Long-Term Infection and Pathogenesis in a Novel Mouse Model of Human Respiratory Syncytial Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Challenge
2.2. Cells and Virus
2.3. Development of the Rag2−/− Mouse Model
2.4. Observation of Clinical Symptoms
2.5. RNA Extraction and Quantification
2.6. TCID50 Assay
2.7. Flow Cytometry
2.8. Cytokine Assay
2.9. Histopathological Analysis
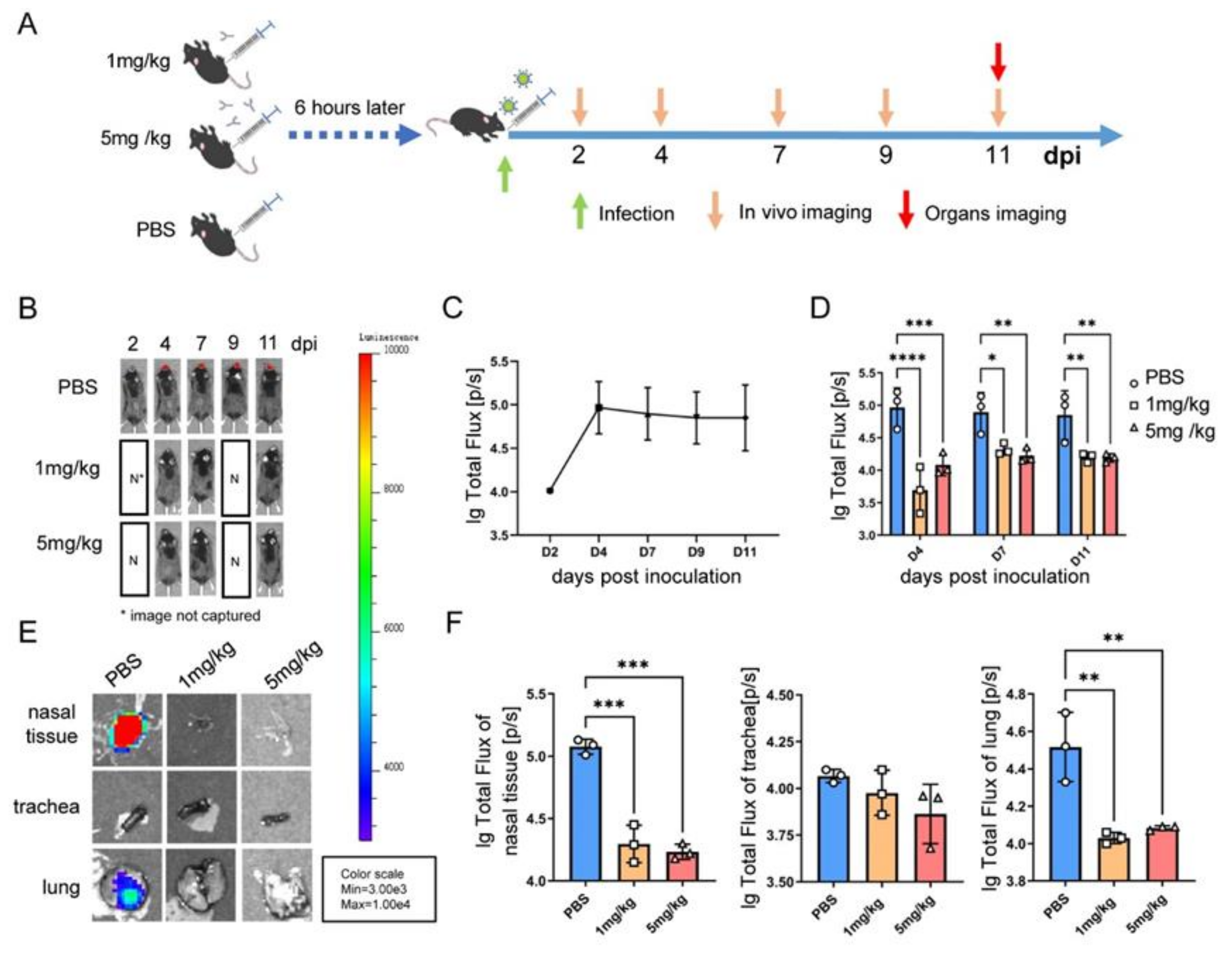
2.10. Evaluation of the In Vivo Effect of the Antibody via Bioluminescence Imaging
2.11. Statistical Analysis
3. Results
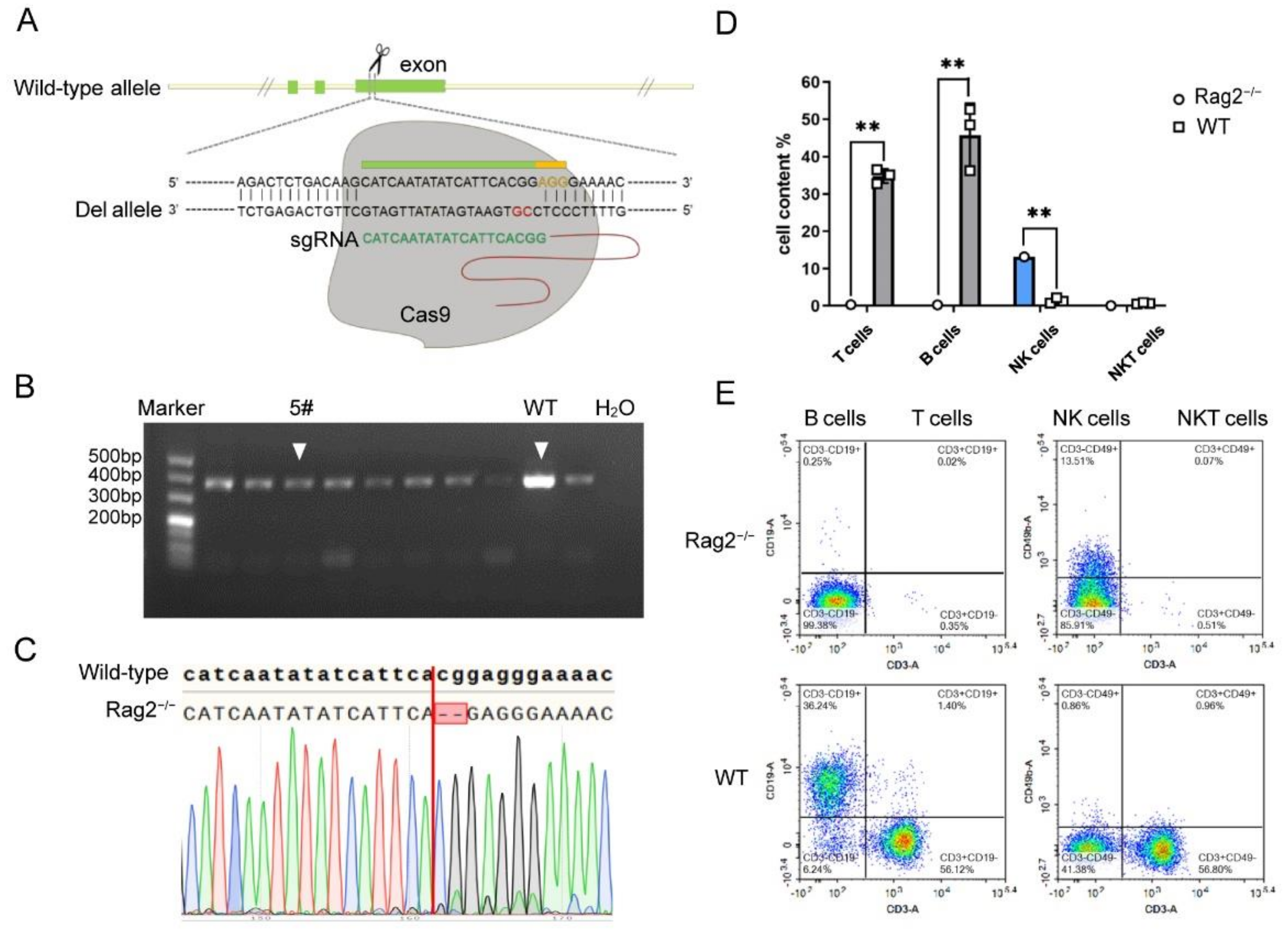
3.1. Establishment of Rag2 Knockout Mice by CRISPR/Cas9 Technology
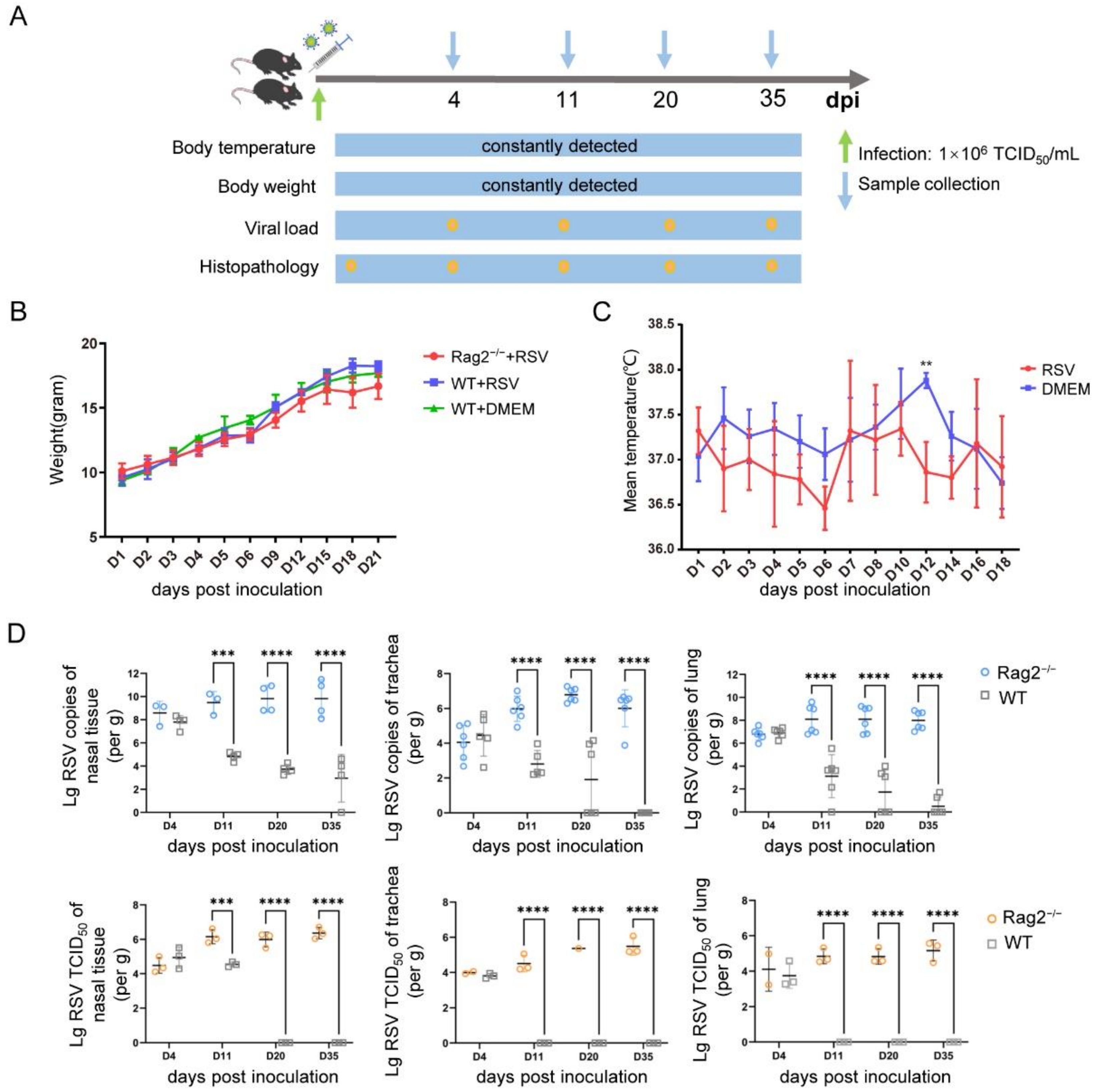
3.2. Establishment of a hRSV Infection Model Using Rag2−/− Mice
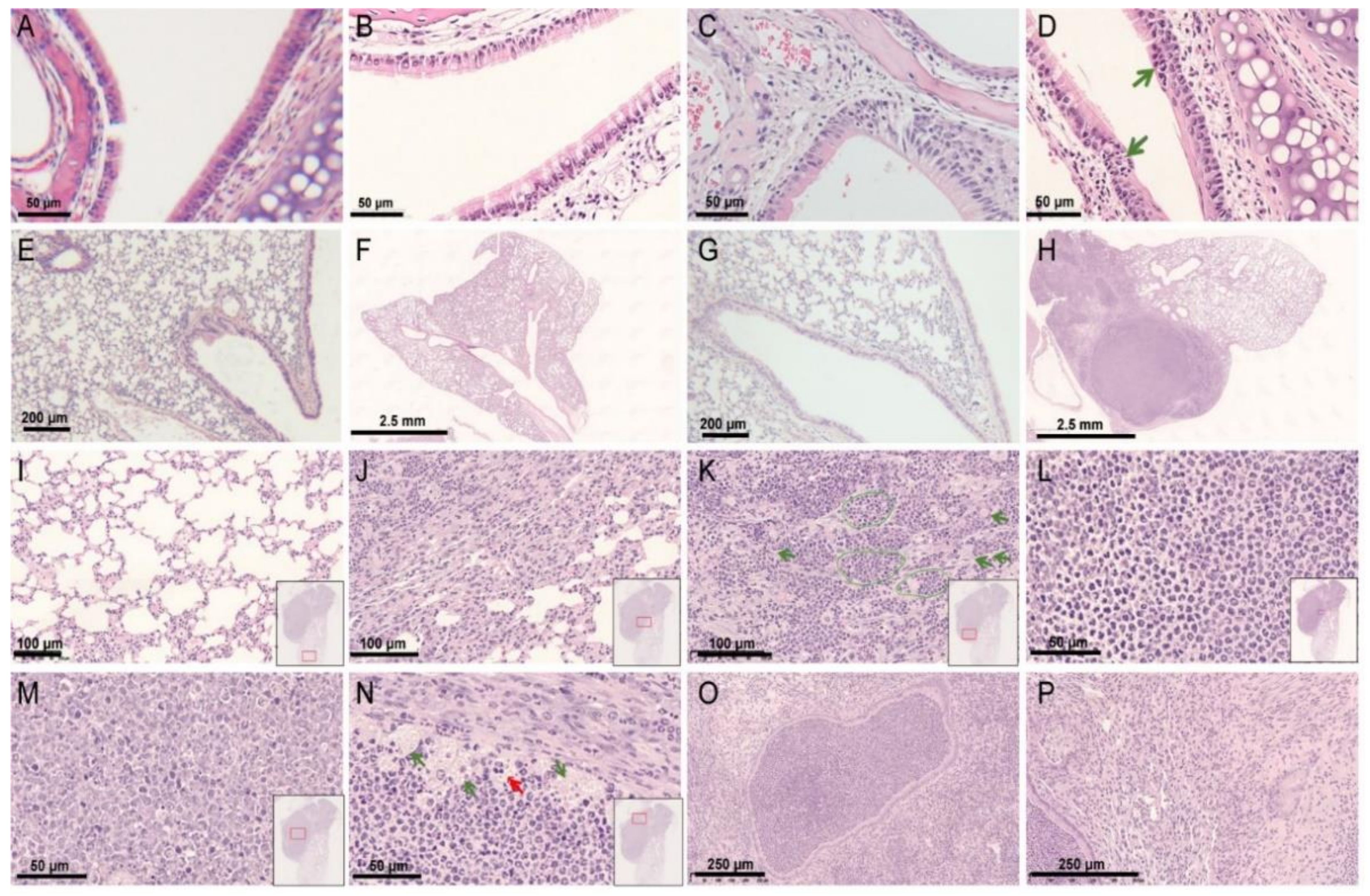
3.3. Histopathological Findings of a hRSV Infection Model Using Rag2−/− Mice
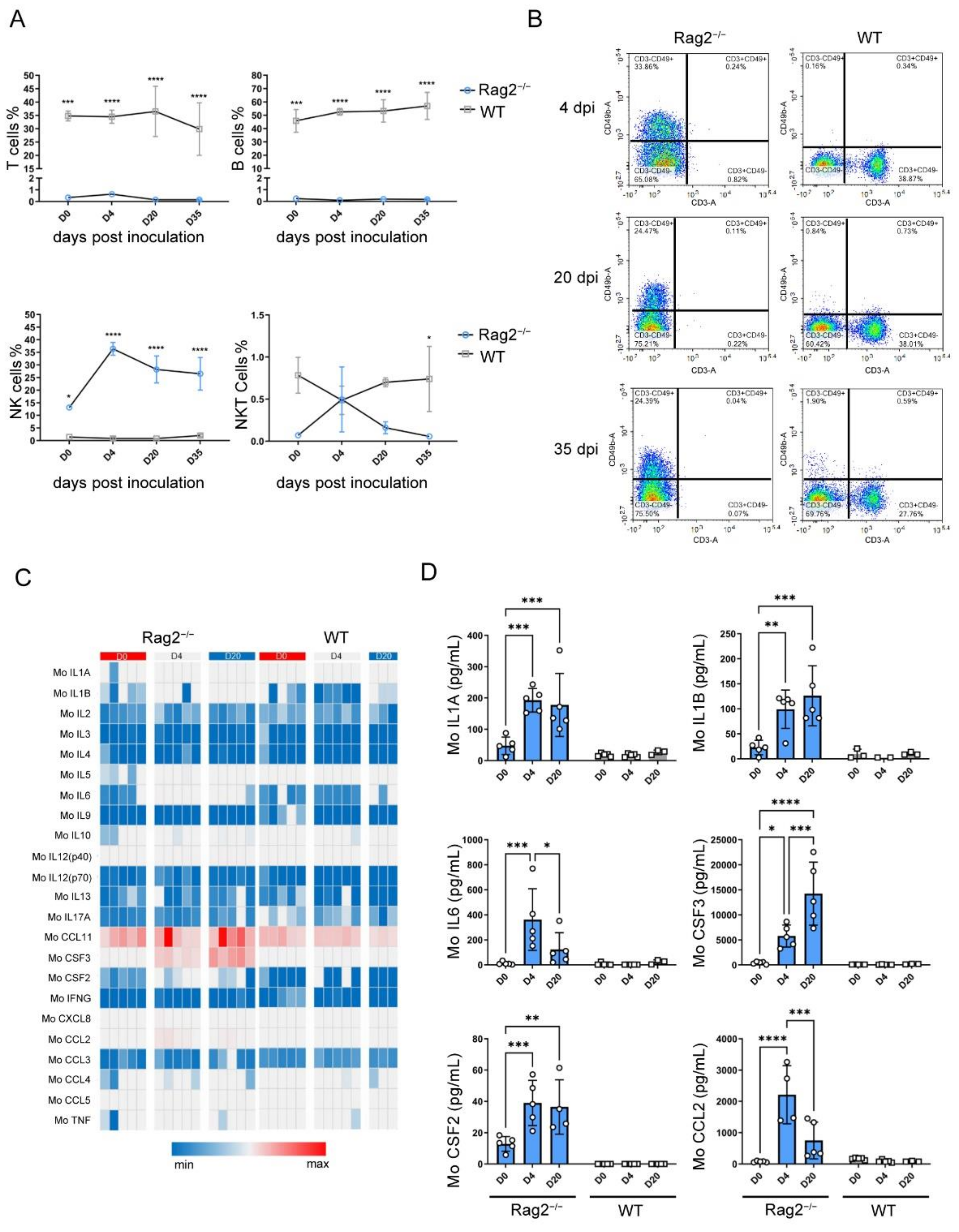
3.4. hRSV Infection Causing Upregulation of NK Cells, Serum Cytokines and Chemokines
3.5. A BLI Rag2−/− Mouse Model Evaluating the In Vivo Antiviral Activity of a Humanized Monoclonal Antibody
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Y.T.; Wertz, G.W. The genome of respiratory syncytial virus is a negative-stranded RNA that codes for at least seven mRNA species. J. Virol. 1982, 43, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, G.L.R.I. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- Glezen, W.P.; Paredes, A.; Allison, J.E.; Taber, L.H.; Frank, A.L. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J. Pediatr. 1981, 98, 708–715. [Google Scholar] [CrossRef]
- Hall, C.B.; Weinberg, G.A.; Iwane, M.K.; Blumkin, A.K.; Edwards, K.M.; Staat, M.A.; Auinger, P.; Griffin, M.R.; Poehling, K.A.; Erdman, D.; et al. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009, 360, 588–598. [Google Scholar] [CrossRef]
- Fretzayas, A.; Moustaki, M. Etiology and clinical features of viral bronchiolitis in infancy. World J. Pediatr. WJP 2017, 13, 293–299. [Google Scholar] [CrossRef]
- Kim, H.W.; Arrobio, J.O.; Brandt, C.D.; Jeffries, B.C.; Pyles, G.; Reid, J.L.; Chanock, R.M.; Parrott, R.H. Epidemiology of respiratory syncytial virus infection in Washington, D.C. I. Importance of the virus in different respiratory tract disease syndromes and temporal distribution of infection. Am. J. Epidemiol. 1973, 98, 216–225. [Google Scholar] [CrossRef]
- McNamara, P.S.; Smyth, R.L. The pathogenesis of respiratory syncytial virus disease in childhood. Br. Med. Bull. 2002, 61, 13–28. [Google Scholar] [CrossRef]
- Blount, R.E., Jr.; Morris, J.A.; Savage, R.E. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc. Soc. Exp. Biol. Med. 1956, 92, 544–549. [Google Scholar] [CrossRef]
- Cardenas, S.; Auais, A.; Piedimonte, G. Palivizumab in the prophylaxis of respiratory syncytial virus infection. Expert Rev. Anti-Infect. Ther. 2005, 3, 719–726. [Google Scholar] [CrossRef]
- Taylor, G. Animal models of respiratory syncytial virus infection. Vaccine 2017, 35, 469–480. [Google Scholar] [CrossRef]
- Belshe, R.B.; Richardson, L.S.; London, W.T.; Sly, D.L.; Lorfeld, J.H.; Camargo, E.; Prevar, D.A.; Chanock, R.M. Experimental respiratory syncytial virus infection of four species of primates. J. Med. Virol. 1977, 1, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.L.; Purcell, R.H.; London, W.T.; Lawrence, L.A.; Chanock, R.M.; Murphy, B.R. Evaluation in chimpanzees of vaccinia virus recombinants that express the surface glycoproteins of human respiratory syncytial virus. Vaccine 1990, 8, 164–168. [Google Scholar] [CrossRef]
- Clarke, C.J.; Watt, N.J.; Meredith, A.; McIntyre, N.; Burns, S.M. Respiratory syncytial virus-associated bronchopneumonia in a young chimpanzee. J. Comp. Pathol. 1994, 110, 207–212. [Google Scholar] [CrossRef]
- Taylor, G.; Stott, E.J.; Bew, M.; Fernie, B.F.; Cote, P.J.; Collins, A.P.; Hughes, M.; Jebbett, J. Monoclonal antibodies protect against respiratory syncytial virus infection in mice. Immunology 1984, 52, 137–142. [Google Scholar] [PubMed]
- Prince, G.A.; Horswood, R.L.; Chanock, R.M. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J. Virol. 1985, 55, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Openshaw, P.J. The mouse model of respiratory syncytial virus disease. Curr. Top. Microbiol. Immunol. 2013, 372, 359–369. [Google Scholar] [CrossRef]
- Graham, B.S.; Perkins, M.D.; Wright, P.F.; Karzon, D.T. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 1988, 26, 153–162. [Google Scholar] [CrossRef]
- Papin, J.F.; Wolf, R.F.; Kosanke, S.D.; Jenkins, J.D.; Moore, S.N.; Anderson, M.P.; Welliver, R.C., Sr. Infant baboons infected with respiratory syncytial virus develop clinical and pathological changes that parallel those of human infants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L530–L539. [Google Scholar] [CrossRef]
- Wong, D.T.; Rosenband, M.; Hovey, K.; Ogra, P.L. Respiratory syncytial virus infection in immunosuppressed animals: Implications in human infection. J. Med. Virol. 1985, 17, 359–370. [Google Scholar] [CrossRef]
- Ottolini, M.G.; Porter, D.D.; Hemming, V.G.; Zimmerman, M.N.; Schwab, N.M.; Prince, G.A. Effectiveness of RSVIG prophylaxis and therapy of respiratory syncytial virus in an immunosuppressed animal model. Bone Marrow Transplant. 1999, 24, 41–45. [Google Scholar] [CrossRef][Green Version]
- Nie, J.; Wu, X.; Ma, J.; Cao, S.; Huang, W.; Liu, Q.; Li, X.; Li, Y.; Wang, Y. Development of in vitro and in vivo rabies virus neutralization assays based on a high-titer pseudovirus system. Sci. Rep. 2017, 7, 42769. [Google Scholar] [CrossRef] [PubMed]
- Oettinger, M.A.; Schatz, D.G.; Gorka, C.; Baltimore, D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 1990, 248, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Held, W.; MacDonald, H.R. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 1994, 179, 1355–1360. [Google Scholar] [CrossRef]
- Wesemann, D.R.; Portuguese, A.J.; Meyers, R.M.; Gallagher, M.P.; Cluff-Jones, K.; Magee, J.M.; Panchakshari, R.A.; Rodig, S.J.; Kepler, T.B.; Alt, F.W. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 2013, 501, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Couëdel, C.; Roman, C.; Jones, A.; Vezzoni, P.; Villa, A.; Cortes, P. Analysis of mutations from SCID and Omenn syndrome patients reveals the central role of the Rag2 PHD domain in regulating V(D)J recombination. J. Clin. Investig. 2010, 120, 1337–1344. [Google Scholar] [CrossRef]
- Liu, Q.; Fan, C.; Zhou, S.; Guo, Y.; Zuo, Q.; Ma, J.; Liu, S.; Wu, X.; Peng, Z.; Fan, T.; et al. Bioluminescent imaging of vaccinia virus infection in immunocompetent and immunodeficient rats as a model for human smallpox. Sci. Rep. 2015, 5, 11397. [Google Scholar] [CrossRef]
- Wickman, J.R.; Luo, X.; Li, W.; Jean-Toussaint, R.; Sahbaie, P.; Sacan, A.; Clark, J.D.; Ajit, S.K. Circulating microRNAs from the mouse tibia fracture model reflect the signature from patients with complex regional pain syndrome. Pain Rep. 2021, 6, e950. [Google Scholar] [CrossRef]
- Boukhvalova, M.S.; Blanco, J.C. The cotton rat Sigmodon hispidus model of respiratory syncytial virus infection. Curr. Top. Microbiol. Immunol. 2013, 372, 347–358. [Google Scholar] [CrossRef]
- Boukhvalova, M.; Blanco, J.C.; Falsey, A.R.; Mond, J. Treatment with novel RSV Ig RI-002 controls viral replication and reduces pulmonary damage in immunocompromised Sigmodon hispidus. Bone Marrow Transplant. 2016, 51, 119–126. [Google Scholar] [CrossRef]
- Johnson, J.E.; Gonzales, R.A.; Olson, S.J.; Wright, P.F.; Graham, B.S. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod. Pathol. 2007, 20, 108–119. [Google Scholar] [CrossRef]
- Everard, M.L.; Swarbrick, A.; Wrightham, M.; McIntyre, J.; Dunkley, C.; James, P.D.; Sewell, H.F.; Milner, A.D. Analysis of cells obtained by bronchial lavage of infants with respiratory syncytial virus infection. Arch. Dis. Child. 1994, 71, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Z.; Xu, H.; Wraith, A.; Bowden, J.J.; Alpers, J.H.; Forsyth, K.D. Neutrophils induce damage to respiratory epithelial cells infected with respiratory syncytial virus. Eur. Respir. J. 1998, 12, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Cillóniz, C.; Dominedò, C.; Magdaleno, D.; Ferrer, M.; Gabarrús, A.; Torres, A. Pure Viral Sepsis Secondary to Community-Acquired Pneumonia in Adults: Risk and Prognostic Factors. J. Infect. Dis. 2019, 220, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.C.; Jacob, S.T.; Banura, P.; Zhang, J.; Stroup, S.; Boulware, D.R.; Scheld, W.M.; Houpt, E.R.; Liu, J. Etiology of Sepsis in Uganda Using a Quantitative Polymerase Chain Reaction-based TaqMan Array Card. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 68, 266–272. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Phipps, S.; Angkasekwinai, P.; Dong, C.; Foster, P.S. NK cell deficiency predisposes to viral-induced Th2-type allergic inflammation via epithelial-derived IL-25. J. Immunol. 2010, 185, 4681–4690. [Google Scholar] [CrossRef]
- Hussell, T.; Openshaw, P.J. Intracellular IFN-gamma expression in natural killer cells precedes lung CD8+ T cell recruitment during respiratory syncytial virus infection. J. Gen. Virol. 1998, 79 Pt 11, 2593–2601. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Domachowske, J.B. Inflammatory responses to respiratory syncytial virus (RSV) infection and the development of immunomodulatory pharmacotherapeutics. Curr. Med. Chem. 2012, 19, 1424–1431. [Google Scholar] [CrossRef]
- Stoppelenburg, A.J.; de Roock, S.; Hennus, M.P.; Bont, L.; Boes, M. Elevated Th17 response in infants undergoing respiratory viral infection. Am. J. Pathol. 2014, 184, 1274–1279. [Google Scholar] [CrossRef]
- Brand, H.K.; Ferwerda, G.; Preijers, F.; de Groot, R.; Neeleman, C.; Staal, F.J.; Warris, A.; Hermans, P.W. CD4+ T-cell counts and interleukin-8 and CCL-5 plasma concentrations discriminate disease severity in children with RSV infection. Pediatr. Res. 2013, 73, 187–193. [Google Scholar] [CrossRef]
- Becker, S.; Soukup, J.M. Airway epithelial cell-induced activation of monocytes and eosinophils in respiratory syncytial viral infection. Immunobiology 1999, 201, 88–106. [Google Scholar] [CrossRef]
- Miller, A.L.; Bowlin, T.L.; Lukacs, N.W. Respiratory syncytial virus-induced chemokine production: Linking viral replication to chemokine production in vitro and in vivo. J. Infect. Dis. 2004, 189, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Schuler, C.F.t.; Malinczak, C.A.; Best, S.K.K.; Morris, S.B.; Rasky, A.J.; Ptaschinski, C.; Lukacs, N.W.; Fonseca, W. Inhibition of uric acid or IL-1β ameliorates respiratory syncytial virus immunopathology and development of asthma. Allergy 2020, 75, 2279–2293. [Google Scholar] [CrossRef]
- Fibbe, W.E.; Van Damme, J.; Billiau, A.; Duinkerken, N.; Lurvink, E.; Ralph, P.; Altrock, B.W.; Kaushansky, K.; Willemze, R.; Falkenburg, J.H. Human fibroblasts produce granulocyte-CSF, macrophage-CSF, and granulocyte-macrophage-CSF following stimulation by interleukin-1 and poly(rI).poly(rC). Blood 1988, 72, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Duncan, M.R.; Berman, B. Stimulation of collagen and glycosaminoglycan production in cultured human adult dermal fibroblasts by recombinant human interleukin 6. J. Investig. Dermatol. 1991, 97, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Standiford, T.J.; Kunkel, S.L.; Phan, S.H.; Rollins, B.J.; Strieter, R.M. Alveolar macrophage-derived cytokines induce monocyte chemoattractant protein-1 expression from human pulmonary type II-like epithelial cells. J. Biol. Chem. 1991, 266, 9912–9918. [Google Scholar] [CrossRef]
- Heydari, F.S.; Zare, S.; Roohbakhsh, A. Inhibition of Interleukin-1 in the Treatment of Selected Cardiovascular Complications. Curr. Rev. Clin. Exp. Pharmacol. 2021, 16, 219–227. [Google Scholar] [CrossRef]
- Boukhvalova, M.S.; Yim, K.C.; Kuhn, K.H.; Hemming, J.P.; Prince, G.A.; Porter, D.D.; Blanco, J.C. Age-related differences in pulmonary cytokine response to respiratory syncytial virus infection: Modulation by anti-inflammatory and antiviral treatment. J. Infect. Dis. 2007, 195, 511–518. [Google Scholar] [CrossRef]
- Bohmwald, K.; Gálvez, N.M.S.; Canedo-Marroquín, G.; Pizarro-Ortega, M.S.; Andrade-Parra, C.; Gómez-Santander, F.; Kalergis, A.M. Contribution of Cytokines to Tissue Damage During Human Respiratory Syncytial Virus Infection. Front. Immunol. 2019, 10, 452. [Google Scholar] [CrossRef]
| Injection Batches | Zygotes Injected | Zygotes Transferred | Newborns | F0 Mice |
|---|---|---|---|---|
| No. 1 | 114 | 53 | 9 | 1 |
| No. 2 | 144 | 82 | 16 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, R.; Fu, R.; Wu, Y.; Wu, X.; Cao, Y.; Qu, Z.; Yang, Y.; Liu, S.; Huo, G.; Wang, S.; et al. Long-Term Infection and Pathogenesis in a Novel Mouse Model of Human Respiratory Syncytial Virus. Viruses 2022, 14, 1740. https://doi.org/10.3390/v14081740
Xiong R, Fu R, Wu Y, Wu X, Cao Y, Qu Z, Yang Y, Liu S, Huo G, Wang S, et al. Long-Term Infection and Pathogenesis in a Novel Mouse Model of Human Respiratory Syncytial Virus. Viruses. 2022; 14(8):1740. https://doi.org/10.3390/v14081740
Chicago/Turabian StyleXiong, Rui, Rui Fu, Yong Wu, Xi Wu, Yuan Cao, Zhe Qu, Yanwei Yang, Susu Liu, Guitao Huo, Sanlong Wang, and et al. 2022. "Long-Term Infection and Pathogenesis in a Novel Mouse Model of Human Respiratory Syncytial Virus" Viruses 14, no. 8: 1740. https://doi.org/10.3390/v14081740
APA StyleXiong, R., Fu, R., Wu, Y., Wu, X., Cao, Y., Qu, Z., Yang, Y., Liu, S., Huo, G., Wang, S., Huang, W., Lyu, J., Zhu, X., Liang, C., Peng, Y., Wang, Y., & Fan, C. (2022). Long-Term Infection and Pathogenesis in a Novel Mouse Model of Human Respiratory Syncytial Virus. Viruses, 14(8), 1740. https://doi.org/10.3390/v14081740






