Functional Analysis of the Cathepsin D Gene Response to SGIV Infection in the Orange-Spotted Grouper, Epinephelus coioides
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish, Cells and Virus
2.2. Cloning of Ec-CD and Bioinformatic Analysis
2.3. Virus Infection Assay
2.4. Ec-CD Expression Patterns
2.5. Plasmid Construction and Cell Transfection
2.6. Cellular Localization Assay
2.7. Ec-CD Knockdown Analysis
2.8. RNA Extraction and qRT-PCR
2.9. Reporter Gene Assay
2.10. Western Blot Analysis
2.11. Cell Apoptosis Analysis
2.12. Statistical Analysis
3. Results
3.1. Sequence Characterization of Ec-CD
3.2. Tissue Distribution, Expression Profiles and Subcellular Distribution of Ec-CD
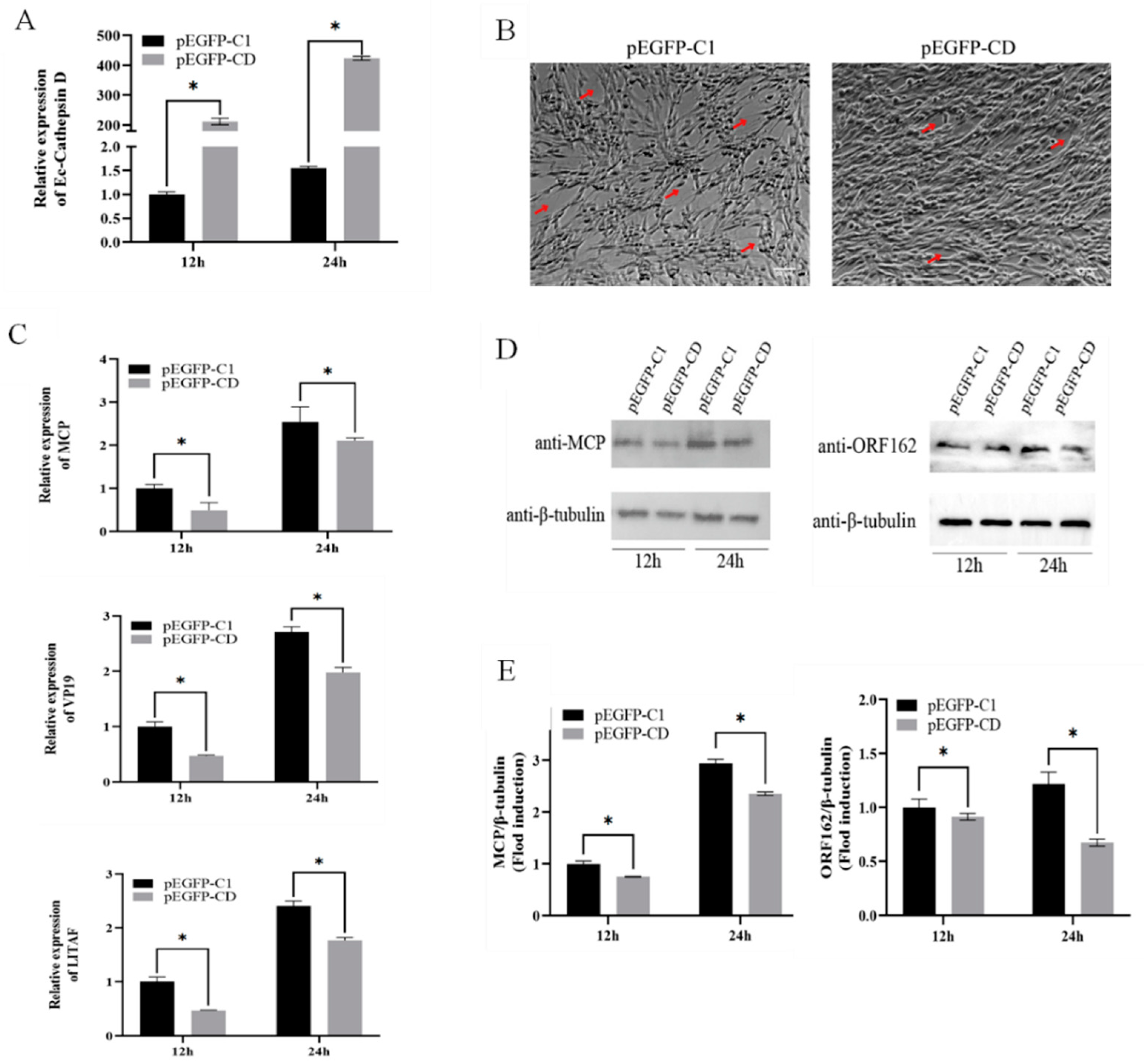
3.3. Overexpression of Ec-CD Inhibited SGIV Infection
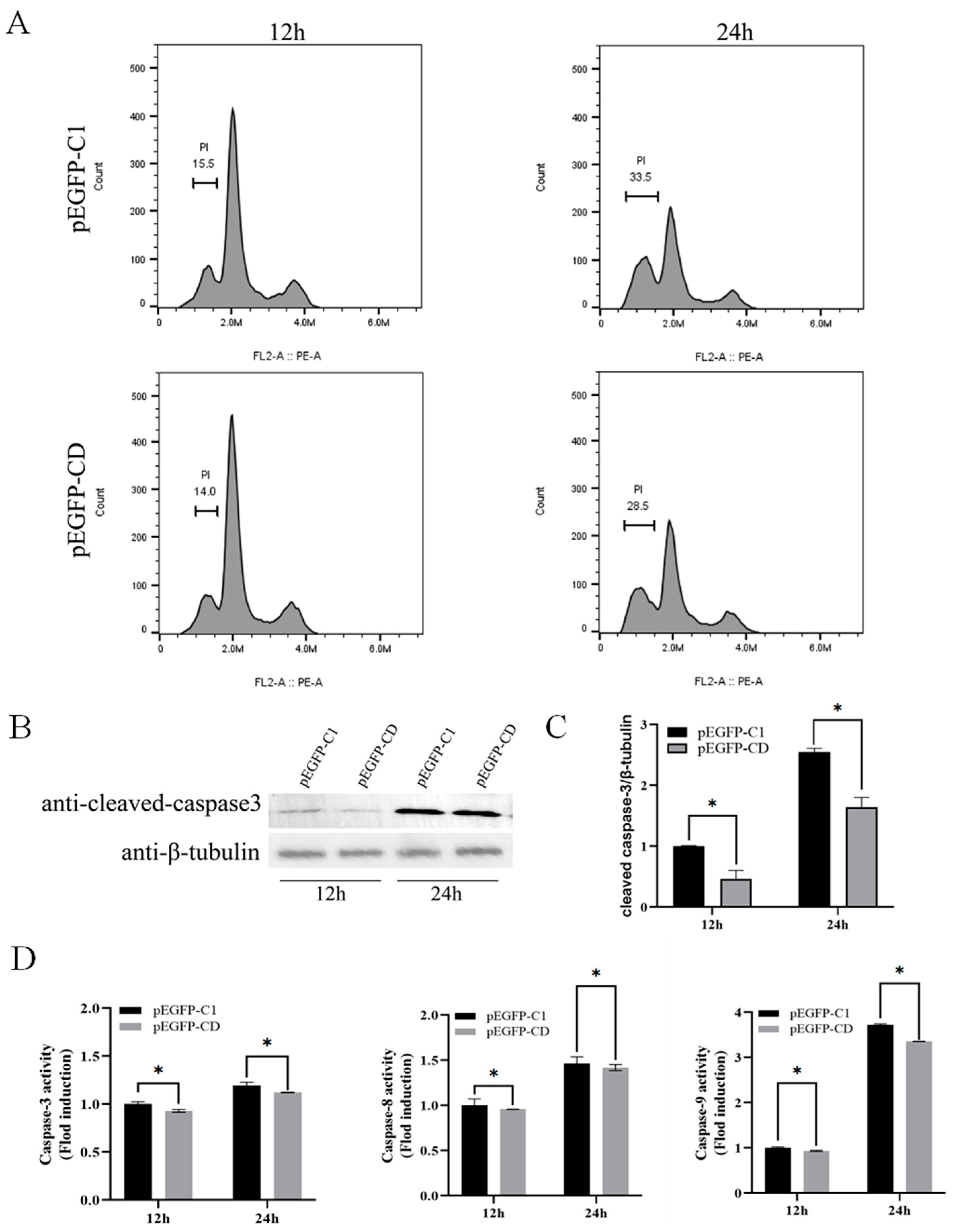
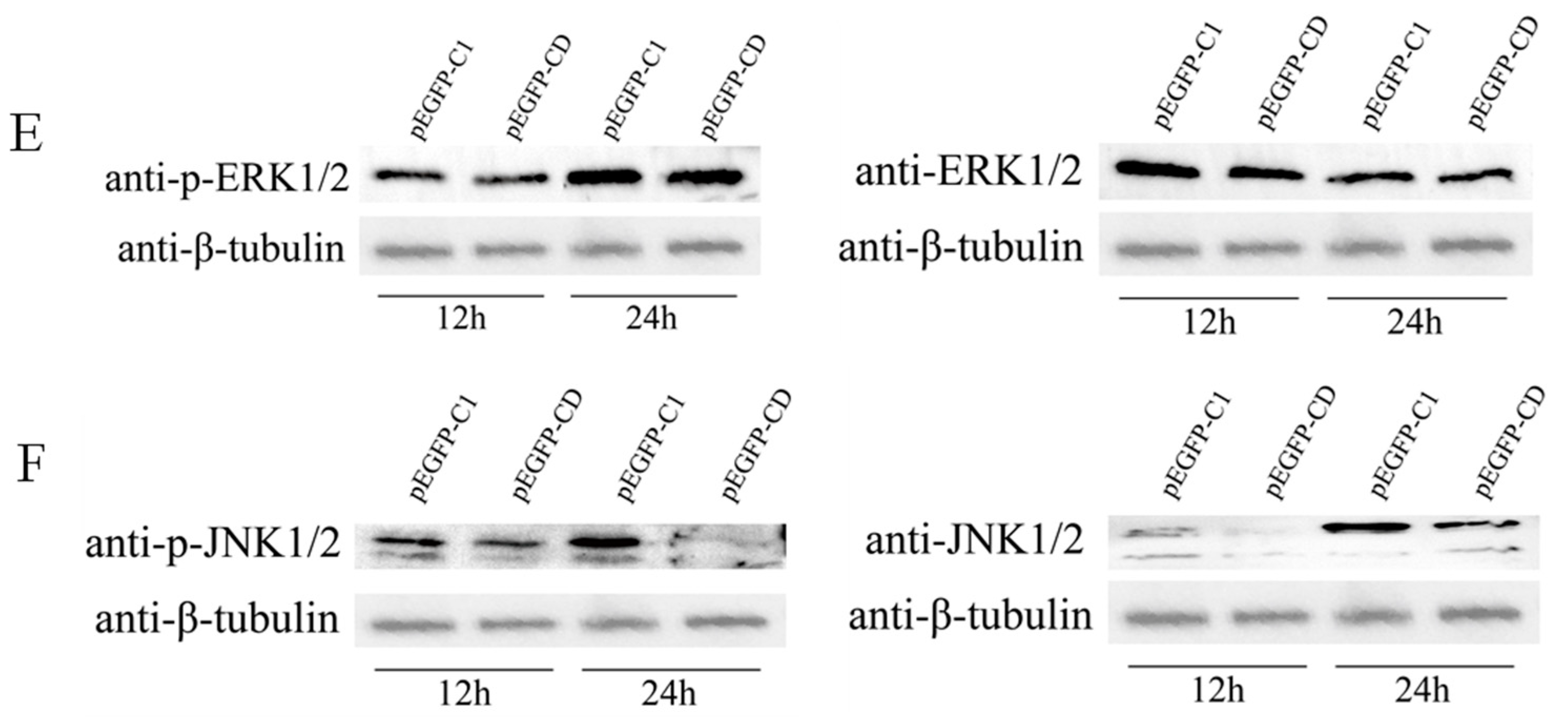
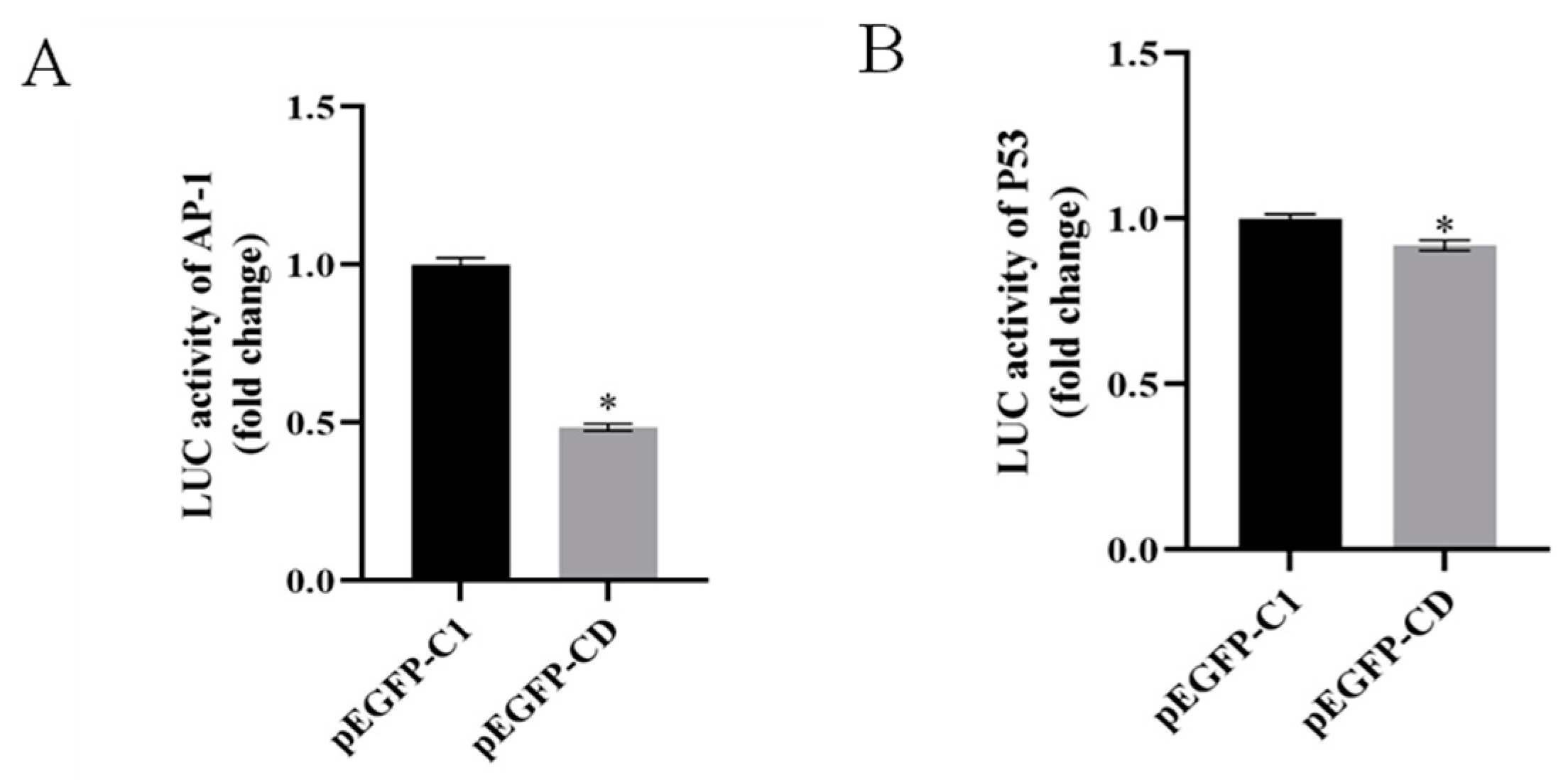
3.4. Overexpression of Ec-CD Hindered SGIV-Induced Cell Apoptosis
3.5. Ec-CD Knockdown Promoted SGIV Infection
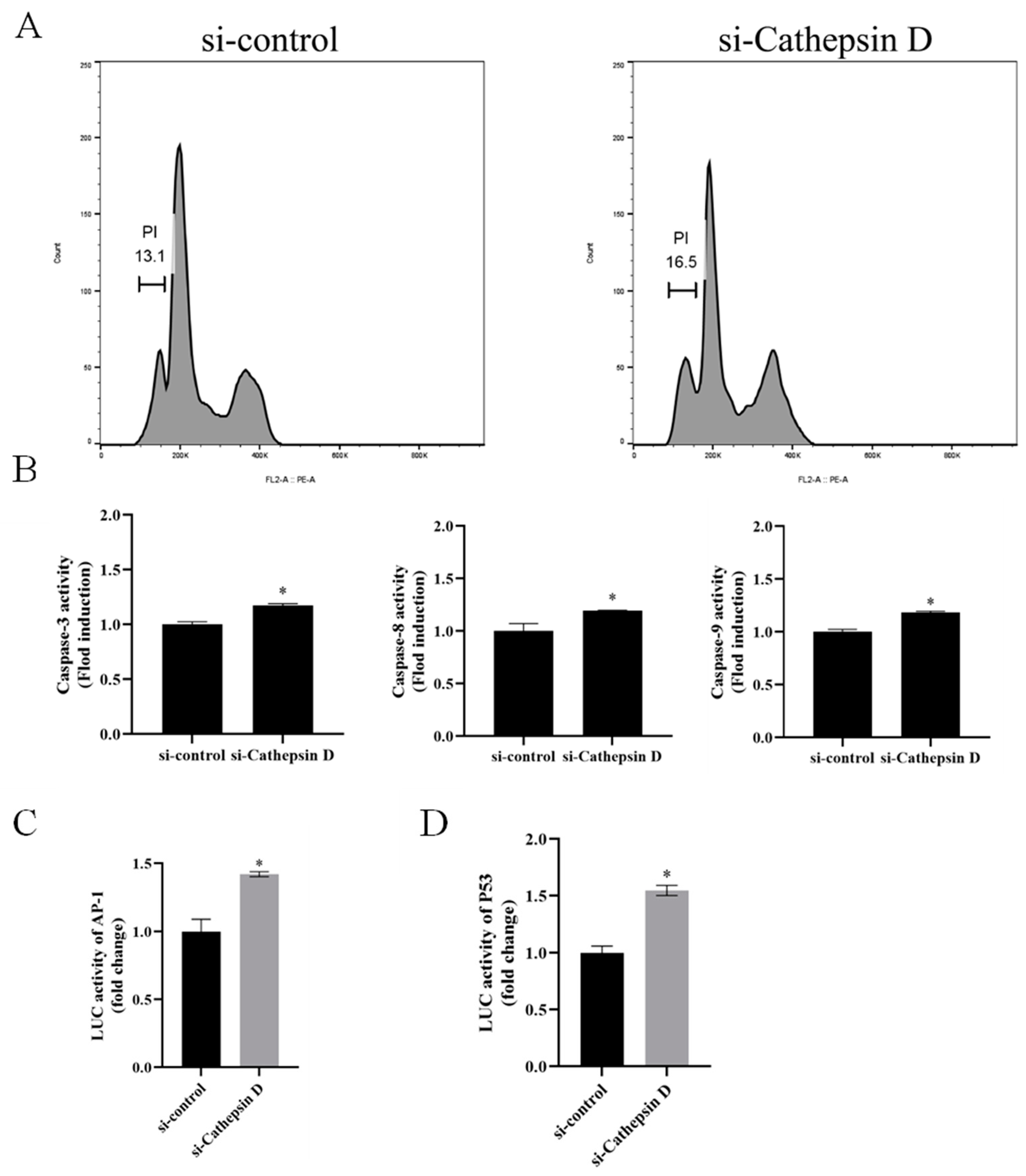
3.6. Ec-CD Knockdown Promoted SGIV-induced Cell Apoptosis
3.7. Ec-CD Knockdown Promoted SGIV-induced Cell Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, L.; Zhang, M.; Sun, L. Identification and expressional analysis of two cathepsins from half-smooth tongue sole (Cynoglossus semilaevis). Fish Shellfish Immunol. 2011, 31, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.F.; Herrington, D.M. The function of cathepsins B, D, and X in atherosclerosis. Am. J. Cardiovasc. Dis. 2016, 6, 163–170. [Google Scholar] [PubMed]
- Benes, P.; Vetvicka, V.; Fusek, M. Cathepsin D--many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 2008, 68, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Baechle, D.; Flad, T.; Cansier, A.; Steffen, H.; Schittek, B.; Tolson, J.; Herrmann, T.; Dihazi, H.; Beck, A.; Mueller, G.A.; et al. Cathepsin D is present in human eccrine sweat and involved in the postsecretory processing of the antimicrobial peptide DCD-1L. J. Biol. Chem. 2006, 281, 5406–5415. [Google Scholar] [CrossRef]
- Miura, Y.; Sakurai, Y.; Hayakawa, M.; Shimada, Y.; Zempel, H.; Sato, Y.; Hisanaga, S.; Endo, T. Translocation of lysosomal cathepsin D caused by oxidative stress or proteasome inhibition in primary cultured neurons and astrocytes. Biol. Pharm. Bull. 2010, 33, 22–28. [Google Scholar] [CrossRef]
- Beaujouin, M.; Baghdiguian, S.; Glondu-Lassis, M.; Berchem, G.; Liaudet-Coopman, E. Overexpression of both catalytically active and -inactive cathepsin D by cancer cells enhances apoptosis-dependent chemo-sensitivity. Oncogene 2006, 25, 1967–1973. [Google Scholar] [CrossRef]
- Rhodes, J.M.; Andersen, A.B. Role of cathepsin D in the degradation of human serum albumin by peritoneal macrophages and veiled cells in antigen presentation. Immunol. Lett. 1993, 37, 103–110. [Google Scholar] [CrossRef]
- Vidoni, C.; Follo, C.; Savino, M.; Melone, M.A.; Isidoro, C. The Role of Cathepsin D in the Pathogenesis of Human Neurodegenerative Disorders. Med. Res. Rev. 2016, 36, 845–870. [Google Scholar] [CrossRef]
- Ashraf, Y.; Mansouri, H.; Laurent-Matha, V.; Alcaraz, L.B.; Roger, P.; Guiu, S.; Derocq, D.; Robin, G.; Michaud, H.A.; Delpech, H.; et al. Immunotherapy of triple-negative breast cancer with cathepsin D-targeting antibodies. J. Immunother. Cancer 2019, 7, 29. [Google Scholar] [CrossRef]
- Di Domenico, F.; Tramutola, A.; Perluigi, M. Cathepsin D as a therapeutic target in Alzheimer’s disease. Expert Opin. Ther. Targets 2016, 20, 1393–1395. [Google Scholar] [CrossRef] [PubMed]
- Marciniszyn, J.J.; Hartsuck, J.A.; Tang, J. Mode of inhibition of acid proteases by pepstatin. J. Biol. Chem. 1976, 251, 7088–7094. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, W.; Li, L.; Salvador, L.A.; Chen, T.; Chen, W.; Felsenstein, K.M.; Ladd, T.B.; Price, A.R.; Golde, T.E.; et al. Cyanobacterial peptides as a prototype for the design of potent beta-secretase inhibitors and the development of selective chemical probes for other aspartic proteases. J. Med. Chem. 2012, 55, 10749–10765. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, W.; Zong, C.; Wang, P.; Li, Y. Total synthesis and stereochemical reassignment of tasiamide B. J. Pept. Sci. 2010, 16, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bao, K.; Xu, H.; Wu, P.; Li, W.; Liu, J.; Zhang, W. Design, synthesis, and bioactivities of tasiamide B derivatives as cathepsin D inhibitors. J. Pept. Sci. 2019, 25, e3154. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Bao, K.; Tang, S.; Ai, J.; Hu, H.; Zhang, W. Cyanobacterial peptides as a prototype for the design of cathepsin D inhibitors. J. Pept. Sci. 2017, 23, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, W.; Xu, Y.; Ren, S.; Zhang, W.; Li, Y. Design, synthesis and biological evaluation of tasiamide B derivatives as BACE1 inhibitors. Bioorg. Med. Chem. 2015, 23, 1963–1974. [Google Scholar] [CrossRef]
- Xiao, R.; Zhang, Z.; Wang, H.; Han, Y.; Gou, M.; Li, B.; Duan, D.; Wang, J.; Liu, X.; Li, Q. Identification and characterization of a cathepsin D homologue from lampreys (Lampetra japonica). Dev. Comp. Immunol. 2015, 49, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Shim, S.H.; An, C.M.; Nam, B.H.; Kim, Y.O.; Kim, J.W.; Park, C.I. Cloning, characterisation, and expression analysis of the cathepsin D gene from rock bream (Oplegnathus fasciatus). Fish Shellfish Immunol. 2014, 40, 253–258. [Google Scholar] [CrossRef]
- Dong, Z.D.; Zhang, J.; Ji, X.S.; Zhou, F.N.; Fu, Y.; Chen, W.; Zeng, Y.Q.; Li, T.M.; Wang, H. Molecular cloning, characterization and expression of cathepsin D from grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2012, 33, 1207–1214. [Google Scholar] [CrossRef]
- Chang, S.F.; Ngoh-Lim, G.H.; Kueh, L.F.; Qin, Q.W.; Seng, E.K.; Sin, Y.M. Initial investigations into two viruses isolated from marine food fish in Singapore. Vet. Rec. 2002, 150, 15–16. [Google Scholar] [CrossRef]
- Qin, Q.W.; Lam, T.J.; Sin, Y.M.; Shen, H.; Chang, S.F.; Ngoh, G.H.; Chen, C.L. Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. J. Virol. Methods 2001, 98, 17–24. [Google Scholar] [CrossRef]
- Wei, S.; Wang, S.; Yang, M.; Huang, Y.; Wei, J.; Huang, X.; Qin, Q. Characterization of cathepsin C from orange-spotted grouper, Epinephelus coioides involved in SGIV infection. Fish Shellfish Immunol. 2019, 84, 423–433. [Google Scholar] [CrossRef]
- Wei, S.; Huang, Y.; Huang, X.; Cai, J.; Yan, Y.; Guo, C.; Qin, Q. Characterization of cathepsin B gene from orange-spotted grouper, Epinephelus coioides involved in SGIV infection. Fish Shellfish Immunol. 2014, 36, 194–205. [Google Scholar] [CrossRef]
- Ni, S.; Yan, Y.; Cui, H.; Yu, Y.; Huang, Y.; Qin, Q. Fish miR-146a promotes Singapore grouper iridovirus infection by regulating cell apoptosis and NF-kappaB activation. J. Gen. Virol. 2017, 98, 1489–1499. [Google Scholar] [CrossRef]
- Gravell, M.; Malsberger, R.G. A permanent cell line from the fathead minnow (Pimephales promelas). Ann. N. Y. Acad. Sci. 1965, 126, 555–565. [Google Scholar] [CrossRef]
- Huang, X.; Huang, Y.; Cai, J.; Wei, S.; Ouyang, Z.; Qin, Q. Molecular cloning, expression and functional analysis of ISG15 in orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 2013, 34, 1094–1102. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, X.H.; Gui, J.F.; Zhang, Q.Y. Mitochondrion-mediated apoptosis induced by Rana grylio virus infection in fish cells. Apoptosis 2007, 12, 1569–1577. [Google Scholar] [CrossRef]
- Huang, X.; Huang, Y.; Ouyang, Z.; Xu, L.; Yan, Y.; Cui, H.; Han, X.; Qin, Q. Singapore grouper iridovirus, a large DNA virus, induces nonapoptotic cell death by a cell type dependent fashion and evokes ERK signaling. Apoptosis 2011, 16, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kalita, J.; Sinha, R.A.; Singh, G.; Shukla, M.; Tiwari, S.; Dhole, T.N.; Misra, U.K. Impaired Autophagy Flux is Associated with Proinflammatory Microglia Activation Following Japanese Encephalitis Virus Infection. Neurochem. Res. 2020, 45, 2184–2195. [Google Scholar] [CrossRef]
- Jia, A.; Zhang, X.H. Molecular cloning, characterization and expression analysis of cathepsin D gene from turbot Scophthalmus maximus. Fish Shellfish Immunol. 2009, 26, 606–613. [Google Scholar] [CrossRef]
- Komai, T.; Kawabata, C.; Amano, M.; Lee, B.R.; Ichishima, E. Todarepsin, a new cathepsin D from hepatopancreas of Japanese common squid (Todarodes pacificus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 137, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Conus, S.; Simon, H.U. Cathepsins and their involvement in immune responses. Swiss Med. Wkly. 2010, 140, w13042. [Google Scholar] [CrossRef] [PubMed]
- Dvornikova, K.A.; Bystrova, E.Y.; Churilov, L.P.; Lerner, A. Pathogenesis of the inflammatory bowel disease in context of SARS-COV-2 infection. Mol. Biol. Rep. 2021, 48, 5745–5758. [Google Scholar] [CrossRef] [PubMed]
- Fara, A.; Mitrev, Z.; Rosalia, R.A.; Assas, B.M. Cytokine storm and COVID-19: A chronicle of pro-inflammatory cytokines. Open Biol. 2020, 10, 200160. [Google Scholar] [CrossRef]
- Conti, P.; Ronconi, G.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Frydas, I.; Kritas, S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J. Biol. Regul. Homeost Agents 2020, 34, 327–331. [Google Scholar]
- Runfeng, L.; Yunlong, H.; Jicheng, H.; Weiqi, P.; Qinhai, M.; Yongxia, S.; Chufang, L.; Jin, Z.; Zhenhua, J.; Haiming, J.; et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. [Google Scholar] [CrossRef]
- Ma, Z.; Damania, B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe 2016, 19, 150–158. [Google Scholar] [CrossRef]
- Vizovisek, M.; Vidak, E.; Javorsek, U.; Mikhaylov, G.; Bratovs, A.; Turk, B. Cysteine cathepsins as therapeutic targets in inflammatory diseases. Expert Opin. Ther. Targets 2020, 24, 573–588. [Google Scholar] [CrossRef]
- Lai, J.L.; Liu, Y.H.; Liu, C.; Qi, M.P.; Liu, R.N.; Zhu, X.F.; Zhou, Q.G.; Chen, Y.Y.; Guo, A.Z.; Hu, C.M. Indirubin Inhibits LPS-Induced Inflammation via TLR4 Abrogation Mediated by the NF-kB and MAPK Signaling Pathways. Inflammation 2017, 40, 1–12. [Google Scholar] [CrossRef]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.; Gao, X.; Liu, W.; Feng, X.; Zhao, Z.; Lai, Y. Lapiferin protects against H1N1 virus-induced pulmonary inflammation by negatively regulating NF-kB signaling. Braz. J. Med. Biol. Res. 2020, 53, e9183. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yu, Y.; Deubel, V. Japanese encephalitis virus NS1’ protein depends on pseudoknot secondary structure and is cleaved by caspase during virus infection and cell apoptosis. Microbes Infect. 2012, 14, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.; Tyler, K.L. Apoptosis in animal models of virus-induced disease. Nat. Rev. Microbiol. 2009, 7, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Morrey, J.D.; Diamond, M.S. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J. Virol. 2007, 81, 2614–2623. [Google Scholar] [CrossRef] [PubMed]
- Cummins, N.W.; Badley, A.D. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis. 2010, 1, e99. [Google Scholar] [CrossRef] [PubMed]
- Badley, A.D.; Pilon, A.A.; Landay, A.; Lynch, D.H. Mechanisms of HIV-associated lymphocyte apoptosis. Blood 2000, 96, 2951–2964. [Google Scholar] [CrossRef]
- Su, Y.L.; Chen, J.P.; Mo, Z.Q.; Zheng, J.Y.; Lv, S.Y.; Li, P.H.; Wei, Y.S.; Liang, Y.L.; Wang, S.W.; Yang, M.; et al. A novel MKK gene (EcMKK6) in Epinephelus coioides: Identification, characterization and its response to Vibrio alginolyticus and SGIV infection. Fish Shellfish Immunol. 2019, 92, 500–507. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Li, C.; Zhang, Y.; Wang, L.; Wei, J.; Qin, Q. Grouper TRADD Mediates Innate Antiviral Immune Responses and Apoptosis Induced by Singapore Grouper Iridovirus (SGIV) Infection. Front. Cell Infect. Microbiol. 2019, 9, 329. [Google Scholar] [CrossRef]
- Guo, M.; Wei, J.; Huang, X.; Zhou, Y.; Yan, Y.; Qin, Q. JNK1 Derived from Orange-Spotted Grouper, Epinephelus coioides, Involving in the Evasion and Infection of Singapore Grouper Iridovirus (SGIV). Front. Microbiol. 2016, 7, 121. [Google Scholar] [CrossRef]
- Conus, S.; Pop, C.; Snipas, S.J.; Salvesen, G.S.; Simon, H.U. Cathepsin D primes caspase-8 activation by multiple intra-chain proteolysis. J. Biol. Chem. 2012, 287, 21142–21151. [Google Scholar] [CrossRef] [PubMed]
- Koike, M.; Shibata, M.; Ohsawa, Y.; Nakanishi, H.; Koga, T.; Kametaka, S.; Waguri, S.; Momoi, T.; Kominami, E.; Peters, C.; et al. Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol. Cell Neurosci. 2003, 22, 146–161. [Google Scholar] [CrossRef]
- Sagulenko, V.; Muth, D.; Sagulenko, E.; Paffhausen, T.; Schwab, M.; Westermann, F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis 2008, 29, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, X.; Cai, J.; Ye, F.; Qin, Q. Involvement of the mitogen-activated protein kinase pathway in soft-shelled turtle iridovirus-induced apoptosis. Apoptosis 2011, 16, 581–593. [Google Scholar] [CrossRef]
- Chitnis, N.S.; D’Costa, S.M.; Paul, E.R.; Bilimoria, S.L. Modulation of iridovirus-induced apoptosis by endocytosis, early expression, JNK, and apical caspase. Virology 2008, 370, 333–342. [Google Scholar] [CrossRef] [PubMed][Green Version]




| Primers | Sequences (5′–3′) |
|---|---|
| NC-F | UUCUUCGAACGUGUCACGUTT |
| NC-R | ACGUGACACGUUCGGAGAATT |
| Si-Cathepsin D-136-F | GCCGACACACACUCCCUUATT |
| Si-Cathepsin D-136-R | UAAGGGAGUGUGUGUCGGCTT |
| Si-Cathepsin D-341-F | GCUGGCUUCACCACAAAUATT |
| Si-Cathepsin D-341-R | UAUUUGUGGUGAAGCCAGCTT |
| Si-Cathepsin D-936-F | GGUGAACUGUGACAAGGUUTT |
| Si-Cathepsin D-936-R | AACCUUGUCACAGUUCACCTT |
| Primers | Sequences (5′–3′) |
|---|---|
| C1-Ec-Cathepsin D-F | ATGAAGCTGTTGCTCCTCTTCGTGT |
| C1-Ec-Cathepsin D-R | TCACTTGGACTTGGCAAAGCCC |
| Ec-Cathepsin D-RT-F | GGTGCCCTCCGTTCACTGCTCCAT |
| Ec-Cathepsin D-RT-R | GCCCGACAAACTGCCACTCCCATA |
| β-Actin-RT-F | TACGAGCTGCCTGACGGACA |
| β-Actin-RT-R | GGCTGTGATCTCCTTCTGCA |
| MCP-RT-F | GCACGCTTCTCTCACCTTCA |
| MCP-RT-R | AACGGCAACGGGAGCACTA |
| VP19-RT-F | TCCAAGGGAGAAACTGTAAG |
| VP19-RT-R | GGGGTAAGCGTGAAGACT |
| LITAF-RT-F | GATGCTGCCGTGTGAACTG |
| LITAF-RT-R | GCACATCCTTGGTGGTGTTG |
| Ec-IL-1β-F | AACCTCATCATCGCCACACA |
| Ec-IL-1β-R | AGTTGCCTCACAACCGAACAC |
| Ec-IL-6-F | GGTTGGTCCAAGGTGTGCTTA |
| Ec-IL-6-R | CTGGGATTGTCGAGGTCCTT |
| Ec-IL-8-F | GCCGTCAGTGAAGGGAGTCTAG |
| Ec-IL-8-R | ATCGCAGTGGGAGTTTGCA |
| Ec-TNFα-F | GTGTCCTGCTGTTTGCTTGGTA |
| Ec-TNFα-R | CAGTGTCCGACTTGATTAGTGCTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Han, H.; Zhu, K.; Xu, S.; Han, C.; Jiang, Y.; Wei, S.; Qin, Q. Functional Analysis of the Cathepsin D Gene Response to SGIV Infection in the Orange-Spotted Grouper, Epinephelus coioides. Viruses 2022, 14, 1680. https://doi.org/10.3390/v14081680
Wang Y, Han H, Zhu K, Xu S, Han C, Jiang Y, Wei S, Qin Q. Functional Analysis of the Cathepsin D Gene Response to SGIV Infection in the Orange-Spotted Grouper, Epinephelus coioides. Viruses. 2022; 14(8):1680. https://doi.org/10.3390/v14081680
Chicago/Turabian StyleWang, Yuexuan, Honglin Han, Kecheng Zhu, Suifeng Xu, Chengzong Han, Yunxiang Jiang, Shina Wei, and Qiwei Qin. 2022. "Functional Analysis of the Cathepsin D Gene Response to SGIV Infection in the Orange-Spotted Grouper, Epinephelus coioides" Viruses 14, no. 8: 1680. https://doi.org/10.3390/v14081680
APA StyleWang, Y., Han, H., Zhu, K., Xu, S., Han, C., Jiang, Y., Wei, S., & Qin, Q. (2022). Functional Analysis of the Cathepsin D Gene Response to SGIV Infection in the Orange-Spotted Grouper, Epinephelus coioides. Viruses, 14(8), 1680. https://doi.org/10.3390/v14081680







