Intergrated Transcriptomic and Proteomic Analysis Revealed the Differential Responses to Novel Duck Reovirus Infection in the Bursa of Fabricius of Cairna moschata
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Sampling Method
2.2. Sampling and Protein Pretreatment
2.3. Trypsin Digestion
2.4. Tandem Mass Tags (TMT) Labeling and HPLC-MS/MS Analysis
2.5. Bioinformatic Analysis
2.6. PPI Network Construction
2.7. Validation of Differentially Accumulated Proteins (DAPs) by Parallel Reaction Monitoring (PRM)
2.8. Library Construction and RNA Sequencing
2.9. Transcriptomic Analysis
2.10. Real-Time PCR
2.11. Statistical Analysis
3. Results
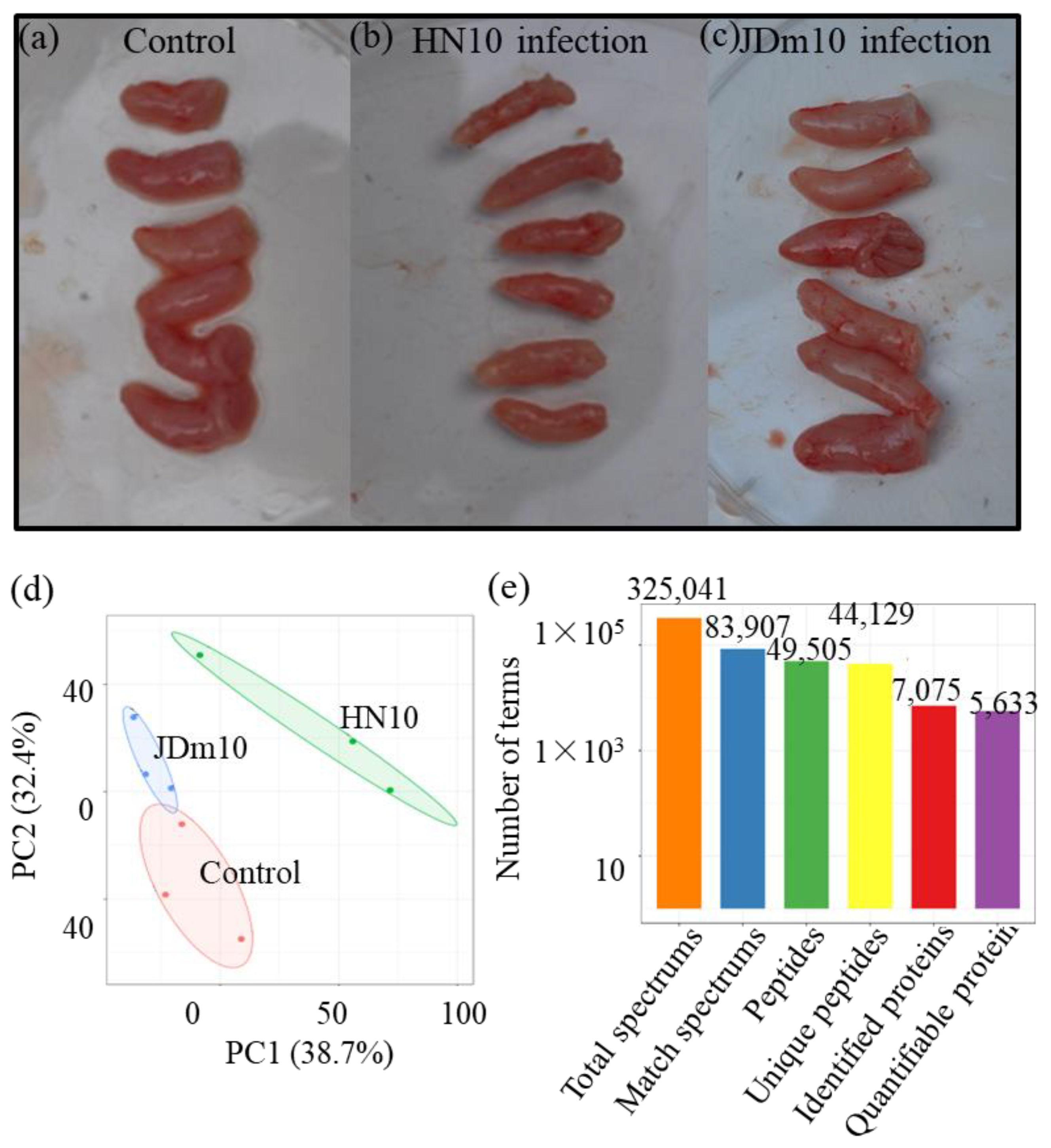
3.1. Overview of the Proteomic Data
3.2. Analysis of the DAPs under HN10/JDm10 Infections
3.3. Classification of the DAPs under the HN10/JDm10 Infections
3.4. Enrichment Analysis of the DAPs under HN10/JDm10 Infections
3.5. PPI Network Analysis of the DAPs under HN10/JDm10 Infections
3.6. Analysis of the DEGs under HN10/JDm10 Infections
3.7. DAPs Related to the Serine Protease System and the Innate and Adaptive Immune Responses
3.8. Comparison of the DAPs among Liver, Spleen, and Bursa of Fabricius Cells
3.9. Verification of the DAPs and DEGs under HN10/JDm10 Infections
3.10. Integrated Proteomic and Transcriptomic Analysis Revealed the Immune Response to HN10/JDm10 Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wozniakowski, G.; Samorek-Salamonowicz, E.; Gawel, A. Occurrence of reovirus infection in Muscovy ducks (Cairna moschata) in south western Poland. Pol. J. Vet. Sci. 2014, 17, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Chen, S.L.; Lin, F.Q.; Wang, S.; Jiang, B.; Cheng, X.X.; Zhu, X.L.; Li, Z.L. The isolation and identification of novel duck reovirus. Bing Du Xue Bao Chin. J. Virol. 2012, 28, 224–230. [Google Scholar]
- Yun, T.; Yu, B.; Ni, Z.; Ye, W.; Chen, L.; Hua, J.; Zhang, C. Genomic characteristics of a novel reovirus from Muscovy duckling in China. Vet. Microbiol. 2014, 168, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Shao, J.W.; Li, X.W.; Mei, M.M.; Guo, J.Y.; Li, W.F.; Huang, W.J.; Chi, S.H.; Yuan, S.; Li, Z.L.; et al. Molecular characterization of two novel reoviruses isolated from Muscovy ducklings in Guangdong, China. BMC Vet. Res. 2019, 15, 143. [Google Scholar] [CrossRef]
- Chen, X.; Zheng, M.; Lin, F.; Cheng, X.; Xiao, S.; Chen, S.; Chen, S. Impacts of novel duck reovirus infection on the composition of intestinal microbiota of Muscovy ducklings. Microb. Pathog. 2019, 137, 103764. [Google Scholar] [CrossRef]
- Chen, S.; Lin, F.; Chen, S.; Hu, Q.; Cheng, X.; Jiang, B.; Zhu, X.; Wang, S.; Zheng, M.; Huang, M. Development of a live attenuated vaccine against Muscovy duck reovirus infection. Vaccine 2018, 36, 8001–8007. [Google Scholar] [CrossRef]
- Jeong, E.; Lee, J.Y. Intrinsic and extrinsic regulation of innate immune receptors. Yonsei Med. J. 2011, 52, 379–392. [Google Scholar] [CrossRef]
- Chapin, J.C.; Hajjar, K.A. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015, 29, 17–24. [Google Scholar] [CrossRef]
- Zheng, C. The emerging roles of NOD-like receptors in antiviral innate immune signaling pathways. Int. J. Biol. Macromol. 2020, 169, 407–413. [Google Scholar] [CrossRef]
- Liu, Q.N.; Yang, T.T.; Wang, C.; Jiang, S.H.; Zhang, D.Z.; Tang, B.P.; Ge, B.M.; Wang, J.L.; Wang, D.; Dai, L.S. A non-mammalian Toll-like receptor 26 (TLR26) gene mediates innate immune responses in yellow catfish Pelteobagrus fulvidraco. Fish Shellfish Immunol. 2019, 95, 491–497. [Google Scholar] [CrossRef]
- Meylan, E.; Tschopp, J. Toll-like receptors and RNA helicases: Two parallel ways to trigger antiviral responses. Mol. Cell 2006, 22, 561–569. [Google Scholar] [CrossRef]
- Amer, L.D.; Saleh, L.S.; Walker, C.; Thomas, S.; Janssen, W.J.; Alper, S.; Bryant, S.J. Inflammation via myeloid differentiation primary response gene 88 signaling mediates the fibrotic response to implantable synthetic poly(ethylene glycol) hydrogels. Acta Biomater. 2019, 100, 105–117. [Google Scholar] [CrossRef]
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39, BSR20181709. [Google Scholar] [CrossRef]
- Ifrah, M.E.; Perelman, B.; Finger, A.; Uni, Z. The role of the bursa of Fabricius in the immune response to vaccinal antigens and the development of immune tolerance in chicks (Gallus domesticus) vaccinated at a very young age. Poult. Sci. 2017, 96, 51–57. [Google Scholar] [CrossRef]
- Taylor, R.L., Jr.; McCorkle, F.M., Jr. A landmark contribution to poultry science—Immunological function of the bursa of Fabricius. Poult. Sci. 2009, 88, 816–823. [Google Scholar] [CrossRef]
- Li, J.P.; Xiong, X.; Gan, X.M.; Pu, F.J.; Ma, S.C.; Bai, L.L.; Mustafa, A.; Li, L.L.; Liu, H.H.; Yang, C.W.; et al. Transcriptome analysis of the bursa of Fabricius and thymus of laying ducks reveals immune gene expression changes underlying the impacts of stocking densities. Br. Poult. Sci. 2021, 62, 820–826. [Google Scholar] [CrossRef]
- Yun, T.; Hua, J.; Ye, W.; Yu, B.; Ni, Z.; Chen, L.; Zhang, C. Comparative proteomic analysis revealed complex responses to classical/novel duck reovirus infections in the spleen tissue of Cairna moschata. J. Proteom. 2019, 193, 162–172. [Google Scholar] [CrossRef]
- Yun, T.; Hua, J.; Ye, W.; Yu, B.; Chen, L.; Ni, Z.; Zhang, C. Comparative proteomic analysis revealed complex responses to classical/novel duck reovirus infections in Cairna moschata. Sci. Rep. 2018, 8, 10079. [Google Scholar] [CrossRef]
- Zhang, M.; Song, K.; Li, C.; Chen, Z.; Ding, C.; Liu, G. Molecular cloning of Peking duck Toll-like receptor 3 (duTLR3) gene and its responses to reovirus infection. Virol. J. 2015, 12, 207. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Leng, L.; Ding, R.; Gong, P.; Liu, C.; Wang, N.; Li, H.; Du, Z.Q.; Cheng, B. Integrated transcriptome and proteome analysis reveals potential mechanisms for differential abdominal fat deposition between divergently selected chicken lines. J. Proteom. 2021, 241, 104242. [Google Scholar] [CrossRef]
- Yan, H.; Xu, G.; Zhu, Y.; Xie, Z.; Zhang, R.; Jiang, S. Isolation and characterization of a naturally attenuated novel duck reovirus strain as a live vaccine candidate. Vet. Microbiol. 2021, 261, 109214. [Google Scholar] [CrossRef]
- Yun, T.; Jiongang, H.; Weicheng, Y.; Zheng, N.; Liu, C.; Cun, Z. Proliferative characteristics of novel duck reovirus strain JDm10 in DF-1 cells. Acta Agric. Zhejiangensis 2019, 31, 716–721. [Google Scholar]
- Yun, T.; Chen, H.; Yu, B.; Zhang, C.; Chen, L.; Ni, Z.; Hua, J.; Ye, W. Development and application of an indirect ELISA for the detection of antibodies to novel duck reovirus. J. Virol. Methods 2015, 220, 55–59. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Yu, C.; Huang, J.; Wu, Q.; Zhang, C.; Li, X.-L.; Xu, X.; Feng, S.; Zhan, X.; Chen, Z.; Wang, H.; et al. Role of female predominant MYB39-bHLH13 complex in sexually dimorphic accumulation of taxol in Taxus media. Hortic. Res. 2022, 9, uhac062. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liang, Y.; Xie, S.B.; Wu, C.H.; Hu, Y.; Zhang, Q.; Li, S.; Fan, Y.G.; Leng, R.X.; Pan, H.F.; Xiong, H.B.; et al. Coagulation cascade and complement system in systemic lupus erythematosus. Oncotarget 2018, 9, 14862–14881. [Google Scholar] [CrossRef]
- Yun, T.; Ye, W.; Ni, Z.; Chen, L.; Yu, B.; Hua, J.; Zhang, Y.; Zhang, C. Complete genomic sequence of goose-origin reovirus from China. J. Virol. 2012, 86, 10257. [Google Scholar] [CrossRef]
- Wang, H.; Gao, B.; Liu, X.; Zhang, S.; Diao, Y.; Tang, Y. Pathogenicity of a variant duck orthoreovirus strain in Cherry Valley Ducklings. Vet. Microbiol. 2020, 242, 108546. [Google Scholar] [CrossRef]
- Korte, J.; Frohlich, T.; Kohn, M.; Kaspers, B.; Arnold, G.J.; Hartle, S. 2D DIGE analysis of the bursa of Fabricius reveals characteristic proteome profiles for different stages of chicken B-cell development. Proteomics 2013, 13, 119–133. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, C.; Xu, L.; Zheng, X.; Liao, M.; Yan, Y.; Jin, Y.; Zhou, J. Proteome dynamics in primary target organ of infectious bursal disease virus. Proteomics 2012, 12, 1844–1859. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Qin, A.; Qian, K.; Chen, X.; Jin, W.; Zhu, Y.; Eltahir, Y.M. Proteomic analysis of the host response in the bursa of Fabricius of chickens infected with Marek’s disease virus. Virus Res. 2010, 153, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Sun, G.; Li, K.; Li, Z.; Su, A.; Liu, X.; Li, G.; Jiang, R.; Han, R.; et al. Transcriptome profile in bursa of Fabricius reveals potential mode for stress-influenced immune function in chicken stress model. BMC Genom. 2018, 19, 918. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Y.; Chen, J.; Hua, G.; Li, J.; Deng, X.; Deng, X. Transcriptome analyses of differential gene expression in the bursa of Fabricius between silky fowl and white leghorn. Sci. Rep. 2017, 7, 45959. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Gao, B.; Zhang, S.; Diao, Y.; Tang, Y. Evidence of vertical transmission of novel duck orthoreovirus in ducks. Vet. Microbiol. 2020, 251, 108861. [Google Scholar] [CrossRef]
- Zhu, Y.Q.; Li, C.F.; Bi, Z.L.; Chen, Z.Y.; Meng, C.C.; Wang, G.J.; Ding, C.; Liu, G.Q. Molecular characterization of a novel reovirus isolated from Pekin ducklings in China. Arch. Virol. 2015, 160, 365–369. [Google Scholar] [CrossRef]
- Kang, Y.; Li, Y.; Yuan, R.; Li, X.; Sun, M.; Wang, Z.; Feng, M.; Jiao, P.; Ren, T. Phylogenetic relationships and pathogenicity variation of two Newcastle disease viruses isolated from domestic ducks in Southern China. Virol. J. 2014, 11, 147. [Google Scholar] [CrossRef]
- Cirino, G.; Napoli, C.; Bucci, M.; Cicala, C. Inflammation-coagulation network: Are serine protease receptors the knot? Trends Pharmacol. Sci. 2000, 21, 170–172. [Google Scholar] [CrossRef]
- Adams, M.N.; Ramachandran, R.; Yau, M.K.; Suen, J.Y.; Fairlie, D.P.; Hollenberg, M.D.; Hooper, J.D. Structure, function and pathophysiology of protease activated receptors. Pharmacol. Ther. 2011, 130, 248–282. [Google Scholar] [CrossRef]
- Markiewski, M.M.; Nilsson, B.; Ekdahl, K.N.; Mollnes, T.E.; Lambris, J.D. Complement and coagulation: Strangers or partners in crime? Trends Immunol. 2007, 28, 184–192. [Google Scholar] [CrossRef]
- Wolberg, A.S. Thrombin generation and fibrin clot structure. Blood Rev. 2007, 21, 131–142. [Google Scholar] [CrossRef]
- Akiyama, M.; Ohtsuki, S.; Berry, G.J.; Liang, D.H.; Goronzy, J.J.; Weyand, C.M. Innate and adaptive immunity in giant cell arteritis. Front. Immunol. 2020, 11, 621098. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Bi, Z.; Zhu, Y.; Chen, Z.; Li, C.; Wang, Y.; Wang, G.; Liu, G. Induction of a robust immunity response against novel duck reovirus in ducklings using a subunit vaccine of sigma C protein. Sci. Rep. 2016, 6, 39092. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, Y.; Li, C.; Liu, G. Outbreak-associated novel duck Reovirus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1209–1211. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, Z.; Ou, C.; Wang, Q.; Zhang, Y.; Guo, F.; Gao, P.; Ma, J. The effect of ghrelin on the fibrosis of chicken bursa of fabricius infected with infectious bursal disease virus. Gen. Comp. Endocrinol. 2021, 303, 113705. [Google Scholar] [CrossRef]
- Lenman, A.; Muller, S.; Nygren, M.I.; Frangsmyr, L.; Stehle, T.; Arnberg, N. Coagulation factor IX mediates serotype-specific binding of species A adenoviruses to host cells. J. Virol. 2011, 85, 13420–13431. [Google Scholar] [CrossRef]






| Protein Accession | Protein Description | Bursa of Fabricius | Liver | Spleen |
|---|---|---|---|---|
| R0JLH6 | Coagulation factor IX | 1.5 | 1.0 | 0.9 |
| U3IBH9 | Coagulation factor XIII | 1.2 | 2.3 | 1.7 |
| R0LYC0 | Coagulation factor II | 1.1 | 1.4 | NA |
| R0JSX9 | Fibrinogen α-chain | 2.4 | 4.2 | 9.3 |
| U3I9E6 | Fibrinogen β-chain | 1.9 | 4.3 | 5.1 |
| U3IA23 | Fibrinogen λ-chain | 2.1 | 3.4 | 5.0 |
| U3J6P0 | Complement C3 | 1.1 | NA | 2.4 |
| U3IKF3 | Complement C8 alpha chain | 1.5 | 2.5 | 1.6 |
| R0JIF4 | Complement C5 | 1.6 | 1.7 | 1.5 |
| B5AG23 | Complement component 3d | 1.0 | 2.0 | NA |
| R0LW73 | Complement C1q subunit B | 1.3 | 2.1 | 2.4 |
| U3IN55 | Complement C9 | 1.1 | 2.1 | 2.7 |
| U3ID07 | Complement C1s | 3.6 | 1.9 | 3.1 |
| U3INH5 | Complement C7 | 1.2 | 3.3 | 1.8 |
| U3IQV8 | Complement C6 | 1.2 | 1.3 | 1.8 |
| U3IKF3 | Complement C8 beta chain | 1.4 | 2.0 | 2.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, T.; Hua, J.; Ye, W.; Ni, Z.; Chen, L.; Zhu, Y.; Zhang, C. Intergrated Transcriptomic and Proteomic Analysis Revealed the Differential Responses to Novel Duck Reovirus Infection in the Bursa of Fabricius of Cairna moschata. Viruses 2022, 14, 1615. https://doi.org/10.3390/v14081615
Yun T, Hua J, Ye W, Ni Z, Chen L, Zhu Y, Zhang C. Intergrated Transcriptomic and Proteomic Analysis Revealed the Differential Responses to Novel Duck Reovirus Infection in the Bursa of Fabricius of Cairna moschata. Viruses. 2022; 14(8):1615. https://doi.org/10.3390/v14081615
Chicago/Turabian StyleYun, Tao, Jionggang Hua, Weicheng Ye, Zheng Ni, Liu Chen, Yinchu Zhu, and Cun Zhang. 2022. "Intergrated Transcriptomic and Proteomic Analysis Revealed the Differential Responses to Novel Duck Reovirus Infection in the Bursa of Fabricius of Cairna moschata" Viruses 14, no. 8: 1615. https://doi.org/10.3390/v14081615
APA StyleYun, T., Hua, J., Ye, W., Ni, Z., Chen, L., Zhu, Y., & Zhang, C. (2022). Intergrated Transcriptomic and Proteomic Analysis Revealed the Differential Responses to Novel Duck Reovirus Infection in the Bursa of Fabricius of Cairna moschata. Viruses, 14(8), 1615. https://doi.org/10.3390/v14081615






