Avian Reovirus σB Interacts with Caveolin-1 in Lipid Rafts during Dynamin-Dependent Caveolae-Mediated Endocytosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Reagents and Antibodies
2.3. Construction of Expression Vectors
2.4. Detergent-Resistant Membranes (DRMs) Extraction by Density Gradient Centrifugation
2.5. Plasmid Transfection
2.6. Indirect Immunofluorescence Assay (IFA) and Confocal Microscopy
2.7. Co-Immunoprecipitation (Co-IP) Assay
2.8. Cell Infection and Drug Treatments
2.9. Real-Time Quantitative Reverse Transcription-PCR (qRT-PCR)
2.10. SDS-PAGE and Western Blot Analyses
2.11. Statistical Analysis
3. Results
3.1. ARV Capsid Protein σB Presented in Cav-1 Rich Fraction in DRMs
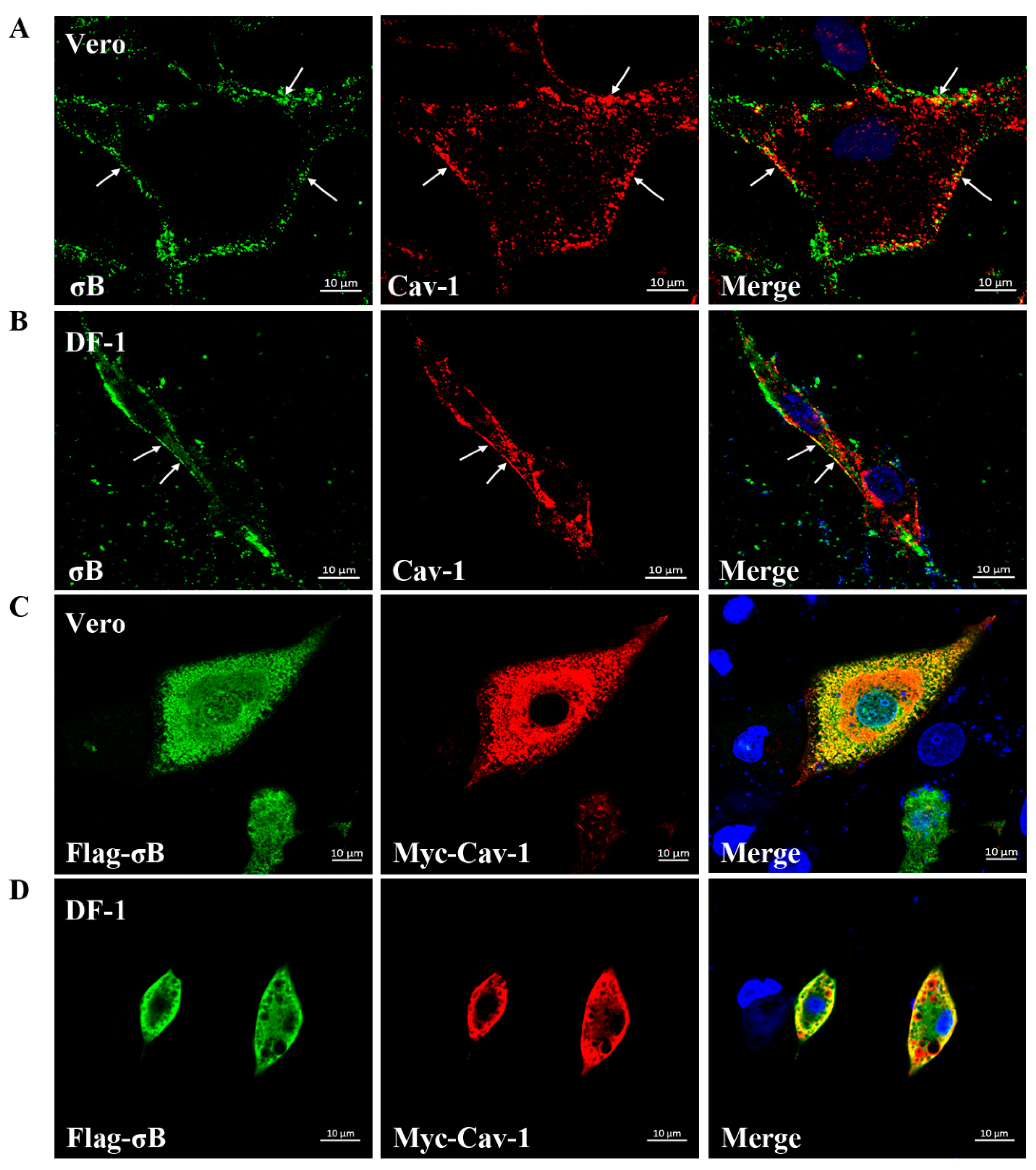
3.2. Co-Localization of σB with Cav-1 during ARV Endocytosis
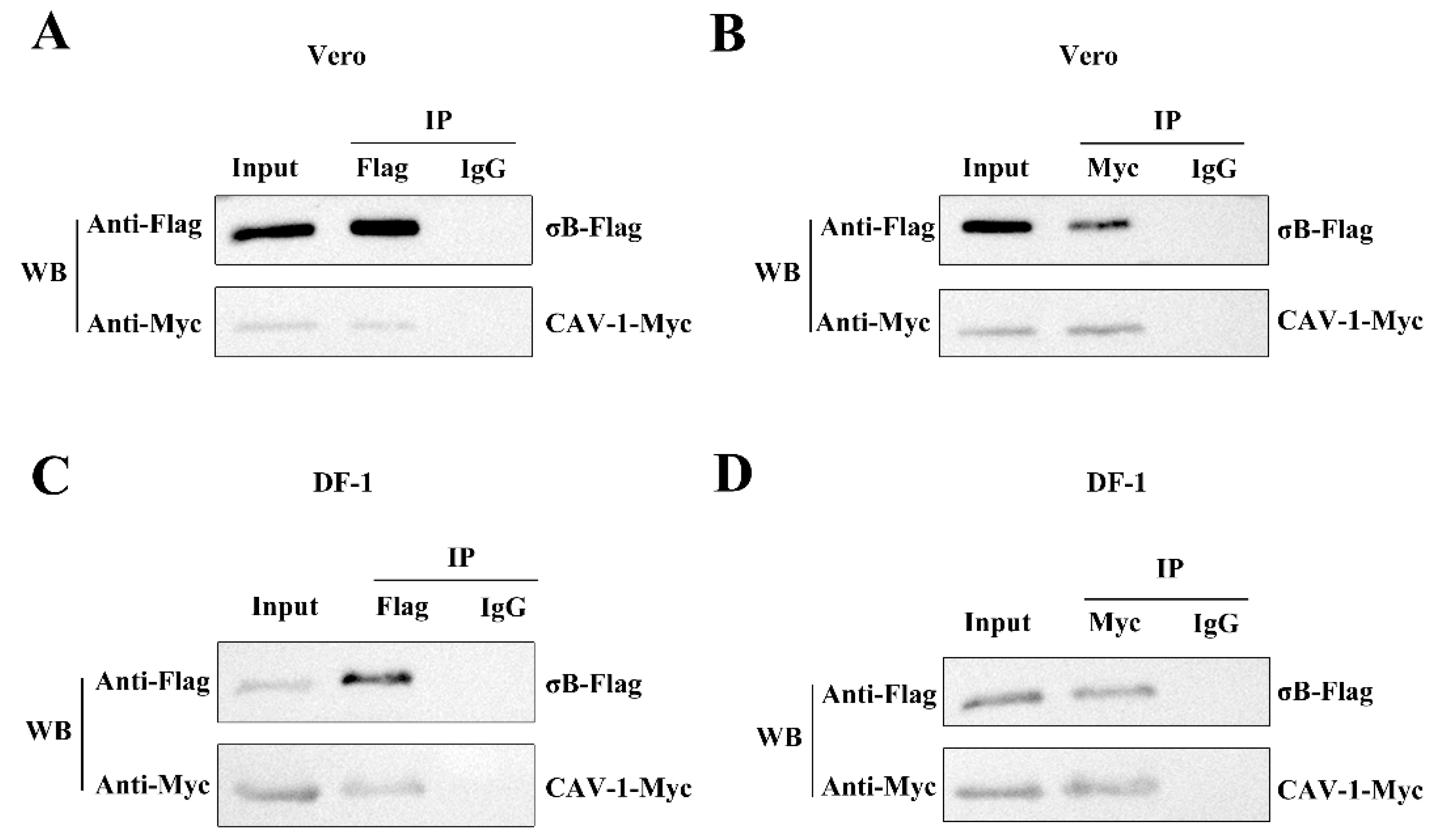
3.3. ARV σB Protein Interacts with Cav-1
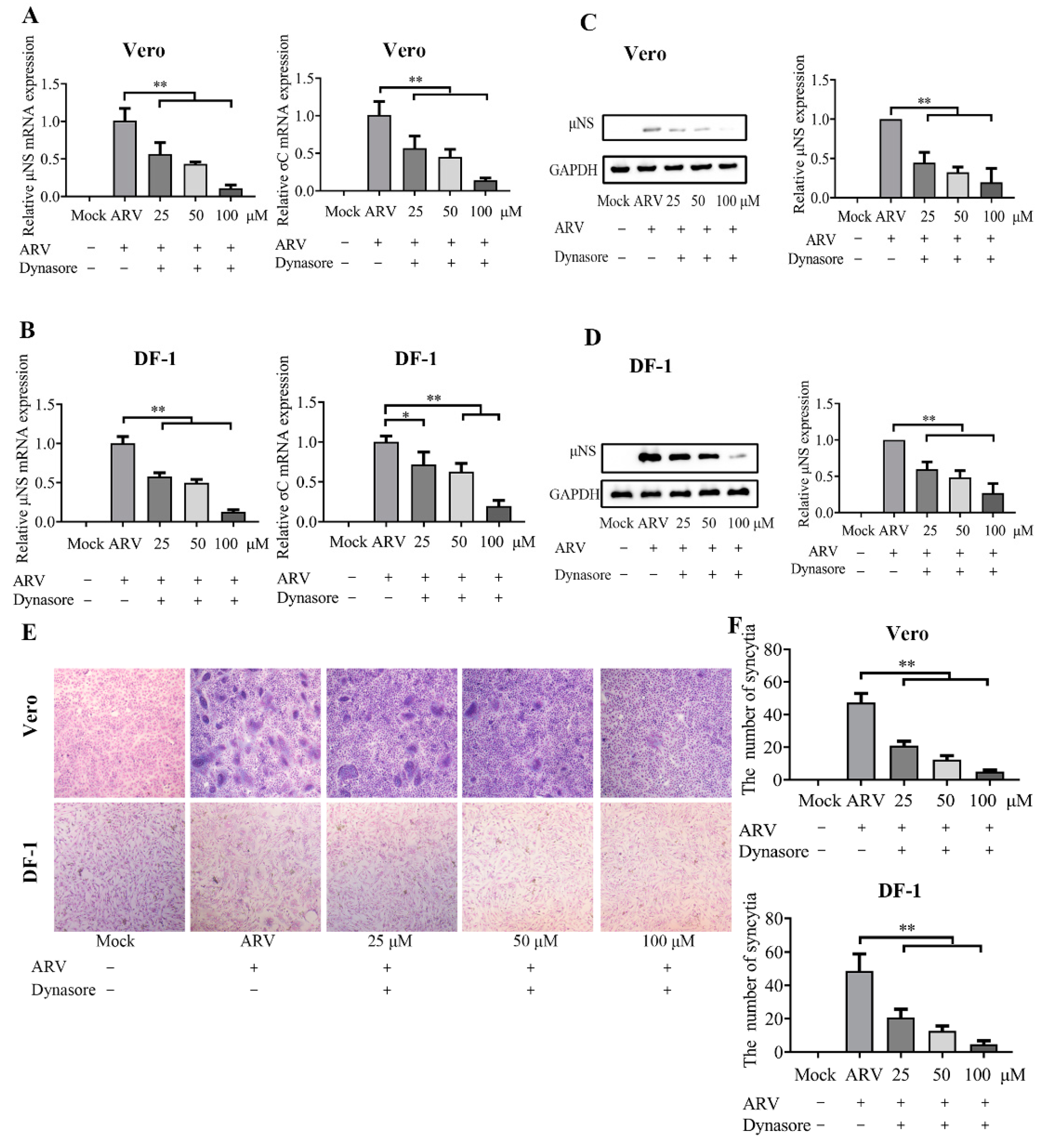
3.4. ARV Are Internalized by Dynamin-Dependent Endocytosis
3.5. The ARV Endocytosis Process Is Clathrin Independent
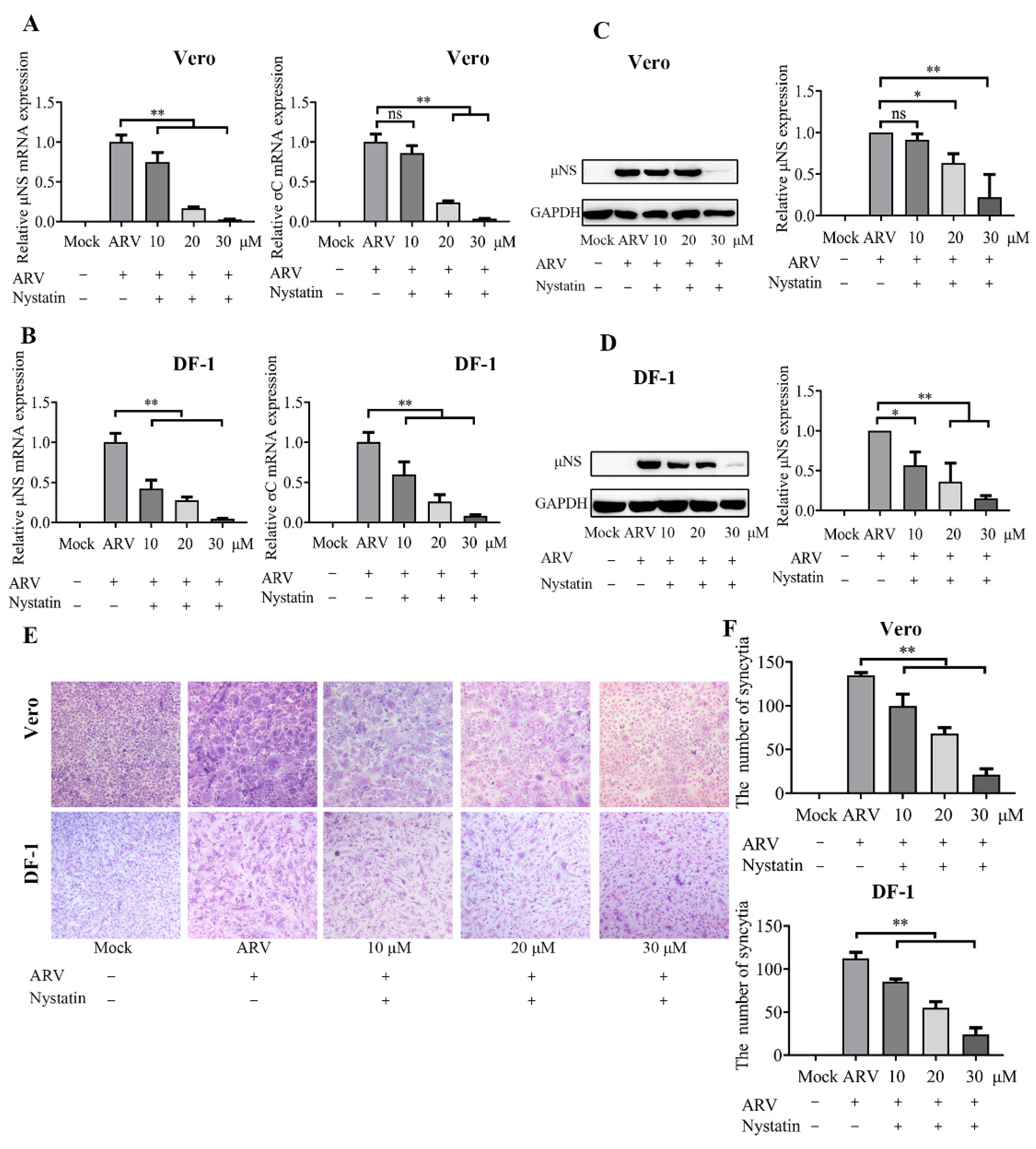
3.6. ARV Entry Involves Caveolae-Mediated Endocytosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, R.C. Avian reovirus infections. Rev. Sci. Tech. 2000, 19, 614–625. [Google Scholar] [CrossRef]
- Neelima, S.; Ram, G.C.; Kataria, J.M.; Goswami, T.K. Avian reovirus induces an inhibitory effect on lymphoproliferation in chickens. Vet. Res. Commun. 2003, 27, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Attoui, H.; Billoir, F.; Biagini, P.; de Micco, P.; de Lamballerie, X. Complete sequence determination and genetic analysis of Banna virus and Kadipiro virus: Proposal for assignment to a new genus (Seadornavirus) within the family Reoviridae. J. Gen. Virol. 2000, 81, 1507–1515. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, J.; Walker, S.B.; O’Hara, D.; Nibert, M.L.; Duncan, R.; Baker, T.S. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 2005, 343, 25–35. [Google Scholar] [CrossRef]
- Benavente, J.; Martinez-Costas, J. Avian reovirus: Structure and biology. Virus Res. 2007, 123, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Getts, D.R.; Chastain, E.M.; Terry, R.L.; Miller, S.D. Virus infection, antiviral immunity, and autoimmunity. Immunol. Rev. 2013, 255, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Cosset, F.L.; Lavillette, D. Cell entry of enveloped viruses. Adv. Genet. 2011, 73, 121–183. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.K.; McMahon, H.T. Snap-shots of clathrin-mediated endocytosis. Trends Biochem. Sci. 2002, 27, 257–263. [Google Scholar] [CrossRef]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef] [PubMed]
- Silvius, J.R. Role of cholesterol in lipid raft formation: Lessons from lipid model systems. Biochim. Biophys. Acta (BBA) Biomembr. 2003, 1610, 174–183. [Google Scholar] [CrossRef]
- Nabi, I.R.; Le, P.U. Caveolae/raft-dependent endocytosis. J. Cell Biol. 2003, 161, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Nichols, B. Caveosomes and endocytosis of lipid rafts. J. Cell Sci. 2003, 116, 4707–4714. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.; Belouzard, S.; Goueslain, L.; Wakita, T.; Dubuisson, J.; Wychowski, C.; Rouille, Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 2006, 80, 6964–6972. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, C. Influenza A virus H5N1 entry into host cells is through clathrin-dependent endocytosis. Sci. China C Life Sci. 2009, 52, 464–469. [Google Scholar] [CrossRef]
- Helenius, A. Virus Entry: Looking Back and Moving Forward. J. Mol. Biol. 2018, 430, 1853–1862. [Google Scholar] [CrossRef]
- Xing, Y.; Wen, Z.; Gao, W.; Lin, Z.; Zhong, J.; Jiu, Y. Multifaceted Functions of Host Cell Caveolae/Caveolin-1 in Virus Infections. Viruses 2020, 12, 487. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Ono, A.; Freed, E.O. Role of Lipid Rafts in Virus Replication. Adv. Virus Res. 2005, 64, 311–358. [Google Scholar] [CrossRef] [PubMed]
- Lorizate, M.; Krausslich, H.G. Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef]
- Huang, W.R.; Wang, Y.C.; Chi, P.I.; Wang, L.; Wang, C.Y.; Lin, C.H.; Liu, H.J. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J. Biol. Chem. 2011, 286, 30780–30794. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yan, P.; Liu, Z.; Cai, D.; Luo, Y.; Wu, X.; Wang, Q.; Huang, Y.; Wu, Y. Muscovy duck reovirus enters susceptible cells via a caveolae-mediated endocytosis-like pathway. Virus Res. 2020, 276, 197806. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Zhang, C.; Hu, M.; Yan, Q.; Zhao, H.; Zhang, X.; Wu, Y. Cholesterol-Rich Lipid Rafts in the Cellular Membrane Play an Essential Role in Avian Reovirus Replication. Front. Microbiol. 2020, 11, 597794. [Google Scholar] [CrossRef]
- Danthi, P.; Guglielmi, K.M.; Kirchner, E.; Mainou, B.; Stehle, T.; Dermody, T.S. From touchdown to transcription: The reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 2010, 343, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.; Isa, P.; Sanchez-San Martin, C.; Perez-Vargas, J.; Espinosa, R.; Arias, C.F.; Lopez, S. Different rotavirus strains enter MA104 cells through different endocytic pathways: The role of clathrin-mediated endocytosis. J. Virol. 2010, 84, 9161–9169. [Google Scholar] [CrossRef]
- Schulz, W.L.; Haj, A.K.; Schiff, L.A. Reovirus uses multiple endocytic pathways for cell entry. J. Virol. 2012, 86, 12665–12675. [Google Scholar] [CrossRef] [PubMed]
- Danthi, P.; Holm, G.H.; Stehle, T.; Dermody, T.S. Reovirus receptors, cell entry, and proapoptotic signaling. Adv. Exp. Med. Biol. 2013, 790, 42–71. [Google Scholar] [CrossRef] [PubMed]
- Le Gall-Recul, G.; Cherbonnel, M.; Arnauld, C.; Blanchard, P.; Jestin, A.; Jestin, V. Molecular characterization and expression of the S3 gene of muscovy duck reovirus strain 89026. J. Gen. Virol. 1999, 80 Pt 1, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tien, P. From endocytosis to membrane fusion: Emerging roles of dynamin in virus entry. Crit. Rev. Microbiol. 2013, 39, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Sieczkarski, S.B.; Whittaker, G.R. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 2003, 4, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, H.; Zhang, J.; Chen, Q.; Fang, Q. Identification of the caveolae/raft-mediated endocytosis as the primary entry pathway for aquareovirus. Virology 2018, 513, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Jiang, K.; Zhang, X.; Zhang, M.; Zhou, Z.; Hu, M.; Yang, R.; Sun, C.; Wu, Y. Avian reovirus triggers autophagy in primary chicken fibroblast cells and Vero cells to promote virus production. Arch. Virol. 2012, 157, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Zhang, C.; Wang, Y.; Guo, M.; Ruan, B.; Wang, X.; Wu, T.; Zhang, X.; Wu, Y. Autophagy induced by avian reovirus enhances viral replication in chickens at the early stage of infection. BMC Vet. Res. 2019, 15, 173. [Google Scholar] [CrossRef] [PubMed]
- McGuinn, K.P.; Mahoney, M.G. Lipid rafts and detergent-resistant membranes in epithelial keratinocytes. Methods Mol. Biol. 2014, 1195, 133–144. [Google Scholar] [CrossRef]
- Read, J.; Clancy, E.K.; Sarker, M.; de Antueno, R.; Langelaan, D.N.; Parmar, H.B.; Shin, K.; Rainey, J.K.; Duncan, R. Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor. PLoS Pathog. 2015, 11, e1004962. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Cheng, J.; Li, C.; Yu, L.; Zhang, X.; Jiang, K.; Wang, Y.; Xu, J.; Wu, Y. Autophagy inhibitors reduce avian-reovirus-mediated apoptosis in cultured cells and in chicken embryos. Arch. Virol. 2015, 160, 1679–1685. [Google Scholar] [CrossRef]
- Grande, A.; Costas, C.; Benavente, J. Subunit composition and conformational stability of the oligomeric form of the avian reovirus cell-attachment protein sigmaC. J. Gen. Virol. 2002, 83, 131–139. [Google Scholar] [CrossRef]
- Shien, J.H.; Yin, H.S.; Lee, L.H. An enzyme-linked immunosorbent assay for the detection of antibody to avian reovirus by using protein sigma B as the coating antigen. Res. Vet. Sci. 2000, 69, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Liemann, S.; Chandran, K.; Baker, T.S.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell 2002, 108, 283–295. [Google Scholar] [CrossRef]
- Snyder, A.J.; Wang, J.C.; Danthi, P. Components of the Reovirus Capsid Differentially Contribute to Stability. J. Virol. 2019, 93, e01894-18. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| σC-F | CGTATCATTCACCCGCGATT |
| σC-R | TGTTCGCTGTACCATCACCT |
| μNS-F | CGTGTGGAAGCGTTAAACCA |
| μNS-R | TCATCACGCTCGTTCAGGTA |
| β-actin-F (chicken) | ATTGTCCACCGCAAATGCTTC |
| β-actin-R (chicken) | AAATAAAGCCATGCCAATCTCGTC |
| β-actin-F (monkey) | GGCCAGGTCATCACCATT |
| β-actin-R (monkey) | ATGTCCACGTCACACTTCATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, Y.; Zuo, W.; Bo, Z.; Zhang, C.; Zhang, X.; Wu, Y. Avian Reovirus σB Interacts with Caveolin-1 in Lipid Rafts during Dynamin-Dependent Caveolae-Mediated Endocytosis. Viruses 2022, 14, 2201. https://doi.org/10.3390/v14102201
Wang Y, Zhang Y, Zuo W, Bo Z, Zhang C, Zhang X, Wu Y. Avian Reovirus σB Interacts with Caveolin-1 in Lipid Rafts during Dynamin-Dependent Caveolae-Mediated Endocytosis. Viruses. 2022; 14(10):2201. https://doi.org/10.3390/v14102201
Chicago/Turabian StyleWang, Yuyang, Yangyang Zhang, Wei Zuo, Zongyi Bo, Chengcheng Zhang, Xiaorong Zhang, and Yantao Wu. 2022. "Avian Reovirus σB Interacts with Caveolin-1 in Lipid Rafts during Dynamin-Dependent Caveolae-Mediated Endocytosis" Viruses 14, no. 10: 2201. https://doi.org/10.3390/v14102201
APA StyleWang, Y., Zhang, Y., Zuo, W., Bo, Z., Zhang, C., Zhang, X., & Wu, Y. (2022). Avian Reovirus σB Interacts with Caveolin-1 in Lipid Rafts during Dynamin-Dependent Caveolae-Mediated Endocytosis. Viruses, 14(10), 2201. https://doi.org/10.3390/v14102201






