Recombinant Bovine Herpesvirus Type I Expressing the Bovine Viral Diarrhea Virus E2 Protein Could Effectively Prevent Infection by Two Viruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Cell Lines
2.2. Antibody and Adjuvant
2.3. Plasmid Construction
2.3.1. Construction of sgRNA Shear Plasmid
2.3.2. Optimization and Synthesis of BVDV−1 E2 Gene
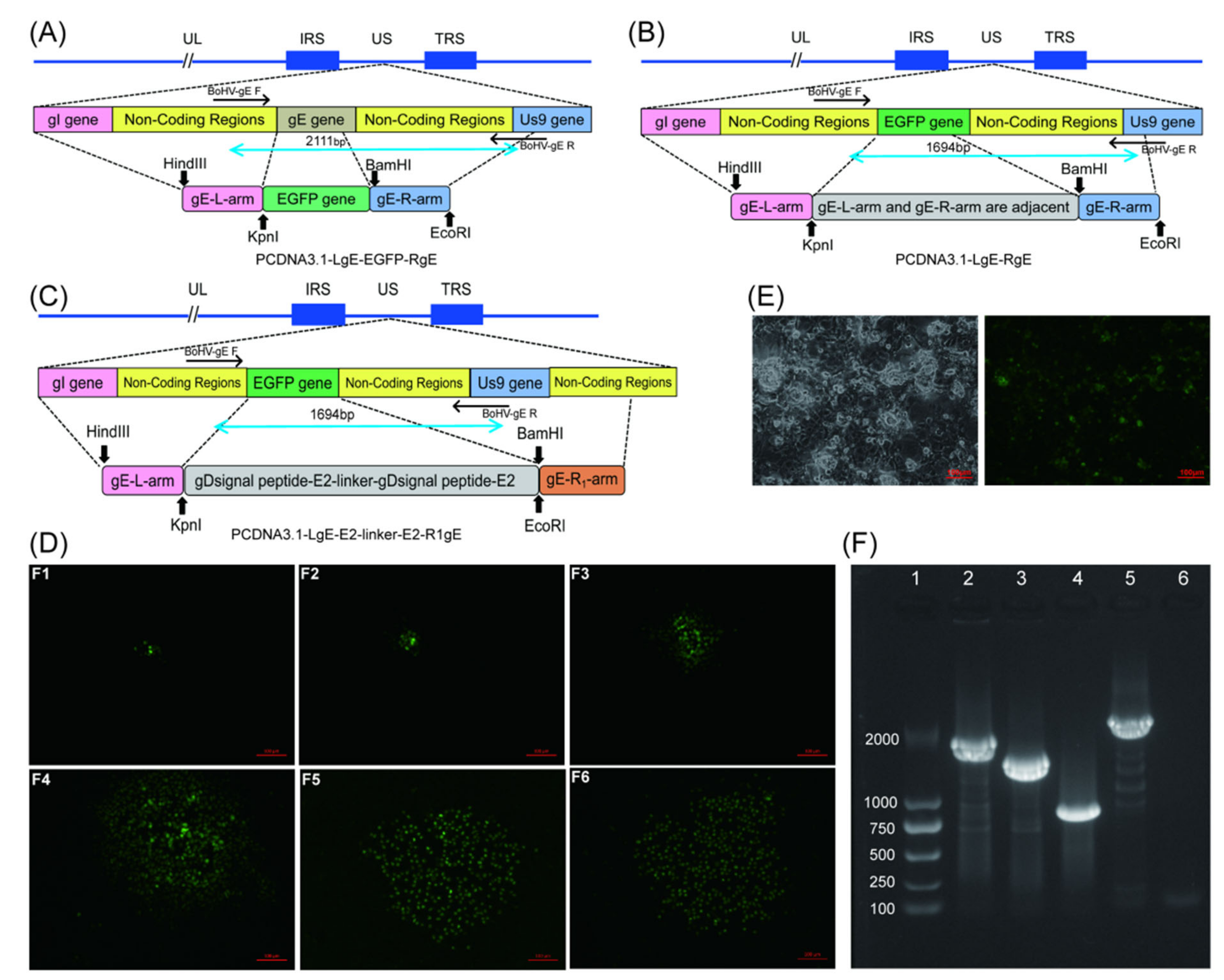
2.3.3. Construction of the Donor Plasmid
2.4. Construction of Recombinant Viruses
2.4.1. Screening for BoHV−1 gE/EGFP+, BoHV−1 ΔgE and BoHV−1 gE/E2−Linker−E2+
2.4.2. PCR Identification of Recombinant Viruses
2.4.3. Viral Titer Determination
2.4.4. Comparison of Replication Kinetics
2.4.5. Virion Structure
2.4.6. Western Blot
2.4.7. RT-PCR
2.5. Vaccine Assessment
2.5.1. Vaccine Preparation
2.5.2. Immunoprotection Assay of Guinea Pigs
2.5.3. Immunoprotective Assay of Calves
2.5.4. Clinical Symptom Score
2.5.5. Histology Score
2.5.6. Hematoxylin and Eosin Staining
2.5.7. Virus Neutralization Assay
2.5.8. ELISA
2.5.9. White Blood Cell Count
2.5.10. Data Analysis
3. Results
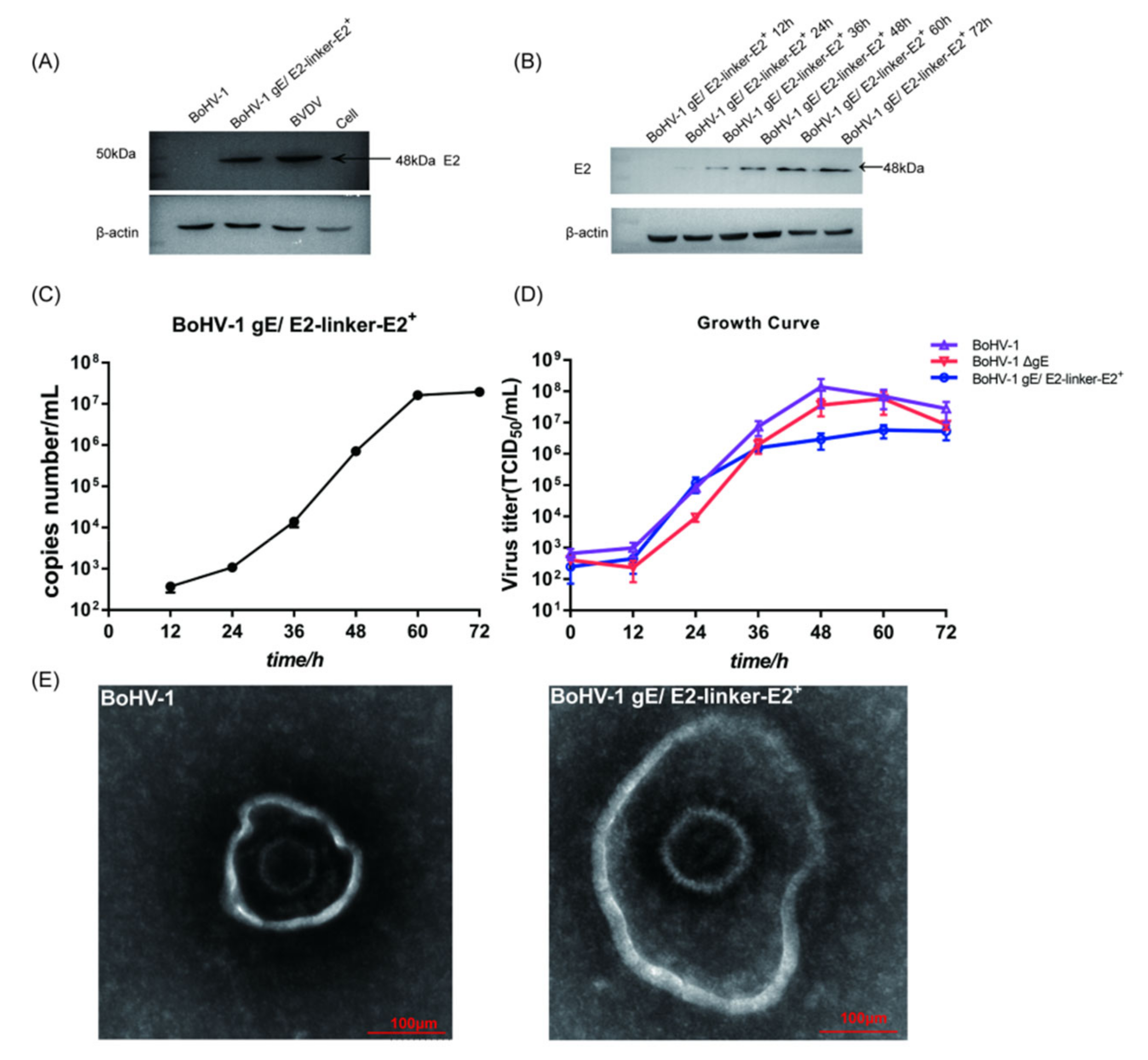
3.1. Screening and Identification of Recombinant Viruses
3.2. In Vitro Characterization of BoHV−1 gE/E2−Linker−E2+ and BoHV−1 ΔgE
3.3. Immunoprotection Assay of Guinea Pigs
3.3.1. BoHV−1 gE/E2−Linker−E2+ Immunization Group Could Produce Specific Serum Neutralizing Antibodies against BVDV−1
3.3.2. After BoHV−1 and BVDV−1 Challenge, the Time and Level of Viral Shedding of Immunized Guinea Pigs Were Less than Those of the Challenge Control Group
3.3.3. After the BVDV−1 Challenge, Mild and Transient Leukopenia Was Observed in the Guinea Pigs in the Challenge Group, While No Leukopenia Occurred in the Immunization Group
3.3.4. According to the Histopathological Lesions of the Guinea Pig Lungs, the Immunization of Inactivated Antigen Groups Showed a Good Protective Effect, and the Lung Lesions Were Reduced Compared with the Challenge Control Group
3.4. Immunogenicity and Protective Experiments of BoHV−1 gE/E2−Linker−E2+ and BoHV−1 ΔgE on Calves
3.4.1. The Average Titer of Neutralizing Antibody in Serum of BoHV−1 in the BoHV−1 Group Was Not Significantly Different from That of BoHV−1 ΔgE and BoHV−1 gE/E2−Linker−E2+ Groups 42 Days after Immunization of Inactivated Antigen, and the BoHV−1 gE/E2−Linker−E2+ Group Could Produce Specific Serum-Neutralizing Antibodies against BVDV−1
3.4.2. After BoHV−1 and BVDV−1 Challenge, the Time and Level of Viral Shedding of Immunized Calves Were Decreased Compared to Those of the Challenge Control Group
3.4.3. After the BVDV−1 Challenge, Leukopenia Was Observed in Calves of the Challenge Control Group, While No Leukopenia Was Observed in the Immunization Groups
3.4.4. Assessment of Muscle Damage
3.4.5. According to the Histopathological Lesions of the Calf Lungs, the Immunization Groups Had a Good Protective Effect, and the Lung Lesions Were Reduced Compared with the Challenge Control Group
3.4.6. According to the Histopathological Lesions of the Small Intestine of Calves, the Immunization Group Showed a Good Protective Effect, and the Lesions of the Small Intestine Were Reduced Compared to the Challenge Control Group
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, B.K.; Step, D.L.; Maxwell, C.L.; Wagner, J.J.; Richards, C.J.; Krehbiel, C.R. Evaluation of multiple ancillary therapies used in combination with an antimicrobial in newly received high-risk calves treated for bovine respiratory disease. J. Anim. Sci. 2015, 93, 3661–3674. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, S.A.; Van Eenennaam, A.L.; Aly, S.S.; Karle, B.M.; Rossitto, P.V.; Overton, M.W.; Lehenbauer, T.W.; Fadel, J.G. Preweaning cost of bovine respiratory disease (BRD) and cost-benefit of implementation of preventative measures in calves on California dairies: The BRD 10K study. J. Dairy Sci. 2020, 103, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Risalde, M.A.; Molina, V.; Sanchez-Cordon, P.J.; Pedrera, M.; Panadero, R.; Romero-Palomo, F.; Gomez-Villamandos, J.C. Response of proinflammatory and anti-inflammatory cytokines in calves with subclinical bovine viral diarrhea challenged with bovine herpesvirus-1. Vet. Immunol. Immunopathol. 2011, 144, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Molina, V.; Risalde, M.A.; Sanchez-Cordon, P.J.; Pedrera, M.; Romero-Palomo, F.; Luzzago, C.; Gomez-Villamandos, J.C. Effect of infection with BHV-1 on peripheral blood leukocytes and lymphocyte subpopulations in calves with subclinical BVD. Res. Vet. Sci. 2013, 95, 115–122. [Google Scholar] [CrossRef]
- Ridpath, J. The contribution of infections with bovine viral diarrhea viruses to bovine respiratory disease. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 335–348. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Verweij, M.C.; Lipinska, A.D.; Wang, Y.; Quinten, E.; Reits, E.A.; Koch, J.; Loch, S.; Marcondes Rezende, M.; Daus, F.; et al. Varicellovirus UL 49.5 proteins differentially affect the function of the transporter associated with antigen processing, TAP. PLoS Pathog. 2008, 4, e1000080. [Google Scholar] [CrossRef]
- Winkler, M.T.; Doster, A.; Jones, C. Bovine herpesvirus 1 can infect CD4(+) T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 1999, 73, 8657–8668. [Google Scholar] [CrossRef]
- Bryant, N.A.; Davis-Poynter, N.; Vanderplasschen, A.; Alcami, A. Glycoprotein G isoforms from some alphaherpesviruses function as broad-spectrum chemokine binding proteins. EMBO J. 2003, 22, 833–846. [Google Scholar] [CrossRef]
- Chase, C.C.; Thakur, N.; Darweesh, M.F.; Morarie-Kane, S.E.; Rajput, M.K. Immune response to bovine viral diarrhea virus--looking at newly defined targets. Anim. Health Res. Rev. 2015, 16, 4–14. [Google Scholar] [CrossRef]
- Inman, M.; Lovato, L.; Doster, A.; Jones, C. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 2001, 75, 8507–8515. [Google Scholar] [CrossRef]
- Iotti, B.; Valdano, E.; Savini, L.; Candeloro, L.; Giovannini, A.; Rosati, S.; Colizza, V.; Giacobini, M. Farm productive contexts and the dynamics of bovine viral diarrhea (BVD) transmission. Prev. Vet. Med. 2019, 165, 23–33. [Google Scholar] [CrossRef]
- Khodakaram-Tafti, A.; Farjanikish, G.H. Persistent bovine viral diarrhea virus (BVDV) infection in cattle herds. Iran. J. Vet. Res. 2017, 18, 154–163. [Google Scholar]
- Jia, S.; Huang, X.; Li, H.; Zheng, D.; Wang, L.; Qiao, X.; Jiang, Y.; Cui, W.; Tang, L.; Li, Y.; et al. Immunogenicity evaluation of recombinant Lactobacillus casei W56 expressing bovine viral diarrhea virus E2 protein in conjunction with cholera toxin B subunit as an adjuvant. Microb. Cell Fact. 2020, 19, 186. [Google Scholar] [CrossRef]
- Richter, V.; Lebl, K.; Baumgartner, W.; Obritzhauser, W.; Kasbohrer, A.; Pinior, B. A systematic worldwide review of the direct monetary losses in cattle due to bovine viral diarrhoea virus infection. Vet. J. 2017, 220, 80–87. [Google Scholar] [CrossRef]
- Vilcek, S.; Paton, D.J.; Durkovic, B.; Strojny, L.; Ibata, G.; Moussa, A.; Loitsch, A.; Rossmanith, W.; Vega, S.; Scicluna, M.T.; et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch. Virol. 2001, 146, 99–115. [Google Scholar] [CrossRef]
- Al-Kubati, A.A.G.; Hussen, J.; Kandeel, M.; Al-Mubarak, A.I.A.; Hemida, M.G. Recent Advances on the Bovine Viral Diarrhea Virus Molecular Pathogenesis, Immune Response, and Vaccines Development. Front. Vet. Sci. 2021, 8, 665128. [Google Scholar] [CrossRef]
- Durantel, D.; Branza-Nichita, N.; Carrouee-Durantel, S.; Butters, T.D.; Dwek, R.A.; Zitzmann, N. Study of the mechanism of antiviral action of iminosugar derivatives against bovine viral diarrhea virus. J. Virol. 2001, 75, 8987–8998. [Google Scholar] [CrossRef]
- Weiland, E.; Stark, R.; Haas, B.; Rumenapf, T.; Meyers, G.; Thiel, H.J. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J. Virol. 1990, 64, 3563–3569. [Google Scholar] [CrossRef] [PubMed]
- El Omari, K.; Iourin, O.; Harlos, K.; Grimes, J.M.; Stuart, D.I. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013, 3, 30–35. [Google Scholar] [CrossRef]
- Callens, N.; Brugger, B.; Bonnafous, P.; Drobecq, H.; Gerl, M.J.; Krey, T.; Roman-Sosa, G.; Rumenapf, T.; Lambert, O.; Dubuisson, J.; et al. Morphology and Molecular Composition of Purified Bovine Viral Diarrhea Virus Envelope. PLoS Pathog. 2016, 12, e1005476. [Google Scholar] [CrossRef]
- Bolin, S.R.; Ridpath, J.F. Glycoprotein E2 of bovine viral diarrhea virus expressed in insect cells provides calves limited protection from systemic infection and disease. Arch. Virol. 1996, 141, 1463–1477. [Google Scholar] [CrossRef] [PubMed]
- Harpin, S.; Hurley, D.J.; Mbikay, M.; Talbot, B.; Elazhary, Y. Vaccination of cattle with a DNA plasmid encoding the bovine viral diarrhoea virus major glycoprotein E2. J. Gen. Virol. 1999, 80, 3137–3144. [Google Scholar] [CrossRef] [PubMed]
- Muylkens, B.; Thiry, J.; Kirten, P.; Schynts, F.; Thiry, E. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet. Res. 2007, 38, 181–209. [Google Scholar] [CrossRef] [PubMed]
- Wirth, U.V.; Gunkel, K.; Engels, M.; Schwyzer, M. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 1989, 63, 4882–4889. [Google Scholar] [CrossRef]
- Chowdhury, S.I.; Coats, J.; Neis, R.A.; Navarro, S.M.; Paulsen, D.B.; Feng, J.M. A bovine herpesvirus type 1 mutant virus with truncated glycoprotein E cytoplasmic tail has defective anterograde neuronal transport in rabbit dorsal root ganglia primary neuronal cultures in a microfluidic chamber system. J. Neurovirol. 2010, 16, 457–465. [Google Scholar] [CrossRef]
- Brum, M.C.; Coats, C.; Sangena, R.B.; Doster, A.; Jones, C.; Chowdhury, S.I. Bovine herpesvirus type 1 (BoHV−1) anterograde neuronal transport from trigeminal ganglia to nose and eye requires glycoprotein E. J. Neurovirol. 2009, 15, 196–201. [Google Scholar] [CrossRef]
- Kaashoek, M.J.; Rijsewijk, F.A.; Ruuls, R.C.; Keil, G.M.; Thiry, E.; Pastoret, P.P.; Van Oirschot, J.T. Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in the gC, gG, gI, gE, or in both the gI and gE gene. Vaccine 1998, 16, 802–809. [Google Scholar] [CrossRef]
- Rebordosa, X.; Pinol, J.; Perez-Pons, J.A.; Lloberas, J.; Naval, J.; Serra-Hartmann, X.; Espuna, E.; Querol, E. Glycoprotein E of bovine herpesvirus type 1 is involved in virus transmission by direct cell-to-cell spread. Virus Res. 1996, 45, 59–68. [Google Scholar] [CrossRef]
- Butchi, N.B.; Jones, C.; Perez, S.; Doster, A.; Chowdhury, S.I. Envelope protein Us9 is required for the anterograde transport of bovine herpesvirus type 1 from trigeminal ganglia to nose and eye upon reactivation. J. Neurovirol. 2007, 13, 384–388. [Google Scholar] [CrossRef]
- Weiss, M.; Brum, M.C.; Anziliero, D.; Weiblen, R.; Flores, E.F. A glycoprotein E gene-deleted bovine herpesvirus 1 as a candidate vaccine strain. Braz. J. Med. Biol. Res. 2015, 48, 843–851. [Google Scholar] [CrossRef]
- Kaashoek, M.J.; van Engelenburg, F.A.; Moerman, A.; Gielkens, A.L.; Rijsewijk, F.A.; van Oirschot, J.T. Virulence and immunogenicity in calves of thymidine kinase- and glycoprotein E-negative bovine herpesvirus 1 mutants. Vet. Microbiol. 1996, 48, 143–153. [Google Scholar] [CrossRef]
- Van Engelenburg, F.A.; Kaashoek, M.J.; Rijsewijk, F.A.; van den Burg, L.; Moerman, A.; Gielkens, A.L.; van Oirschot, J.T. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J. Gen. Virol. 1994, 75, 2311–2318. [Google Scholar] [CrossRef]
- El-Kholy, A.A.; Rady, D.I.; Abdou, E.R.; Elseafy, M.M.; Abdelrahman, K.A.; Soliman, H. Construction, characterization and immunogenicity of a glycoprotein E negative bovine herpesvirus-1.1 Egyptian strain “Abu-Hammad”. J. Virol. Methods 2013, 194, 74–81. [Google Scholar] [CrossRef]
- Bosch, J.C.; Kaashoek, M.J.; Kroese, A.H.; van Oirschot, J.T. An attenuated bovine herpesvirus 1 marker vaccine induces a better protection than two inactivated marker vaccines. Vet. Microbiol. 1996, 52, 223–234. [Google Scholar] [CrossRef]
- Muylkens, B.; Meurens, F.; Schynts, F.; de Fays, K.; Pourchet, A.; Thiry, J.; Vanderplasschen, A.; Antoine, N.; Thiry, E. Biological characterization of bovine herpesvirus 1 recombinants possessing the vaccine glycoprotein E negative phenotype. Vet. Microbiol. 2006, 113, 283–291. [Google Scholar] [CrossRef]
- Kaashoek, M.J.; Moerman, A.; Madic, J.; Rijsewijk, F.A.; Quak, J.; Gielkens, A.L.; van Oirschot, J.T. A conventionally attenuated glycoprotein E-negative strain of bovine herpesvirus type 1 is an efficacious and safe vaccine. Vaccine 1994, 12, 439–444. [Google Scholar] [CrossRef]
- Lemaire, M.; Schynts, F.; Meyer, G.; Georgin, J.P.; Baranowski, E.; Gabriel, A.; Ros, C.; Belak, S.; Thiry, E. Latency and reactivation of a glycoprotein E negative bovine herpesvirus type 1 vaccine: Influence of virus load and effect of specific maternal antibodies. Vaccine 2001, 19, 4795–4804. [Google Scholar] [CrossRef]
- Kerkhofs, P.; Renjifo, X.; Toussaint, J.F.; Letellier, C.; Vanopdenbosch, E.; Wellemans, G. Enhancement of the immune response and virological protection of calves against bovine herpesvirus type 1 with an inactivated gE-deleted vaccine. Vet. Rec. 2003, 152, 681–686. [Google Scholar] [CrossRef]
- Bosch, J.C.; De Jong, M.C.; Franken, P.; Frankena, K.; Hage, J.J.; Kaashoek, M.J.; Maris-Veldhuis, M.A.; Noordhuizen, J.P.; Van der Poel, W.H.; Verhoeff, J.; et al. An inactivated gE-negative marker vaccine and an experimental gD-subunit vaccine reduce the incidence of bovine herpesvirus 1 infections in the field. Vaccine 1998, 16, 265–271. [Google Scholar] [CrossRef]
- Chowdhury, S.I.; Pannhorst, K.; Sangewar, N.; Pavulraj, S.; Wen, X.; Stout, R.W.; Mwangi, W.; Paulsen, D.B. BoHV−1-Vectored BVDV−2 Subunit Vaccine Induces BVDV Cross-Reactive Cellular Immune Responses and Protects against BVDV−2 Challenge. Vaccines 2021, 9, 46. [Google Scholar] [CrossRef]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef] [PubMed]
- Muylkens, B.; Meurens, F.; Schynts, F.; Farnir, F.; Pourchet, A.; Bardiau, M.; Gogev, S.; Thiry, J.; Cuisenaire, A.; Vanderplasschen, A.; et al. Intraspecific bovine herpesvirus 1 recombinants carrying glycoprotein E deletion as a vaccine marker are virulent in cattle. J. Gen. Virol. 2006, 87, 2149–2154. [Google Scholar] [CrossRef] [PubMed]
- Raaperi, K.; Orro, T.; Viltrop, A. Epidemiology and control of bovine herpesvirus 1 infection in Europe. Vet. J. 2014, 201, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Engels, M. Pro and contra IBR-eradication. Vet. Microbiol. 2006, 113, 293–302. [Google Scholar] [CrossRef]
- Castrucci, G.; Frigeri, F.; Salvatori, D.; Ferrari, M.; Sardonini, Q.; Cassai, E.; Lo, D.M.; Rotola, A.; Angelini, R. Vaccination of calves against bovine herpesvirus-1: Assessment of the protective value of eight vaccines. Comp. Immunol. Microbiol. Infect. Dis. 2002, 25, 29–41. [Google Scholar] [CrossRef]
- Van Drunen Littel-van den Hurk, S.; Tikoo, S.K.; van den Hurk, J.V.; Babiuk, L.A.; Van Donkersgoed, J. Protective immunity in cattle following vaccination with conventional and marker bovine herpesvirus-1 (BHV1) vaccines. Vaccine 1997, 15, 36–44. [Google Scholar] [CrossRef]
- Ran, X.; Chen, X.; Ma, L.; Wen, X.; Zhai, J.; Wang, M.; Tong, X.; Hou, G.; Ni, H. A systematic review and meta-analysis of the epidemiology of bovine viral diarrhea virus (BVDV) infection in dairy cattle in China. Acta Trop. 2019, 190, 296–303. [Google Scholar] [CrossRef]
- Wernike, K.; Michelitsch, A.; Aebischer, A.; Schaarschmidt, U.; Konrath, A.; Nieper, H.; Sehl, J.; Teifke, J.P.; Beer, M. The Occurrence of a Commercial N(pro) and E(rns) Double Mutant BVDV−1 Live-Vaccine Strain in Newborn Calves. Viruses 2018, 10, 274. [Google Scholar] [CrossRef]
- Lanyon, S.R.; Hill, F.I.; Reichel, M.P.; Brownlie, J. Bovine viral diarrhoea: Pathogenesis and diagnosis. Vet. J. 2014, 199, 201–209. [Google Scholar] [CrossRef]
- Mahmoodi, P.; Shapouri, M.R.; Ghorbanpour, M.; Ekhtelat, M.; Hajikolaei, M.R.; Lotfi, M.; Boroujeni, M.P.; Daghari, M. Epitope mapping of bovine viral diarrhea virus nonstructural protein 3. Vet. Immunol. Immunopathol. 2014, 161, 232–239. [Google Scholar] [CrossRef]
- Stahl, K.; Alenius, S. BVDV control and eradication in Europe--an update. Jpn. J. Vet. Res. 2012, 60, S31–S39. [Google Scholar]
- Presi, P.; Heim, D. BVD eradication in Switzerland--a new approach. Vet. Microbiol. 2010, 142, 137–142. [Google Scholar] [CrossRef]
- Bachofen, C.; Stalder, H.; Vogt, H.R.; Wegmuller, M.; Schweizer, M.; Zanoni, R.; Peterhans, E. Bovine viral diarrhea (BVD): From biology to control. Berl. Und Munch. Tierarztl. Wochenschr. 2013, 126, 452–461. [Google Scholar]
- Chung, Y.C.; Cheng, L.T.; Zhang, J.Y.; Wu, Y.J.; Liu, S.S.; Chu, C.Y. Recombinant E2 protein enhances protective efficacy of inactivated bovine viral diarrhea virus 2 vaccine in a goat model. BMC Vet. Res. 2018, 14, 194. [Google Scholar] [CrossRef]
- Schmitt, J.; Becher, P.; Thiel, H.J.; Keil, G.M. Expression of bovine viral diarrhoea virus glycoprotein E2 by bovine herpesvirus-1 from a synthetic ORF and incorporation of E2 into recombinant virions. J. Gen. Virol. 1999, 80, 2839–2848. [Google Scholar] [CrossRef]
- Bhuyan, A.A.; Memon, A.M.; Bhuiyan, A.A.; Zhonghua, L.; Zhang, B.; Ye, S.; Mengying, L.; He, Q.G. The construction of recombinant Lactobacillus casei expressing BVDV E2 protein and its immune response in mice. J. Biotechnol. 2018, 270, 51–60. [Google Scholar] [CrossRef]
- Pecora, A.; Malacari, D.A.; Perez Aguirreburualde, M.S.; Bellido, D.; Nunez, M.C.; Dus Santos, M.J.; Escribano, J.M.; Wigdorovitz, A. Development of an APC-targeted multivalent E2-based vaccine against Bovine Viral Diarrhea Virus types 1 and 2. Vaccine 2015, 33, 5163–5171. [Google Scholar] [CrossRef][Green Version]
| Primers | Nucleotide Sequences (5′–3′) | Genome Position | Restriction Sites |
|---|---|---|---|
| LgE−F | aagcttTGCTCTTCTCCATCGCCCATC | 120663–120683 | Hind III |
| LgE−R | ggtaccCATTGCCAAATGCCCTTTTCGA | 121695–121716 | Kpn I |
| EGFP−F | ggtaccATGGTGAGCAAGGGCGA | 1–17 | Kpn I |
| EGFP−R | ggatccCTTGTACAGCTCGTCC | 702–717 | BamH I |
| RgE−F | ggatccAGTCGTTACTTCGGACCGTTTGGTGC | 122847–122872 | BamH I |
| RgE−R | gaattcTCAGCGCCTCGATAGTTTTCGTTGAC | 123569–123594 | EcoR I |
| R1gE−F | ggatccCTCAAGTCCATCCTCCGCTAG | 123421–123441 | BamH I |
| R1gE−R | gaattcGCCCTTGTCATATTTTTTTAA | 124486–124506 | EcoR I |
| BoHV−1 gE−F | CGCCGGGTTGTTAAATGGGTCTCG | 121573–121596 | |
| BoHV−1 gE−R | CGGGCGCGTCCTCGATGGTG | 123664–123683 | |
| pX459−EGFP−sgRNA1−F | caccgGTCGCCCTCGAACTTCACCT | 335–354 | Bbs I |
| pX459−EGFP−sgRNA1−R | aaacAGGTGAAGTTCGAGGGCGACc | 335–354 | Bbs I |
| pX459−EGFP−sgRNA2−F | caccgGTGGTTGTCGGGCAGCAGCA | 581–600 | Bbs I |
| pX459−EGFP−sgRNA2−R | aaacTGCTGCTGCCCGACAACCACCc | 581–600 | Bbs I |
| pX459−gE−sgRNA1−F | caccgCGGCGACGAGGAGACGCAGTTGG | 122217–122239 | Bbs I |
| pX459−gE−sgRNA1−R | aaacCCAACTGCGTCTCCTCGTCGCCGc | 122217–122239 | Bbs I |
| pX459−gE−sgRNA2−F | caccgCGCCGATGAGCCGGTCGTACAGG | 122190–122212 | Bbs I |
| pX459−gE−sgRNA2−R | aaacCCTGTACGACCGGCTCATCGGCGc | 122190–122212 | Bbs I |
| E2−F | cggggtaccATGCAAGGGCCGACATTGGCCG | 1–22 | Kpn I |
| E2−Rm | GCCGCTGCCGCCGCTGCCCTACCCGGCCACGACCA | 1194–1128 | |
| E2−Fm | GGCAGCGGCGGCAGCGGCATGCAAGGGCCGACATT | 1111–1145 | |
| E2−R | cgcggatccCTACCCGGCCACGACCACCAC | 2218–2238 | BamH I |
| BVDV−F | GGGNAGTCGTCARTGGTTCG | 177–196 | |
| BVDV−R | GTGCCATGTACAGCAGAGWTTTT | 354–376 | |
| Probe | FAM-CCAYGTGGACGAGGGCAYGC-TAMRA | 224–243 | |
| gB−F | CGTGACGGTAGCCTGGGACT | 56473–56492 | |
| gB−R | CGTCTCGCAGCATTTCGTC | 56543–56561 | |
| E2−E2−F | GCGTTTCAGATGGTGTGC | 373–390 | |
| E2−E2−R | CTTGCTGCGGCGGTAGGT | 463–480 | |
| 5′UTR−F | TAGCCATGCCCTTAGTAGGAC | 93–113 | |
| 5′UTR−R | CTCCATGTGCCATGTACAGCA | 362–382 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-Y.; Guo, H.; Zhao, H.-Z.; Hou, L.-N.; Wen, Y.-J.; Wang, F.-X. Recombinant Bovine Herpesvirus Type I Expressing the Bovine Viral Diarrhea Virus E2 Protein Could Effectively Prevent Infection by Two Viruses. Viruses 2022, 14, 1618. https://doi.org/10.3390/v14081618
Liu C-Y, Guo H, Zhao H-Z, Hou L-N, Wen Y-J, Wang F-X. Recombinant Bovine Herpesvirus Type I Expressing the Bovine Viral Diarrhea Virus E2 Protein Could Effectively Prevent Infection by Two Viruses. Viruses. 2022; 14(8):1618. https://doi.org/10.3390/v14081618
Chicago/Turabian StyleLiu, Chun-Yu, Hao Guo, Hong-Zhe Zhao, Li-Na Hou, Yong-Jun Wen, and Feng-Xue Wang. 2022. "Recombinant Bovine Herpesvirus Type I Expressing the Bovine Viral Diarrhea Virus E2 Protein Could Effectively Prevent Infection by Two Viruses" Viruses 14, no. 8: 1618. https://doi.org/10.3390/v14081618
APA StyleLiu, C.-Y., Guo, H., Zhao, H.-Z., Hou, L.-N., Wen, Y.-J., & Wang, F.-X. (2022). Recombinant Bovine Herpesvirus Type I Expressing the Bovine Viral Diarrhea Virus E2 Protein Could Effectively Prevent Infection by Two Viruses. Viruses, 14(8), 1618. https://doi.org/10.3390/v14081618






