung, C.Y.; Li, P.H.; Lee, H.H.; Daëron, M.; Peiris, J.S.; Bruzzone, R.; Jaume, M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014, 11, 82. [Google Scholar] [CrossRef] [Green Version]
- Hotez, P.J. COVID19 meets the antivaccine movement. Microbes Infect. 2020, 22, 162–164. [Google Scholar] [CrossRef]
- Ulrich, H.; Pillat, M.M.; Tarnok, A. Dengue fever, COVID-19 (SARS-CoV-2), and antibody-dependent enhancement (ADE): A perspective. Cytom. A 2020, 97, 662–667. [Google Scholar] [CrossRef]

| Viruses | Ig Types | Fc Receptors | Viral Proteins Responsible for ADE | Mechanisms Underlying the ADE | References |
|---|---|---|---|---|---|
| DENV | IgG | FcγRI/FcγRIIa/FcγRIIIa | prM and E proteins | Facilitating virus entry into target cells Inhibiting innate immunity Changing the transcriptional levels of host molecules | [39,40] |
| ZIKV | IgG | FcγR | prM and E proteins | Facilitating virus entry into target cells | [41,42] |
| WNV | IgM | FcμR/CR | prM and E proteins | Facilitating virus entry into target cells | [43,44] |
| MERS-CoV/SARS-CoV | IgG | FcγRIIa | S protein | Mimicking the viral receptor using the MAb against the S protein to mediate viral invasion | [45] |
| FIPV | IgG | FcγRI/FcγRII | S and M proteins | Enhancing the production of inflammatory cytokines, such as IL-1β and TNF-α | [46] |
| PEDV | IgG | FcR | S protein | Enhancing viral infection in target cells | [47,48] |
| RSV | IgG | FcγR | G and F proteins | Stimulating poor Toll-like receptor (TLR) and producing non-protective antibodies | [49] |
| PRRSV | IgG | FcγRI/FcγRIIb/FcγRIII/FcεRI | GP5 and N proteins | Inhibiting the antiviral responses of host cells | [50,51] |
| HIV | IgG/IgA | FcαR/FcγRIII/CR | GP160 protein | Promoting membrane fusion through FcR and CR to facilitate virus entry | [52] |
| IV | IgG | FcR | HA protein | Increasing IV fusion dynamics and promoting IV infection | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Zhang, X.; Zhao, X.; Yuan, M.; Zhang, K.; Dai, J.; Guan, X.; Qiu, H.-J.; Li, Y. Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections. Viruses 2022, 14, 1739. https://doi.org/10.3390/v14081739
Yang X, Zhang X, Zhao X, Yuan M, Zhang K, Dai J, Guan X, Qiu H-J, Li Y. Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections. Viruses. 2022; 14(8):1739. https://doi.org/10.3390/v14081739
Chicago/Turabian StyleYang, Xiaoke, Xin Zhang, Xiaotian Zhao, Mengqi Yuan, Kehui Zhang, Jingwen Dai, Xiangyu Guan, Hua-Ji Qiu, and Yongfeng Li. 2022. "Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections" Viruses 14, no. 8: 1739. https://doi.org/10.3390/v14081739
APA StyleYang, X., Zhang, X., Zhao, X., Yuan, M., Zhang, K., Dai, J., Guan, X., Qiu, H.-J., & Li, Y. (2022). Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections. Viruses, 14(8), 1739. https://doi.org/10.3390/v14081739






