Treatment Response and Drug Resistance Profiling of Genotype 6 of Hepatitis C Virus in HCV/HIV Co-Infected Patients: A Pilot Study from INDIA
Abstract
1. Introduction
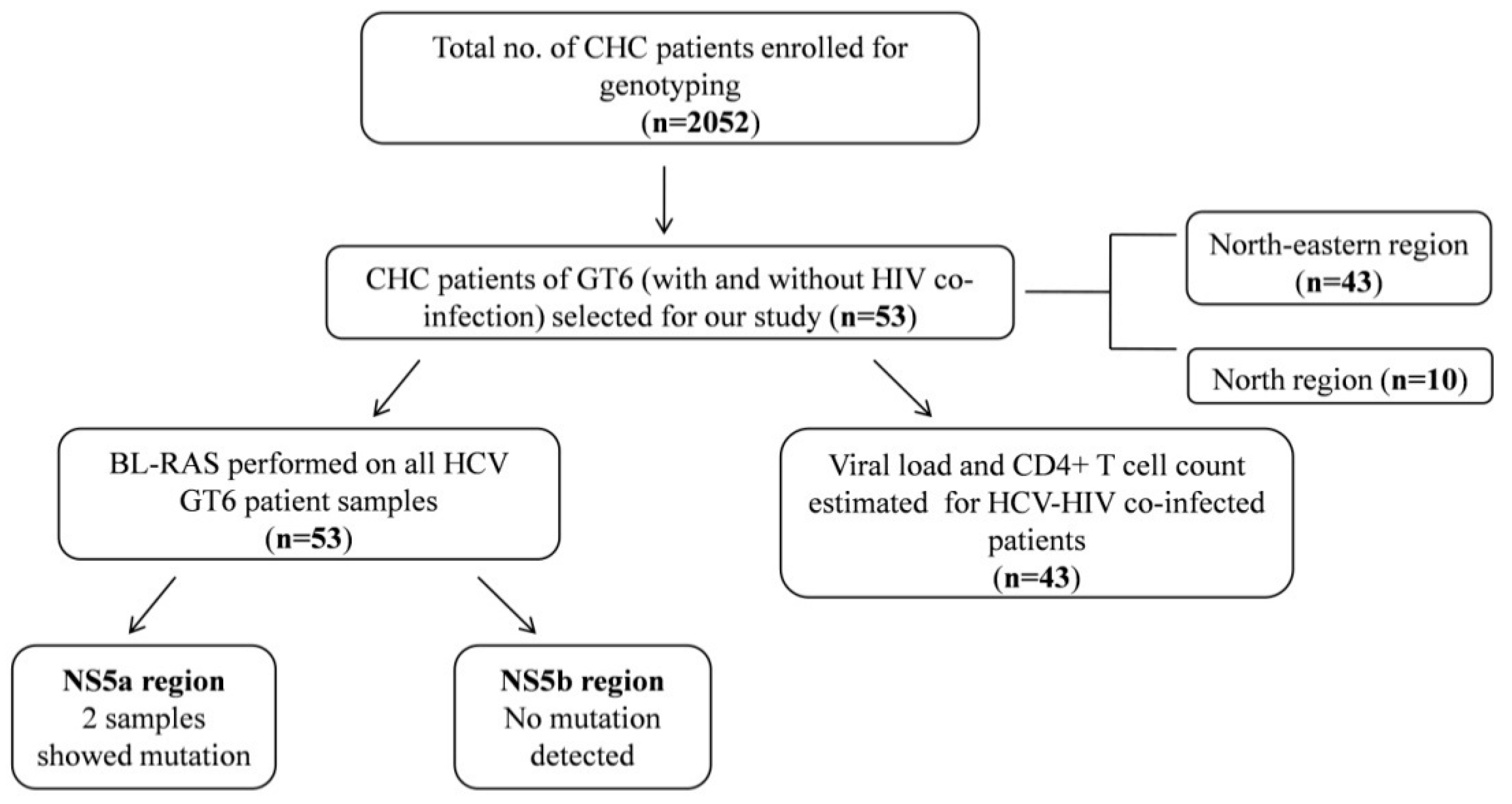
2. Materials and Methods
3. Results
3.1. Distribution of HCV GT6 and Its Subtypes
Phylogenetic Analysis of HCV GT6 Subtypes
3.2. Correlation of HCV GT6 Subtypes with Viral Load and CD4+ T Cell Count
3.3. Occurrence of BL-RAS and Its Association with the Treatment Response
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petruzziello, A.; Marigliano, S.; Loquercio, G.; Cozzolino, A.; Cacciapuoti, C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016, 22, 7824–7840. [Google Scholar] [CrossRef] [PubMed]
- López-Huertas, M.R.; Palladino, C.; Garrido-Arquero, M.; Esteban-Cartelle, B.; Sánchez-Carrillo, M.; Martínez-Román, P.; Martín-Carbonero, L.; Ryan, P.; Domínguez-Domínguez, L.; Santos, I.L.; et al. Multidisciplinary Group of viral coinfection HIV/Hepatitis (COVIHEP). HCV-coinfection is related to an increased HIV-1 reservoir size in cART-treated HIV patients: A cross-sectional study. Sci. Rep. 2019, 9, 5606. [Google Scholar] [CrossRef] [PubMed]
- Leyna, G.H.; Makyao, N.; Mwijage, A.; Ramadhan, A.; Likindikoki, S.; Mizinduko, M.; Leshabari, M.T.; Moen, K.; Mmbaga, E.J. HIV/HCV co-infection and associated risk factors among injecting drug users in Dar es Salaam, Tanzania: Potential for HCV elimination. Harm Reduct. J. 2019, 16, 68. [Google Scholar] [CrossRef]
- Ngwaga, T.; Kong, L.; Lin, D.; Schoborg, C.; Taylor, L.E.; Mayer, K.H.; Klein, R.S.; Celentano, D.D.; Sobel, J.D.; Jamieson, D.J.; et al. Diversity of the hepatitis C virus NS5B gene during HIV co-infection. PLoS ONE 2020, 15, e0237162. [Google Scholar] [CrossRef] [PubMed]
- Navaneethan, U.; Kemmer, N.; Neff, G.W. Predicting the probable outcome of treatment in HCV patients. Therap. Adv. Gastroenterol. 2009, 2, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Panyala, B.R.; Mukherjee, R.M.; Devarakonda, H.; Tadivaka, S.; Padaki, N.R.; Sharma, M.; Duvvuru, N.R. Genotype distribution in relation to viral load in a large cohort of Indian patients with chronic hepatitis C virus infection: A retrospective analysis. Indian J. Gastroenterol. 2019, 38, 110–116. [Google Scholar] [CrossRef]
- Christdas, J.; Sivakumar, J.; David, J.; Daniel, H.D.; Raghuraman, S.; Abraham, P. Genotypes of hepatitis C virus in the Indian sub-continent: A decade-long experience from a tertiary care hospital in South India. Indian J. Med. Microbiol. 2013, 31, 349–353. [Google Scholar] [CrossRef]
- Medhi, S.; Goswami, B.; Das, A.K.; Singh, T.B.; Husain, S.A.; Sehgal, A.; Kar, P. New insights into hepatitis C virus infection in the tribal-dominant part of Northeast India. Arch. Virol. 2012, 157, 2083–2093. [Google Scholar] [CrossRef]
- Mettikanont, P.; Bunchorntavakul, C.; Reddy, K.R. Systematic review: Epidemiology and response to direct-acting antiviral therapy in genotype 6 chronic hepatitis C virus infection. Aliment. Pharmacol. Ther. 2019, 49, 492–505. [Google Scholar] [CrossRef]
- Jia, Y.; Yue, W.; Gao, Q.; Tao, R.; Zhang, Y.; Fu, X.; Liu, Y.; Liu, L.; Feng, Y.; Xia, X. Characterization of a Novel Hepatitis C Subtype, 6xj, and Its Consequences for Direct-Acting Antiviral Treatment in Yunnan, China. Microbiol. Spectr. 2021, 9, e0029721. [Google Scholar] [CrossRef]
- Thong, V.D.; Akkarathamrongsin, S.; Poovorawan, K.; Tangkijvanich, P.; Poovorawan, Y. Hepatitis C virus genotype 6: Virology, epidemiology, genetic variation and clinical implication. World J. Gastroenterol. 2014, 20, 2927–2940. [Google Scholar] [CrossRef] [PubMed]
- Gupta, E.; Agarwal, R.; Rastogi, A.; Rani, N.; Jindal, A. Naturally Occurring Resistance Associated Substitutions in Non-Cirrhotic, Treatment Naive HCV-HIV Co-Infected Patients Does Not Affect the Treatment Response for Anti-HCV Antiviral Therapy. Infect. Drug Resist. 2021, 14, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for the Care and Treatment of Persons Diagnosed with Chronic Hepatitis C Virus Infection; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Sistayanarain, A.; Chaiwong, S. Molecular characterization of hepatitis C virus genotype 6 subtypes in Thai blood donors. J. Microbiol. Immunol. Infect. 2017, 50, 26–31. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bukowska-Ośko, I.; Pawełczyk, A.; Perlejewski, K.; Kubisa, N.; Caraballo Cortés, K.; Rosińska, M.; Płoski, R.; Fic, M.; Kaźmierczak, J.; Popiel, M.; et al. Genetic Variability of Hepatitis C Virus (HCV) 5′ Untranslated Region in HIV/HCV Coinfected Patients Treated with Pegylated Interferon and Ribavirin. PLoS ONE 2015, 10, e0125604. [Google Scholar] [CrossRef]
- Barman, B.; Bora, K.; Lynrah, K.G.; Lyngdoh, W.V.; Jamil, M. Hepatitis C virus and its genotypes in chronic liver disease patients from Meghalaya, Northeast India. Indian J. Med. Microbiol. 2018, 36, 376–380. [Google Scholar] [CrossRef]
- Lwin, A.A.; Shinji, T.; Khin, M.; Win, N.; Obika, M.; Okada, S.; Koide, N. Hepatitis C virus genotype distribution in Myanmar: Predominance of genotype 6 and existence of new genotype 6 subtype. Hepatol. Res. 2007, 37, 337–345. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, C.; Yin, X.; Guo, X.; Wu, S.; Hou, J. Hepatitis C virus genotypes and subtypes circulating in Mainland China. Emerg. Microbes Infect. 2017, 6, e95. [Google Scholar] [CrossRef]
- Akkarathamrongsin, S.; Praianantathavorn, K.; Hacharoen, N.; Theamboonlers, A.; Tangkijvanich, P.; Tanaka, Y.; Mizokami, M.; Poovorawan, Y. Geographic distribution of hepatitis C virus genotype 6 subtypes in Thailand. J. Med. Virol. 2010, 82, 257–262. [Google Scholar] [CrossRef]
- Kermode, M.; Nuken, A.; Medhi, G.K.; Akoijam, B.S.; Sharma, H.U.; Mahanta, J. High burden of hepatitis C & HIV co-infection among people who inject drugs in Manipur, Northeast India. Indian J. Med. Res. 2016, 143, 348–356. [Google Scholar]
- Chelleng, P.K.; Borkakoty, B.J.; Chetia, M.; Das, H.K.; Mahanta, J. Risk of hepatitis C infection among injection drug users in Mizoram, India. Indian J. Med. Res. 2008, 128, 640–646. [Google Scholar]
- Malandris, K.; Kalopitas, G.; Theocharidou, E.; Germanidis, G. The Role of RASs/RVs in the Current Management of HCV. Viruses 2021, 13, 2096. [Google Scholar] [CrossRef] [PubMed]
- Zarębska-Michaluk, D.; Jaroszewicz, J.; Parfieniuk-Kowerda, A.; Pawłowska, M.; Janczewska, E.; Berak, H.; Janocha-Litwin, J.; Klapaczyński, J.; Tomasiewicz, K.; Piekarska, A.; et al. Pangenotypic and Genotype-Specific Antivirals in the Treatment of HCV Genotype 4 Infected Patients with HCV Monoinfection and HIV/HCV Coinfection. J. Clin. Med. 2022, 11, 389. [Google Scholar] [CrossRef] [PubMed]
- Flower, B.; McCabe, L.; Le Ngoc, C.; Le Manh, H.; Le Thanh, P.; Dang Trong, T.; Vo Thi, T.; Vu Thi Kim, H.; Nguyen Tat, T.; Phan Thi Hong, D.; et al. High Cure Rates for Hepatitis C Virus Genotype 6 in Advanced Liver Fibrosis with 12 Weeks Sofosbuvir and Daclatasvir: The Vietnam SEARCH Study. Open Forum Infect. Dis. 2021, 8, ofab267. [Google Scholar] [CrossRef] [PubMed]


| Variables | Count | p-Value | Significance |
|---|---|---|---|
| Demographics | |||
| Age | 44 ± 12.40 | ||
| Gender | Male 37/53 (69.8%) | ||
| Female 16/53 (30.2%) | |||
| Patient Characteristics | |||
| Total subjects | 53/2052 (2.5%) | ||
| HCV GT6 co-infection with HIV | 43/53 (81.1%) | ||
| HCV GT6 infection alone | 10/53 (18.9%) | ||
| HCV GT6 subtypes distribution | |||
| 6a, 6f, 6i, 6w | 1/53 (1.9%) | ||
| 6b, 6u, 6v | 2/53 (3.8%) | ||
| 6n | 21/53 (39.6%) | ||
| 6xa | 22/53 (41.5%) | ||
| Gender | |||
| Viral Load | 0.263 | NS | |
| Subtype | 0.077 | NS | |
| Treatment Outcome | 0.123 | NS | |
| HCV GT6 subtypes | |||
| Viral Load | 0.006 | S | |
| CD4+ T Cell Count | 0.036 | S | |
| Treatment Outcome | 0.755 | NS | |
| Treatment Outcome | |||
| Viral Load | 0.171 | NS | |
| CD4+ T Cell Count | 0.652 | NS | |
| Choice of Therapy | 0.066 | NS | |
| Sustained Virological Response (SVR) | |||
| Viral Load | 0.015 | S | |
| CD4+ T Cell Count | 0.197 | NS | |
| Choice of Therapy | 0.182 | NS | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, E.; Samal, J.; Pandey, A.; Singh, G.; Gupta, H.A.S.; Agarwal, R.; Sharma, M.K. Treatment Response and Drug Resistance Profiling of Genotype 6 of Hepatitis C Virus in HCV/HIV Co-Infected Patients: A Pilot Study from INDIA. Viruses 2022, 14, 944. https://doi.org/10.3390/v14050944
Gupta E, Samal J, Pandey A, Singh G, Gupta HAS, Agarwal R, Sharma MK. Treatment Response and Drug Resistance Profiling of Genotype 6 of Hepatitis C Virus in HCV/HIV Co-Infected Patients: A Pilot Study from INDIA. Viruses. 2022; 14(5):944. https://doi.org/10.3390/v14050944
Chicago/Turabian StyleGupta, Ekta, Jasmine Samal, Amit Pandey, Gaurav Singh, Hajra A. S. Gupta, Reshu Agarwal, and Manoj Kumar Sharma. 2022. "Treatment Response and Drug Resistance Profiling of Genotype 6 of Hepatitis C Virus in HCV/HIV Co-Infected Patients: A Pilot Study from INDIA" Viruses 14, no. 5: 944. https://doi.org/10.3390/v14050944
APA StyleGupta, E., Samal, J., Pandey, A., Singh, G., Gupta, H. A. S., Agarwal, R., & Sharma, M. K. (2022). Treatment Response and Drug Resistance Profiling of Genotype 6 of Hepatitis C Virus in HCV/HIV Co-Infected Patients: A Pilot Study from INDIA. Viruses, 14(5), 944. https://doi.org/10.3390/v14050944





