Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020
Abstract
:1. Introduction
2. Materials and Methods
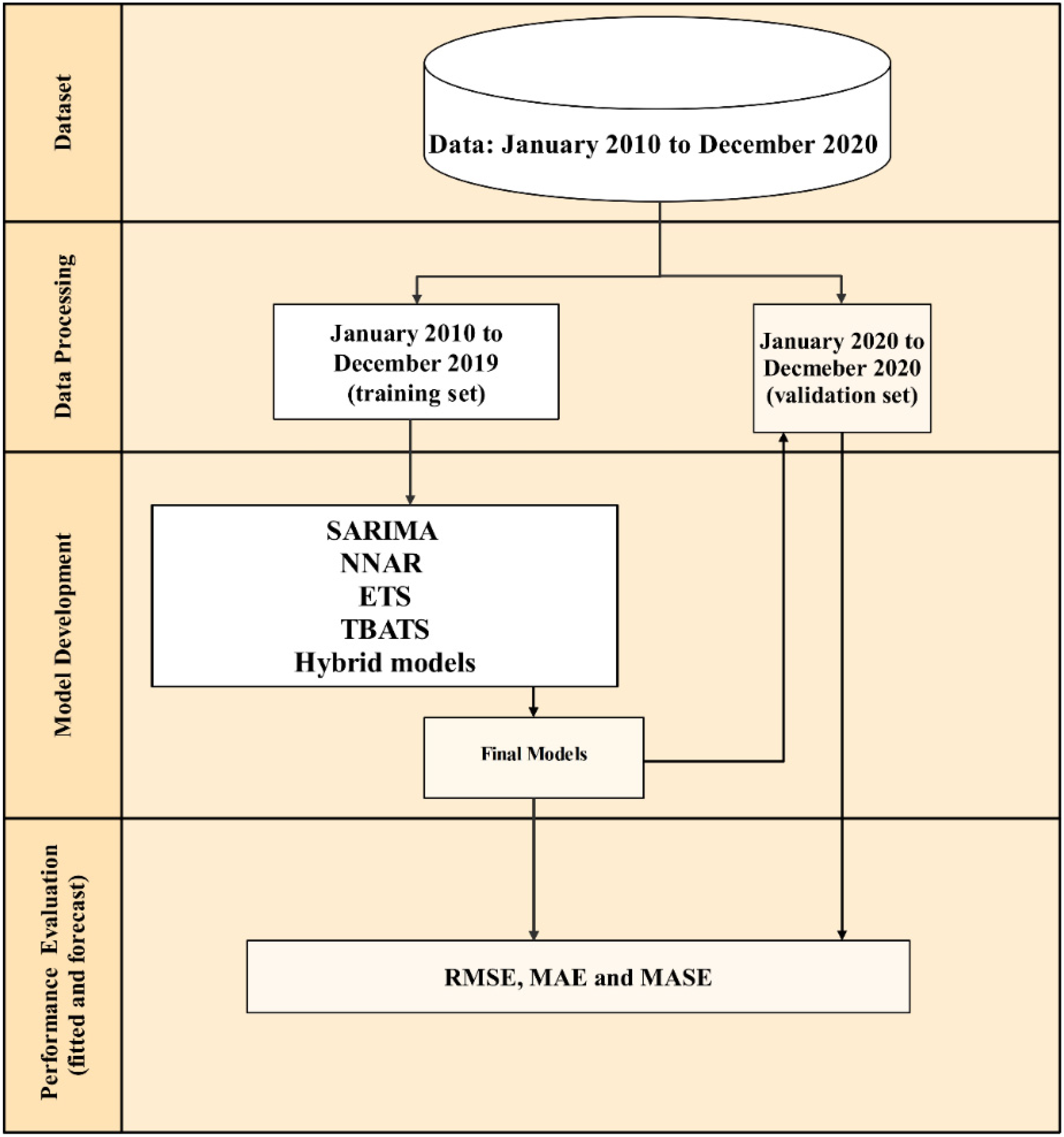
2.1. Outbreak Episode Definition, Data, and Data Analysis Steps
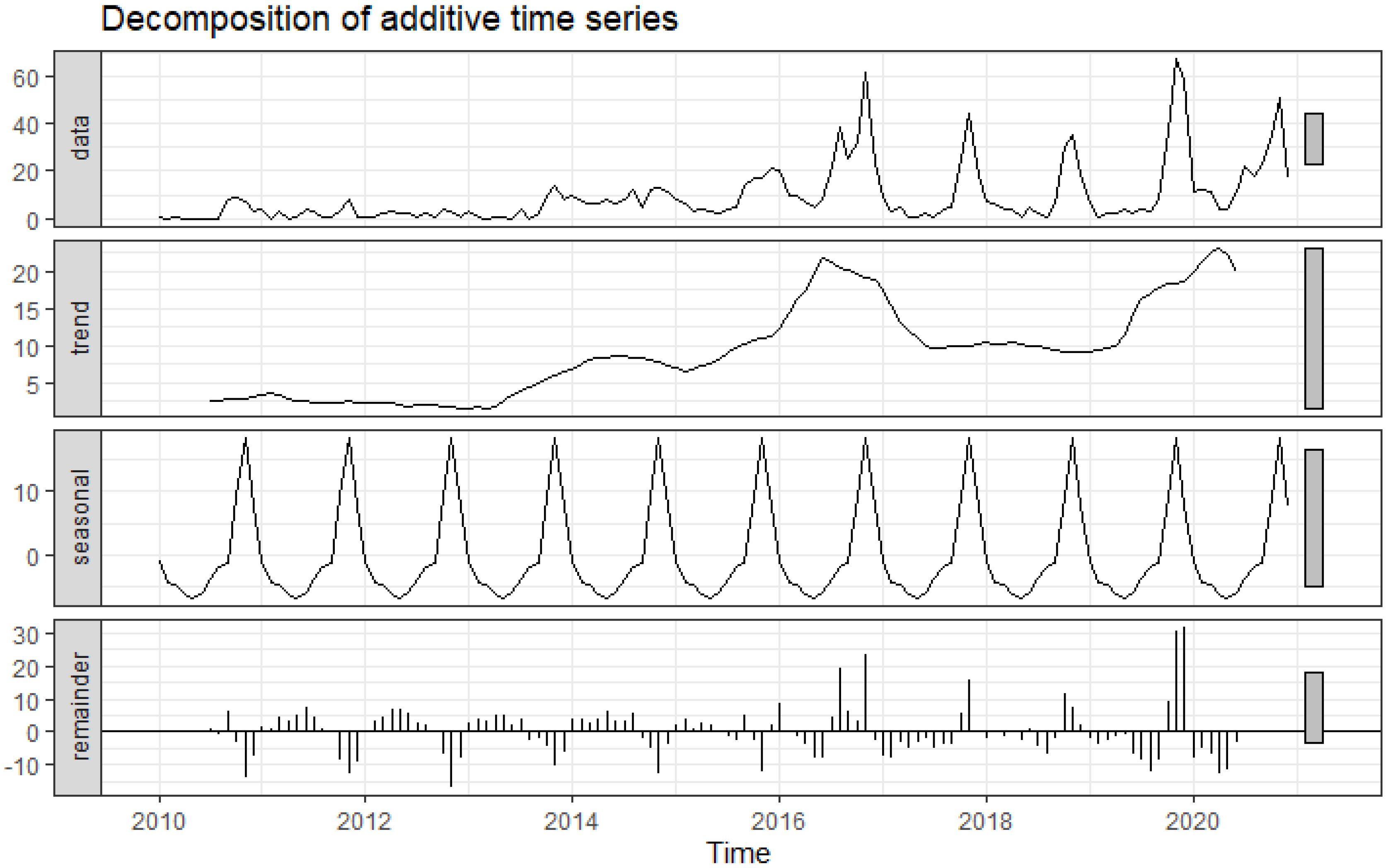
2.2. Decomposition of Time-Series Data
2.3. Model Development
2.3.1. SARIMA Model
2.3.2. NNAR Model
2.3.3. ETS Model
2.3.4. TBATS Model
2.3.5. Hybrid Model
2.4. Forecast and Model Performances
3. Results
3.1. Trends and Seasonality
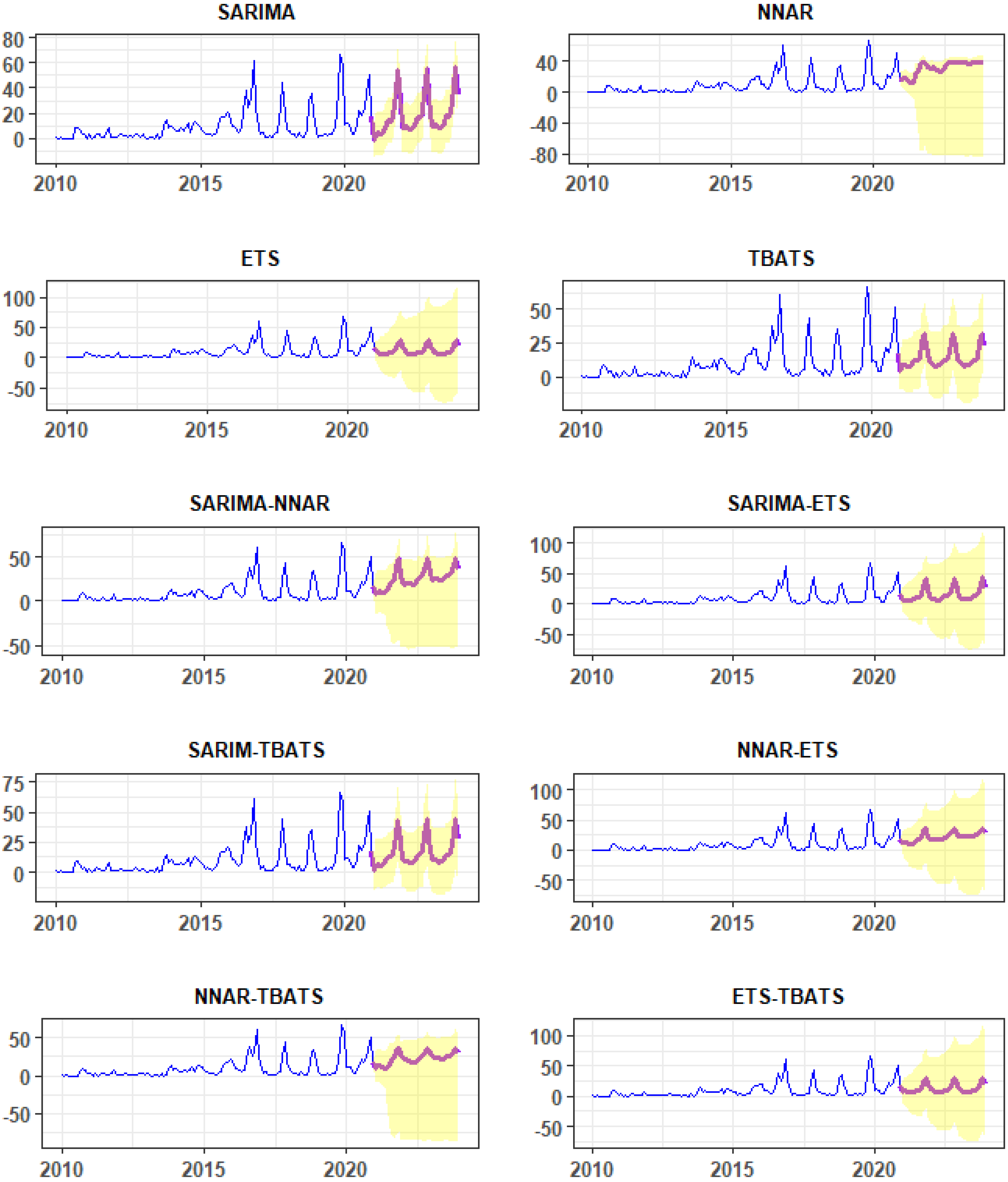
3.2. Fitted Time-Series Models, Model Performances, and Forecasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- OIE. Foot and Mouth Disease. 2022. Available online: https://www.oie.int/en/disease/foot-and-mouth-disease/ (accessed on 23 December 2021).
- Brito, B.; Rodriguez, L.; Hammond, J.; Pinto, J.; Perez, A. Review of the global distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound. Emerg. Dis. 2017, 64, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj, G.; Krishnamohan, A.; Hegde, R.; Kumar, N.; Prabhakaran, K.; Wadhwan, V.M.; Kakker, N.; Lokhande, T.; Sharma, K.; Kanani, A. Foot and Mouth Disease (FMD) incidence in cattle and buffaloes and its associated farm-level economic costs in endemic India. Prev. Vet. Med. 2021, 190, 105318. [Google Scholar]
- Knight-Jones, T.J.; Robinson, L.; Charleston, B.; Rodriguez, L.; Gay, C.; Sumption, K.J.; Vosloo, W. Global Foot-and-Mouth Disease Research Update and Gap Analysis: 1-Overview of Global Status and Research Needs. Transbound. Emerg. Dis. 2016, 63, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Alhaji, N.; Amin, J.; Aliyu, M.; Mohammad, B.; Babalobi, O.; Wungak, Y.; Odetokun, I. Economic impact assessment of foot-and-mouth disease burden and control in pastoral local dairy cattle production systems in northern Nigeria: A cross-sectional survey. Prev. Vet. Med. 2020, 177, 104974. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, A.S.; Boj, E. Application of Neural Network Time Series (NNAR) and ARIMA to Forecast Infection Fatality Rate (IFR) of COVID-19 in Brazil. JOIV Int. J. Inform. Vis. 2021, 5, 8–10. [Google Scholar] [CrossRef]
- Jemberu, W.; Mourits, M.; Sahle, M.; Siraw, B.; Vernooij, J.; Hogeveen, H. Epidemiology of foot and mouth disease in E thiopia: A retrospective analysis of district level outbreaks, 2007–2012. Transbound. Emerg. Dis. 2016, 63, 246–259. [Google Scholar] [CrossRef]
- Gunasekera, U.C.; Sivasothy, A.; Wedasingha, N.; Thayaparan, S.; Rotewewa, B.; Muralithas, M.; Baumann, M.P.O.; Punyapornwithaya, V. Analyzing the Foot and Mouth Disease outbreak as from 2008 to 2014 in cattle and buffaloes in Sri Lanka. Prev. Vet. Med. 2017, 148, 78–88. [Google Scholar] [CrossRef]
- Madin, B. An evaluation of Foot-and-Mouth Disease outbreak reporting in mainland South-East Asia from 2000 to 2010. Prev. Vet. Med. 2011, 102, 230–241. [Google Scholar] [CrossRef]
- Picado, A.; Guitian, F.; Pfeiffer, D. Space–time interaction as an indicator of local spread during the 2001 FMD outbreak in the UK. Prev. Vet. Med. 2007, 79, 3–19. [Google Scholar] [CrossRef]
- Bouma, A.; Elbers, A.; Dekker, A.; De Koeijer, A.; Bartels, C.; Vellema, P.; Van der Wal, P.; Van Rooij, E.; Pluimers, F.; De Jong, M. The foot-and-mouth disease epidemic in The Netherlands in 2001. Prev. Vet. Med. 2003, 57, 155–166. [Google Scholar] [CrossRef]
- Upadhyaya, S.; Mahapatra, M.; Mioulet, V.; Parida, S. Molecular Basis of Antigenic Drift in Serotype O Foot-and-Mouth Disease Viruses (2013–2018) from Southeast Asia. Viruses 2021, 13, 1886. [Google Scholar] [CrossRef] [PubMed]
- Gunasekara, U.; Bertram, M.R.; Dung, D.H.; Hoang, B.H.; Phuong, N.T.; Hung, V.V.; Long, N.V.; Minh, P.Q.; Vu, L.T.; Dong, P.V. Use of Slaughterhouses as Sentinel Points for Genomic Surveillance of Foot-and-Mouth Disease Virus in Southern Vietnam. Viruses 2021, 13, 2203. [Google Scholar] [CrossRef] [PubMed]
- Bo, L.L.; Lwin, K.S.; Ungvanijban, S.; Knowles, N.J.; Wadsworth, J.; King, D.P.; Abila, R.; Qiu, Y. Foot-and-mouth disease outbreaks due to an exotic serotype Asia 1 virus in Myanmar in 2017. Transbound. Emerg. Dis. 2019, 66, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Blacksell, S.D.; Siengsanan-Lamont, J.; Kamolsiripichaiporn, S.; Gleeson, L.J.; Windsor, P.A. A history of FMD research and control programmes in Southeast Asia: Lessons from the past informing the future. Epidemiol. Infect. 2019, 147, e171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arjkumpa, O.; Yano, T.; Prakotcheo, R.; Sansamur, C.; Punyapornwithaya, V. Epidemiology and National Surveillance System for Foot and Mouth Disease in Cattle in Thailand during 2008–2019. Vet Sci. 2020, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Seeyo, K.B.; Nishi, T.; Kawaguchi, R.; Ungvanijban, S.; Udon, R.; Fukai, K.; Yamakawa, M.; Rukkwamsuk, T. Evolution of antigenic and genetic characteristics of foot-and-mouth disease virus serotype A circulating in Thailand, 2007–2019. Virus Res. 2020, 290, 198166. [Google Scholar] [CrossRef]
- Arjkumpa, O.; Picasso-Risso, C.; Perez, A.; Punyapornwithaya, V. Subdistrict-Level Reproductive Number for Foot and Mouth Disease in Cattle in Northern Thailand. Front. Vet. Sci. 2021, 8, 757132. [Google Scholar] [CrossRef]
- Sangrat, W.; Thanapongtharm, W.; Poolkhet, C. Identification of risk areas for foot and mouth disease in Thailand using a geographic information system-based multi-criteria decision analysis. Prev. Vet. Med. 2020, 185, 105183. [Google Scholar] [CrossRef]
- Chanchaidechachai, T.; de Jong, M.C.; Fischer, E.A. Spatial model of foot-and-mouth disease outbreak in an endemic area of Thailand. Prev. Vet. Med. 2021, 195, 105468. [Google Scholar] [CrossRef]
- Arjkumpa, O.; Sansamur, C.; Sutthipankul, P.; Inchaisri, C.; Na Lampang, K.; Charoenpanyanet, A.; Punyapornwithaya, V. Spatiotemporal analyses of foot and mouth disease outbreaks in cattle farms in Chiang Mai and Lamphun, Thailand. BMC Vet. Res. 2020, 16, 170. [Google Scholar] [CrossRef]
- Gleeson, L. A review of the status of foot and mouth disease in South-East Asia and approaches to control and eradication. Rev. Sci. Tech. Off. Int. Epiz. 2002, 21, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.S.; Her, M.; Levine, M.; Moore, G.E. Time series analysis of human and bovine brucellosis in South Korea from 2005 to 2010. Prev. Vet. Med. 2013, 110, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Sato, R.C. Disease management with ARIMA model in time series. Einstein 2013, 11, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, M.; Iglesias, R.; Brookes, V.J. Autoregressive models applied to time-series data in veterinary science. Front. Vet. Sci. 2020, 7, 604. [Google Scholar] [CrossRef]
- Tian, C.; Wang, H.; Luo, X. Time-series modelling and forecasting of hand, foot and mouth disease cases in China from 2008 to 2018. Epidemiol. Infect. 2019, 147, e82. [Google Scholar] [CrossRef] [Green Version]
- Weng, R.; Fu, H.; Zhang, C.; Ye, J.; Hong, F.; Chen, X.; Cai, Y. Time series analysis and forecasting of chlamydia trachomatis incidence using surveillance data from 2008 to 2019 in Shenzhen, China. Epidemiol. Infect. 2020, 148. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Deng, C.; Li, J.; Zhang, Y.; Li, X.; Wu, M. Hybrid methodology for tuberculosis incidence time-series forecasting based on ARIMA and a NAR neural network. Epidemiol. Infect. 2017, 145, 1118–1129. [Google Scholar] [CrossRef] [Green Version]
- Hyndman, R.J.; Athanasopoulos, G. Forecasting: Principles and Practice; OTexts: Melbourne, Australia, 2018. [Google Scholar]
- Perone, G. Comparison of ARIMA, ETS, NNAR, TBATS and hybrid models to forecast the second wave of COVID-19 hospitalizations in Italy. Eur. J. Health Econ. 2021, 4, 1–24. [Google Scholar] [CrossRef]
- Yu, C.; Xu, C.; Li, Y.; Yao, S.; Bai, Y.; Li, J.; Wang, L.; Wu, W.; Wang, Y. Time series analysis and forecasting of the hand-foot-mouth disease morbidity in China using an advanced exponential smoothing state space TBATS model. Infect. Drug Resist. 2021, 14, 2809. [Google Scholar] [CrossRef]
- Azeez, A.; Obaromi, D.; Odeyemi, A.; Ndege, J.; Muntabayi, R. Seasonality and trend forecasting of tuberculosis prevalence data in Eastern Cape, South Africa, using a hybrid model. Int. J. Environ. Res. Public Health 2016, 13, 757. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, C.; Li, Y.; Wu, W.; Gui, L.; Ren, J.; Yao, S. An advanced data-driven hybrid model of SARIMA-NNNAR for tuberculosis incidence time series forecasting in Qinghai Province, China. Infect. Drug Resist. 2020, 13, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Xia, J.; Yu, L.; Wang, Y.; Shi, Y.; Cai, S.; Nie, S. Using a hybrid model to forecast the prevalence of schistosomiasis in humans. Int. J. Environ. Res. Public Health 2016, 13, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Han, Y.; So, S.-S. Overview of artificial neural networks. In Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2008; pp. 14–22. [Google Scholar]
- Abotaleb, M.; Makarovskikh, T.; Yonar, A.; Badr, A.; Mishra, P.; Williams, A.; Yonar, H. State of the art in wind speed in England using BATS, TBATS, Holt’s Linear and ARIMA model: Wind speed in England using BATS, TBATS, Holt’s Linear and ARIMA model. Mausam 2022, 73, 129–138. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, L.; Wang, Y.; Lu, Z.; Tian, L.; Tan, L.; Shi, Y.; Nie, S.; Liu, L. A hybrid model for predicting the prevalence of schistosomiasis in humans of Qianjiang City, China. PLoS ONE 2014, 9, e104875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, E.A.; Carpenter, T.E.; Farver, T.B. The application of time series analysis to determine the pattern of foot-and-mouth disease in cattle in Paraguay. Prev. Vet. Med. 1982, 1, 27–36. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Donnelly, C.A.; Anderson, R.M. The foot-and-mouth epidemic in Great Britain: Pattern of spread and impact of interventions. Science 2001, 292, 1155–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souley Kouato, B.; De Clercq, K.; Abatih, E.; Dal Pozzo, F.; King, D.P.; Thys, E.; Marichatou, H.; Saegerman, C. Review of epidemiological risk models for foot-and-mouth disease: Implications for prevention strategies with a focus on Africa. PLoS ONE 2018, 13, e0208296. [Google Scholar] [CrossRef]
- Gallego, M.; Perez, A.; Thurmond, M. Temporal and spatial distributions of foot-and-mouth disease under three different strategies of control and eradication in Colombia (1982–2003). Vet. Res. Commun. 2007, 31, 819–834. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Azad, A.S.; Sokkalingam, R.; Daud, H.; Adhikary, S.K.; Khurshid, H.; Mazlan, S.N.A.; Rabbani, M.B.A. Water Level Prediction through Hybrid SARIMA and ANN Models Based on Time Series Analysis: Red Hills Reservoir Case Study. Sustainability 2022, 14, 1843. [Google Scholar] [CrossRef]
- Yusof, F.; Kane, I. Modelling monthly rainfall time series using ETS state space and SARIMA models. Int. J. Curr. Res. 2012, 4, 195–200. [Google Scholar]
- Sun, Z. Comparison of Trend Forecast Using ARIMA and ETS Models for S&P500 Close Price. In Proceedings of the 2020 the 4th International Conference on E-Business and Internet, Singapore, 9–11 October 2020; pp. 57–60. [Google Scholar]
- De Livera, A.M.; Hyndman, R.J.; Snyder, R.D. Forecasting time series with complex seasonal patterns using exponential smoothing. J. Am. Stat. Assoc. 2011, 106, 1513–1527. [Google Scholar] [CrossRef] [Green Version]
- Shaub, D. Fast and accurate yearly time series forecasting with forecast combinations. Int. J. Forecast. 2020, 36, 116–120. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Yano, T.; Premashthira, S.; Dejyong, T.; Tangtrongsup, S.; Salman, M.D. The Effectiveness of a Foot and Mouth Disease Outbreak Control Programme in Thailand 2008–2015: Case Studies and Lessons Learned. Vet. Sci. 2018, 5, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuel, A.; Ouldridge, E.; Arrowsmith, A.; Kitching, R.; Knowles, N. Antigenic analysis of serotype O foot-and-mouth disease virus isolates from the Middle East, 1981 to 1988. Vaccine 1990, 8, 390–396. [Google Scholar] [CrossRef]
- Cleland, P.C.; Baldock, F.C.; Chamnanpood, P. Village level risk factors for foot-and-mouth disease in northern Thailand. Prev. Vet. Med. 1996, 26, 253–261. [Google Scholar] [CrossRef]
- Yano, T. Study of foot and mouth disease outbreak in Chiang Mai and Lamphoon. ChiangMai V. J. 2011, 11, 277–287. [Google Scholar]
- Wang, Y.; Xu, C.; Wu, W.; Ren, J.; Li, Y.; Gui, L.; Yao, S. Time series analysis of temporal trends in hemorrhagic fever with renal syndrome morbidity rate in China from 2005 to 2019. Sci. Rep. 2020, 10, 9609. [Google Scholar] [CrossRef]
- Xiao, Y.; Xiao, J.; Wang, S. A hybrid model for time series forecasting. Hum. Syst. Manag. 2012, 31, 133–143. [Google Scholar] [CrossRef]
- Zhang, G.P. Time series forecasting using a hybrid ARIMA and neural network model. Neurocomputing 2003, 50, 159–175. [Google Scholar] [CrossRef]
- Sansamur, C.; Wiratsudakul, A.; Charoenpanyanet, A.; Punyapornwithaya, V. Estimating the number of farms experienced foot and mouth disease outbreaks using capture-recapture methods. Trop. Anim. Health Prod. 2020, 53, 12. [Google Scholar] [CrossRef] [PubMed]
- Gunarathne, A.; Kubota, S.; Kumarawadu, P.; Karunagoda, K.; Kon, H. Is hiding foot and mouth disease sensitive behavior for farmers? A survey study in Sri Lanka. Asian-Australas. J. Anim. Sci. 2016, 29, 280. [Google Scholar] [CrossRef] [PubMed]
- OIE. SEACFMD Roadmap. 2016. Available online: https://rr-asia.oie.int/en/projects/foot-and-mouth-disease-fmd/seacfmd-campaign/seacfmd-roadmap/ (accessed on 23 December 2021).
- Al Khatib, A.M.G.; Yonar, H.; Abotaleb, M.; Mishra, P.; Yonar, A.; Karakaya, K.; Badr, A.; Dhaka, V. Modeling and forecasting of egg production in India using time series models. Eurasian J. Vet. Sci. 2021, 37, 265–273. [Google Scholar] [CrossRef]
- Mishra, P.; Al Khatib, A.M.G.; Sardar, I.; Mohammed, J.; Ray, M.; Manish, K.; Rawat, D.; Pandey, S.A.; Dubey, A.; Feys, J.; et al. Modelling and forecasting of COVID-19 in India. J. Infect. Dis. Epidemiol. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Mishra, P.; Alakkari, K.M.; Lama, A.; Ray, S.; Singh, M.; Shoko, C.; Abotaleb, M.; Al khatib, A.M.G.; Karakaya, K. Modeling and Forecasting of Sugarcane Production in South Asian Countries. Curr. Appl. Sci. Technol. 2023, 23, 1–15. [Google Scholar] [CrossRef]
- Raghav, Y.S.; Mishra, P.; Alakkari, K.M.; Singh, M.; Al Khatib, A.M.G.; Balloo, R. Modelling and forecasting of pulses production in south asian countries and its role in nutritional security. Legume Res.-Int. J. 2022, 45, 454–461. [Google Scholar] [CrossRef]
- Rahman, U.H.; Ray, S.; Al Khatib, A.M.G.; Lal, P.; Mishra, P.; Fatih, C.; Williams, A.J.; Karakaya, K.; Shrivastri, S.; Alakkari, K. State of Art of SARIMA Model in Second Wave on COVID-19 in India. Int. J. Agric. Stat. Sci. 2022. Available online: https://connectjournals.com/03899.2022.18.141 (accessed on 28 March 2022).
- Schreiber, M.A.; Cavanaugh, C.H. The Application of Machine Learning Methods to Time Series Forecasting. 2020. Available online: https://research-api.cbs.dk/ws/portalfiles/portal/66025537/Cavanaugh_and_Schreiber.pdf (accessed on 28 March 2022).
- Yonar, H.; Yonar, A.; Mishra, P.; Abotaleb, M.; Al Khatib, A.M.G.; Makarovskikh, T.; Cam, M. Modeling and forecasting of milk production in different breeds in Turkey. Indian J. Anim. Sci. 2022, 92, 105–111. [Google Scholar]
- Abotaleb, M.; Ray, S.; Mishra, P.; Karakaya, K.; Shoko, C.; Al khatib, A.M.G.; Balloo, R. Modelling and forecasting of rice production in south Asian countries. AMA Agric. Mech. Asia Afr. Lat. Am. 2021, 51, 1611–1627. [Google Scholar]
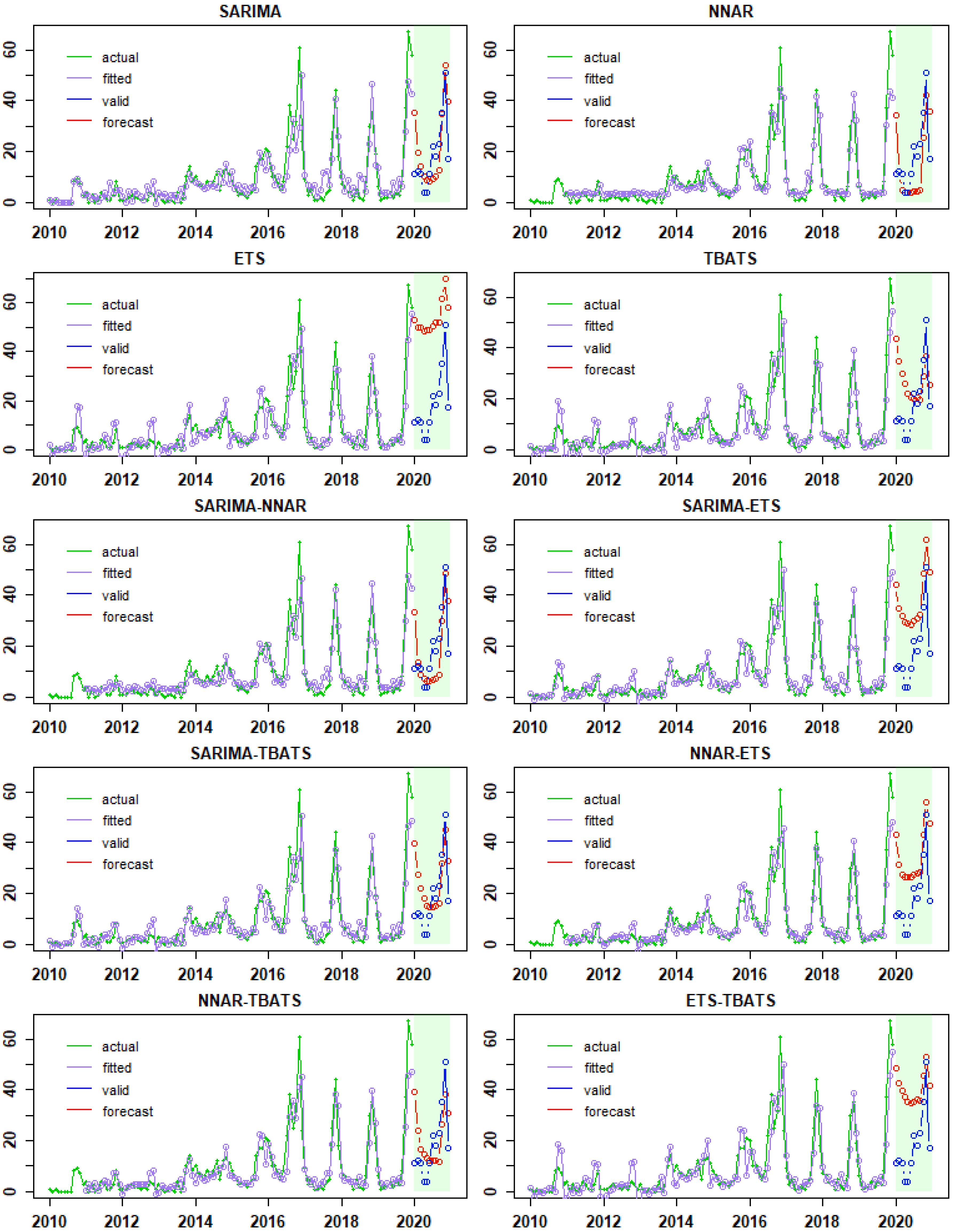
| Model 1 | Training Data (Data: 2010–2019) | Validation Data (Data: 2020) | ||||
|---|---|---|---|---|---|---|
| RMSE 2 | MAE | MASE | RMSE | MAE | MASE | |
| SARIMA | 6.21 | 3.81 | 0.64 | 11.47 | 8.75 | 1.47 |
| NNAR | 5.21 | 3.39 | 0.56 | 12.61 | 10.46 | 1.76 |
| ETS | 6.59 | 4.44 | 0.75 | 36.11 | 35.26 | 5.94 |
| TBATS | 6.38 | 4.26 | 0.89 | 16.24 | 13.38 | 2.25 |
| SARIMA-NNAR | 5.59 | 3.63 | 0.61 | 11.43 | 8.79 | 1.48 |
| SARIMA-ETS | 5.94 | 3.82 | 0.64 | 20.89 | 19.2 | 3.23 |
| SARIMA-TBATS | 5.91 | 3.75 | 0.63 | 12.56 | 10.4 | 1.75 |
| NNAR-ETS | 5.38 | 3.52 | 0.60 | 17.49 | 15.17 | 2.60 |
| NNAR-TBATS | 5.40 | 3.53 | 0.60 | 11.41 | 10.02 | 1.69 |
| ETS-TBATS | 6.42 | 4.29 | 0.72 | 24.44 | 22.14 | 3.73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Punyapornwithaya, V.; Mishra, P.; Sansamur, C.; Pfeiffer, D.; Arjkumpa, O.; Prakotcheo, R.; Damrongwatanapokin, T.; Jampachaisri, K. Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020. Viruses 2022, 14, 1367. https://doi.org/10.3390/v14071367
Punyapornwithaya V, Mishra P, Sansamur C, Pfeiffer D, Arjkumpa O, Prakotcheo R, Damrongwatanapokin T, Jampachaisri K. Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020. Viruses. 2022; 14(7):1367. https://doi.org/10.3390/v14071367
Chicago/Turabian StylePunyapornwithaya, Veerasak, Pradeep Mishra, Chalutwan Sansamur, Dirk Pfeiffer, Orapun Arjkumpa, Rotchana Prakotcheo, Thanis Damrongwatanapokin, and Katechan Jampachaisri. 2022. "Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020" Viruses 14, no. 7: 1367. https://doi.org/10.3390/v14071367
APA StylePunyapornwithaya, V., Mishra, P., Sansamur, C., Pfeiffer, D., Arjkumpa, O., Prakotcheo, R., Damrongwatanapokin, T., & Jampachaisri, K. (2022). Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020. Viruses, 14(7), 1367. https://doi.org/10.3390/v14071367









