Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Analyzed during the Routine Diagnostic Activity in Northeast Italy
2.2. Detection and Characterization of CPV-2 in Dog Tissues
2.2.1. Sample Preparation and DNA Extraction
2.2.2. Real Time PCR
2.3. VP2 Sanger Sequencing
2.4. Phylogenetic Analysis of the VP2 Gene
2.5. Identification of Unique Nucleotide and Amino Acid Mutations in Recent Italian CPV-2 Strains
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pollock, R.V.H.; Coyne, M.J. Canine Parvovirus. Vet. Clin. 1993, 23, 555–568. [Google Scholar] [CrossRef]
- Kumar, S.N.M. Canine Parvovirus: Current Perspective. Indian J. Virol. 2010, 21, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Decaro, N.; Buonavoglia, C. Canine Parvovirus-A Review of Epidemiological and Diagnostic Aspects, with Emphasis on Type 2c. Vet. Microbiol. 2012, 155, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Day, M.J.; Horzinek, M.C.; Schultz, R.D.; Squires, R.A. Guidelines for the Vaccination of Dogs and Cats. J. Small Anim. Pract. 2016, 57, E1–E45. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Blanco, B.; Catala-López, F. Are Licensed Canine Parvovirus (CPV2 and CPV2b) Vaccines Able to Elicit Protection against CPV2c Subtype in Puppies?: A Systematic Review of Controlled Clinical Trials. Vet. Microbiol. 2015, 180, 1–9. [Google Scholar] [CrossRef]
- Reed, A.P.; Jones, E.V.; Miller, T.J. Nucleotide Sequence and Genome Organization of Canine Parvovirus. J. Virol. 1988, 62, 266–276. [Google Scholar] [CrossRef] [Green Version]
- Khatri, R.; Poonam; Mohan, H.; Minakshi; Pundir, C.S. Epidemiology, Pathogenesis, Diagnosis and Treatment of Canine Parvovirus Disease in Dogs: A Mini Review. J. Vet. Sci. Med. Diagn. 2017, 16, 3. [Google Scholar] [CrossRef]
- Tsao, J.; Chapman, M.S.; Agbandje, M.; Keller, W.; Smith, K.; Luo, M.; Smith, T.J.; Rossmann, M.G.; Compans, R.W.; Colin, R.; et al. The Structure of Canine Parvovirus and Its Functional Implications. Science 1991, 251, 1456–1464. [Google Scholar] [CrossRef] [Green Version]
- Mira, F.; Canuti, M.; Purpari, G.; Cannella, V.; Di Bella, S.; Occhiogrosso, L.; Schirò, G.; Chiaramonte, G.; Barreca, S.; Pisano, P.; et al. Molecular Characterization and Evolutionary Analyses of Carnivore Protoparvovirus 1 NS1 Gene. Viruses 2019, 11, 308. [Google Scholar] [CrossRef] [Green Version]
- Mira, F.; Dowgier, G.; Purpari, G.; Vicari, D.; Di Bella, S.; Macaluso, G.; Gucciardi, F.; Randazzo, V.; Decaro, N.; Guercio, A. Molecular Typing of a Novel Canine Parvovirus Type 2a Mutant Circulating in Italy. Infect. Genet. Evol. 2018, 61, 67–73. [Google Scholar] [CrossRef]
- Tucciarone, C.M.; Franzo, G.; Mazzetto, E.; Legnardi, M.; Caldin, M.; Furlanello, T.; Cecchinato, M.; Drigo, M. Molecular Insight into Italian Canine Parvovirus Heterogeneity and Comparison with the Worldwide Scenario. Infect. Genet. Evol. 2018, 66, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Battilani, M.; Modugno, F.; Mira, F.; Purpari, G.; Di Bella, S.; Guercio, A.; Balboni, A. Molecular Epidemiology of Canine Parvovirus Type 2 in Italy from 1994 to 2017: Recurrence of the CPV-2b Variant. BMC Vet. Res. 2019, 15, 393. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, J.; Chen, Z.; Niu, G.; Liu, G. A Divergent Canine Parvovirus Type 2c (CPV-2c) Isolate Circulating in China. Infect. Genet. Evol. 2019, 73, 242–247. [Google Scholar] [CrossRef]
- Inthong, N.; Kaewmongkol, S.; Meekhanon, N.; Sirinarumitr, K.; Sirinarumitr, T. Dynamic Evolution of Canine Parvovirus in Thailand. Vet. World 2020, 13, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Parrish, C.R. Emergence, Natural History, and Variation of Canine, Mink, and Feline Parvoviruses. Adv. Virus Res. 1990, 38, 403–450. [Google Scholar] [CrossRef]
- Parrish, C.R.; Aquadro, C.F.; Strassheim, M.L.; Evermann, J.F.; Sgro, J.Y.; Mohammed, H.O. Rapid Antigenic-Type Replacement and DNA Sequence Evolution of Canine Parvovirus. J. Virol. 1991, 65, 6544–6552. [Google Scholar] [CrossRef] [Green Version]
- Allison, A.B.; Kohler, D.J.; Fox, K.A.; Brown, J.D.; Gerhold, R.W.; Shearn-bochsler, V.I.; Dubovi, E.J.; Parrish, C.R.; Holmes, C.; Carolina, S. Frequent Cross-Species Transmission of Parvoviruses among Diverse Carnivore Hosts. J. Virol. 2013, 87, 2342–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High Rate of Viral Evolution Associated with the Emergence of Carnivore Parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef] [Green Version]
- Parrish, C.R.; O’Connell, P.H.; Evermann, J.F.; Carmichael, L.E. Natural Variation of Canine Parvovirus. Science 1985, 230, 1046–1048. [Google Scholar] [CrossRef]
- Parrish, C.R.; Have, P.; Foreyt, W.J.; Evermann, J.F.; Senda, M.; Carmichael, L.E. The Global Spread and Replacement of Canine Parvovirus Strains. J. Gen. Virol. 1988, 69 Pt 5, 1111–1116. [Google Scholar] [CrossRef]
- Buonavoglia, C.; Martella, V.; Pratella, A.; Tempesta, M.; Cavalli, A.; Buonavoglia, D.; Bozzo, G.; Elia, G.; Decaro, N.; Carmichael, L. Evidence for Evolution of Canine Parvovirus Type 2 in Italy. J. Gen. Virol. 2001, 82, 3021–3025. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Cavalli, A.; Pratelli, A.; Bozzo, G.; Camero, M.; Buonavoglia, D.; Narcisi, D.; Tempesta, M.; Buonavoglia, C. A Canine Parvovirus Mutant Is Spreading in Italy. J. Clin. Microbiol. 2004, 42, 1333–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truyen, U. Emergence and Recent Evolution of Canine Parvovirus. Vet. Microbiol. 1999, 69, 47–50. [Google Scholar] [CrossRef]
- Battilani, M.; Scagliarini, A.; Tisato, E.; Turilli, C.; Jacoboni, I.; Casadio, R.; Prosperi, S. Analysis of Canine Parvovirus Sequences from Wolves and Dogs Isolated in Italy. J. Gen. Virol. 2001, 82, 1555–1560. [Google Scholar] [CrossRef]
- Altman, K.D.; Kelman, M.; Ward, M.P. Are Vaccine Strain, Type or Administration Protocol Risk Factors for Canine Parvovirus Vaccine Failure? Vet. Microbiol. 2017, 210, 8–16. [Google Scholar] [CrossRef]
- Calatayud, O.; Esperón, F.; Roman, S.C.; Biek, J.K.; Eblate, E.; Neves, E.; Lembo, T.; Lankester, F. Carnivore Parvovirus Ecology in the Serengeti Ecosystem: Vaccine Strains Circulating and New Host Species Identified. J. Virol. 2019, 93, e02220-18. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.; Jeong, W.; Kim, H.; An, D. Molecular Insights into the Phylogeny of Canine Parvovirus 2 (CPV-2) with Emphasis on Korean Isolates: A Bayesian Approach. Arch. Virol. 2009, 2, 1353–1360. [Google Scholar] [CrossRef]
- Truyen, U.W.E.; Evermann, J.F.; Vieler, E.; Parrish, C.R. Evolution of Canine Parvovirus Involved Loss and Gain of Feline Host Range. Virology 1996, 189, 186–189. [Google Scholar] [CrossRef] [Green Version]
- Miranda, C.; Thompson, G. Canine Parvovirus: The Worldwide Occurrence of Antigenic Variants. J. Gen. Virol. 2016, 97, 2043–2057. [Google Scholar] [CrossRef]
- Purpari, G.; Mira, F.; Di Bella, S.; Di Pietro, S.; Giudice, E.; Guercio, A. Investigation on Canine Parvovirus Circulation in Dogs from Sicily (Italy) by Biomolecular Assay. Acta Vet. Brno 2018, 68, 80–94. [Google Scholar] [CrossRef] [Green Version]
- Dei Giudici, S.; Cubeddu, T.; Giagu, A.; Sanna, G.; Rocca, S.; Oggiano, A. First Molecular Characterization of Canine Parvovirus Strains in Sardinia, Italy. Arch. Virol. 2017, 162, 3481–3486. [Google Scholar] [CrossRef] [PubMed]
- Cocchi, M.; Danesi, P.; De Zan, G.; Leati, M.; Gagliazzo, L.; Ruggeri, M.; Palei, M.; Bremini, A.; Rossmann, M.; Lippert-petscharnig, M.; et al. A Three-Year Biocrime Sanitary Surveillance on Illegally Imported Companion Animals. Pathogens 2021, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- Campalto, M.; Carrino, M.; Tassoni, L.; Rizzo, G.; Rossmann, M.C.; Cocchi, M.; De Benedictis, P.; Beato, M.S. Divergent Minute Virus of Canines Strains Identified in Illegally Imported Puppies in Italy. Arch. Virol. 2020, 165, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Pérez, R.; Calleros, L.; Marandino, A.; Sarute, N.; Iraola, G.; Grecco, S.; Blanc, H.; Vignuzzi, M.; Isakov, O.; Shomron, N.; et al. Phylogenetic and Genome-Wide Deep-Sequencing Analyses of Canine Parvovirus Reveal Co-Infection with Field Variants and Emergence of a Recent Recombinant Strain. PLoS ONE 2014, 9, e111779. [Google Scholar] [CrossRef] [Green Version]
- Bingga, G.; Liu, Z.; Zhang, J.; Zhu, Y.; Lin, L.; Ding, S.; Guo, P. High Resolution Melting Curve Analysis as a New Tool for Rapid Identification of Canine Parvovirus Type 2 Strains. Mol. Cell. Probes 2014, 28, 271–278. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Nguyen, L.; Schmidt, H.A.; Haeseler, A.; Von Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Voorhees, I.E.H.; Lee, H.; Allison, A.B.; Lopez-Astacio, R.; Goodman, L.B.; Oyesola, O.O.; Omobowale, O.; Fagbohun, O.; Dubovi, E.J.; Hafenstein, S.L.; et al. Limited Intrahost Diversity and Background Evolution Accompany 40 Years of Canine Parvovirus Host Adaptation and Spread. J. Virol. 2019, 94, e01162-19. [Google Scholar] [CrossRef]
- Battilani, M.; Ciulli, S.; Tisato, E.; Prosperi, S. Genetic Analysis of Canine Parvovirus Isolates ( CPV-2 ) from Dogs in Italy. Virus Res. 2002, 83, 149–157. [Google Scholar] [CrossRef]
- Decaro, N.; Desario, C.; Parisi, A.; Martella, V.; Lorusso, A.; Miccolupo, A.; Mari, V.; Loredana, M.; Cavalli, A.; Di, L.; et al. Genetic Analysis of Canine Parvovirus Type 2c. Virology 2009, 385, 5–10. [Google Scholar] [CrossRef]
- Kang, B.-K.; Song, D.-S.; Lee, C.-S.; Jung, K.-I.; Park, S.-J.; Kim, E.-M.; Park, B.-K. Prevalence and Genetic Characterization of Canine Parvoviruses in Korea. Virus Genes 2008, 2, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Maya, L.; Callegaros, L.; Francia, L.; Hernandez, M.; Iraola, G.; Panzera, Y.; Sosa, K.; Perez, R. Phylodynamics Analysis of Canine Parvovirus in Uruguay: Evidence of Two Successive Invasions by Different Variants. Arch. Virol. 2013, 158, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jin, H.; Li, Q.; Zhao, G.; Cheng, N. Isolation and Sequence Analysis of the Complete NS1 and VP2 Genes of Canine Parvovirus from Domestic Dogs in 2013 and 2014 in China. Arch. Virol. 2016, 161, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Mittal, M.; Chakravarti, S.; Mohapatra, J.K.; Chug, P.K.; Dubey, R.; Upmanuyu, V.; Narwal, P.S.; Kumar, A.; Churamani, C.P.; Kanwar, N.S. Molecular Typing of Canine Parvovirus Strains Circulating from 2008 to 2012 in an Organized Kennel in India Reveals the Possibility of Vaccination Failure. Infect. Genet. Evol. 2014, 23, 1–6. [Google Scholar] [CrossRef]
- Phromnoi, S.; Sirinarumitr, K.; Sirinarumitr, T. Sequence Analysis of VP2 Gene of Canine Parvovirus Isolates in Thailand. Virus Genes 2010, 41, 23–29. [Google Scholar] [CrossRef]
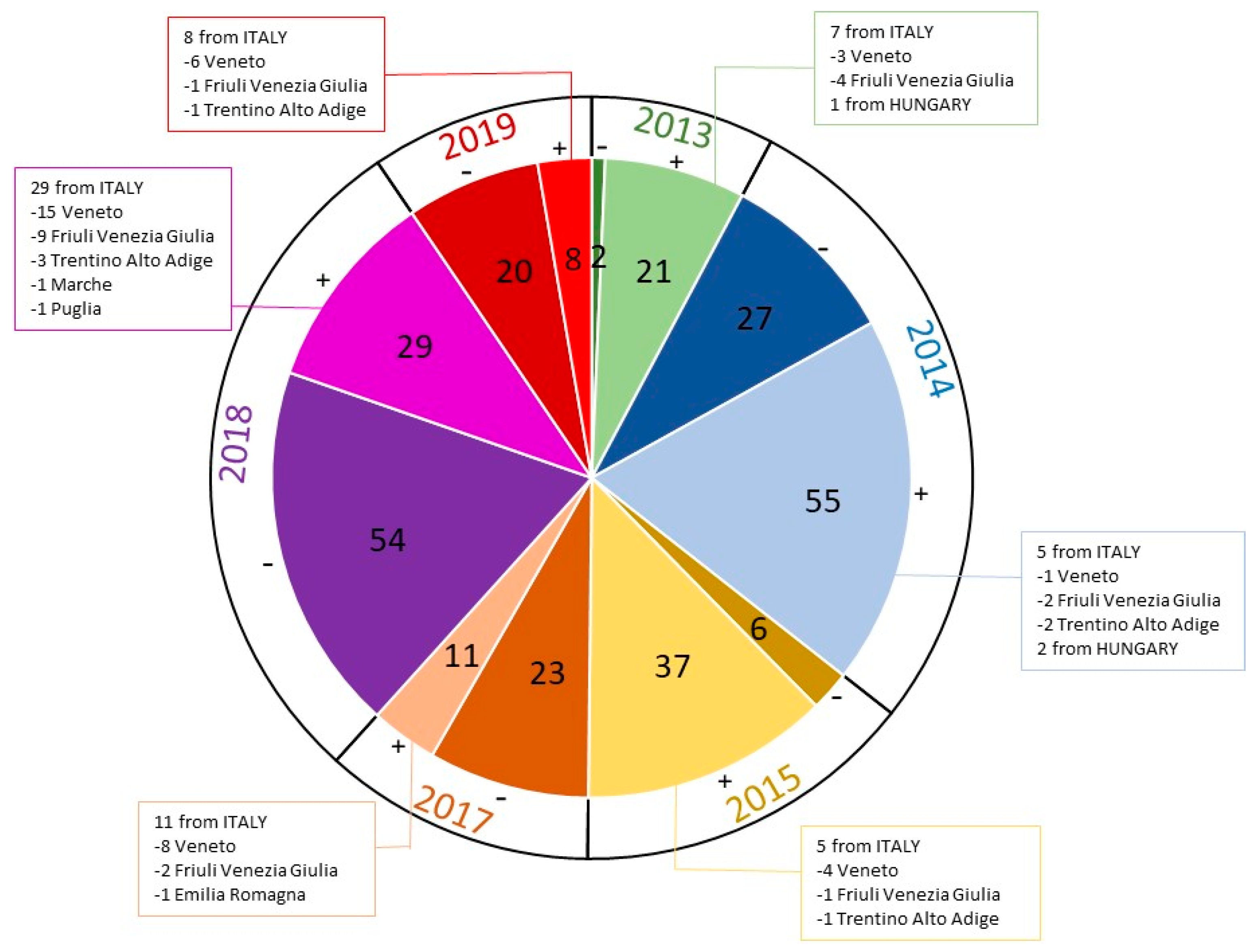
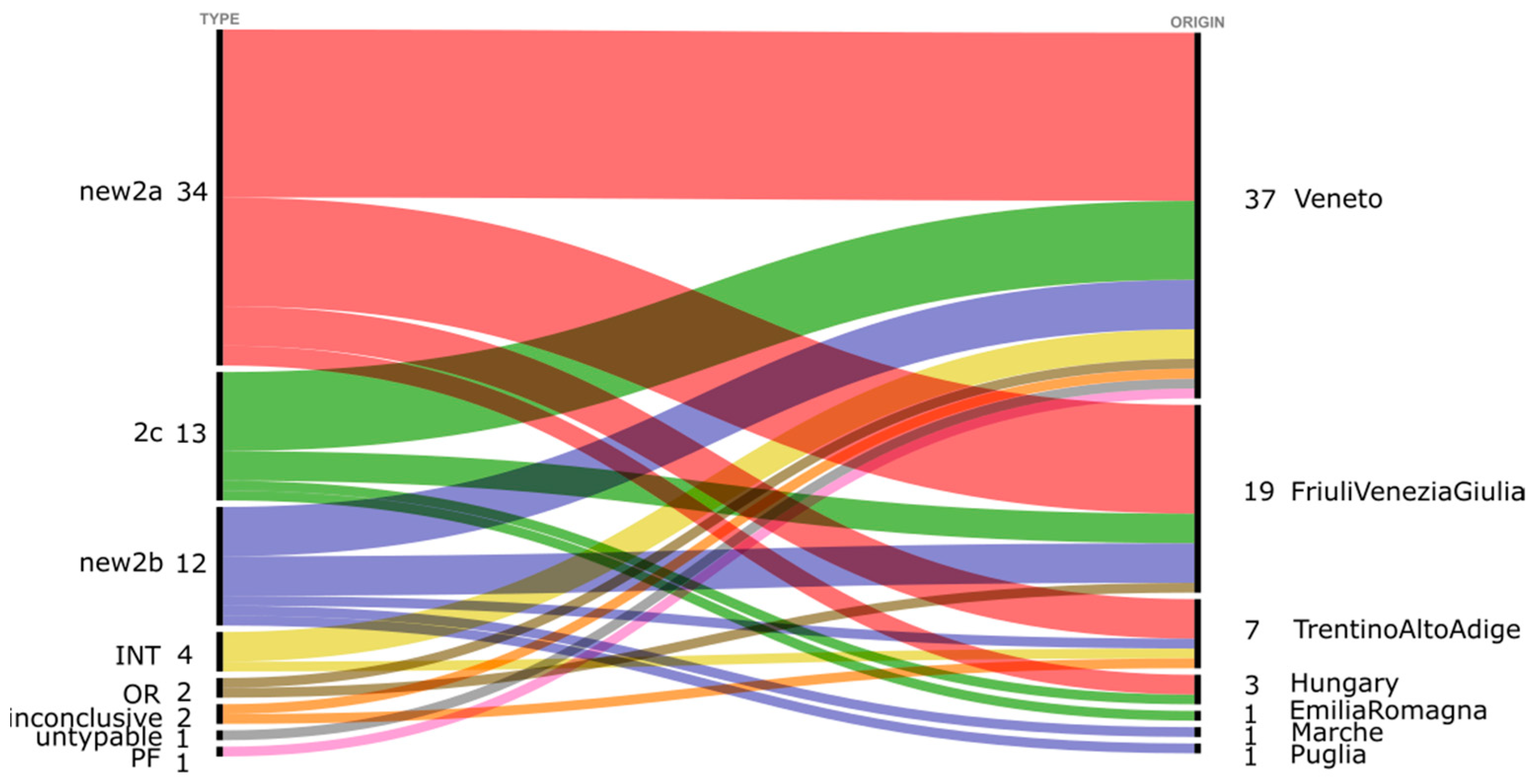



| Sample | Year | Origin of the Sample | Breed | M/F | Puppy/Adult | Age | Intestinal Lesions | Vaccination |
|---|---|---|---|---|---|---|---|---|
| VIR-2391/6 | 2013 | Veneto | Chihuahua | M | P | 5 m | Y | N/A |
| VIR-2697/8 | 2013 | Veneto | Rottweiler | F | A | 6 m | N | N/A |
| VIR-5519/4 | 2013 | Hungary | Yorkshire Terrier | M | P | 1 m | N/A | N/A |
| VIR-5997/4 | 2013 | Friuli Venezia Giulia | N/A | M | P | N/A | N | N/A |
| VIR-6066/4 | 2013 | Friuli Venezia Giulia | N/A | M | P | N/A | N | N/A |
| VIR-6067/4 | 2013 | Friuli Venezia Giulia | Dachshund | M | P | N/A | Y | N/A |
| VIR-6542/3 | 2013 | Friuli Venezia Giulia | N/A | M | P | N/A | Y | N/A |
| VIR-6927/7 | 2013 | Veneto | Maltese | F | P | 1 m | Y | N/A |
| VIR-1263/3 | 2014 | Hungary | Pincher | M | P | 4 m | Y | N/A |
| VIR-3058/2 | 2014 | Trentino Alto Adige | N/A | F | P | 3 m | Y | N/A |
| VIR-3493/3 | 2014 | Trentino Alto Adige | N/A | N/A | N/A | N/A | Y | N/A |
| VIR-5175/9 | 2014 | Hungary | Siberian Husky | F | P | 2 m | N | N/A |
| VIR-6046/5 | 2014 | Friuli Venezia Giulia | Chow-Chow | M | P | 4 m | Y | N/A |
| VIR-6666/2 | 2014 | Friuli Venezia Giulia | N/A | N/A | N/A | N/A | N/A | N/A |
| VIR-7758/9 | 2014 | Veneto | Yorkshire Terrier | F | P | 2 m | Y | N/A |
| VIR-1684/6 | 2015 | Veneto | N/A | F | N/A | N/A | Y | N/A |
| VIR-1838/3 | 2015 | Veneto | N/A | F | N/A | N/A | Y | N/A |
| VIR-2037/1 | 2015 | Veneto | N/A | N/A | N/A | N/A | N/A | N/A |
| VIR-6238/10 | 2015 | Veneto | Mix breed | N/A | P | 3.5 m | Y | N/A |
| VIR-6378/5 | 2015 | Friuli Venezia Giulia | Maltese | M | P | 2 m | Y | N/A |
| VIR-6478/4 | 2015 | Trentino Alto Adige | N/A | N/A | N/A | N/A | N/A | N/A |
| DIAPD-1387/7 | 2017 | Veneto | American Staffordshire Terrier | N/A | P | 3 m | Y | N/A |
| DIAPD-1751/2 | 2017 | Emilia Romagna | Labrador | M | P | 50 d | Y | N/A |
| DIAPD-2046/16 | 2017 | Veneto | Golden Retriever | N/A | P | 4 m | Y | N/A |
| DIAPD-53054/6 | 2017 | Veneto | Mix breed | M | A | 2 y | Y | N/A |
| DIAPD-53504/6 | 2017 | Veneto | Mix breed | M | P | 65 d | Y | N/A |
| DIAPD-55092/2 | 2017 | Friuli Venezia Giulia | N/A | F | N/A | N/A | N/A | N/A |
| DIAPD-56192/2 | 2017 | Veneto | Siberian Husky | F | A | N/A | N/A | N/A |
| DIAPD-56643/4 | 2017 | Veneto | Beagle | N/A | P | 3 m | N | N/A |
| DIAPD-57033/8 | 2017 | Veneto | Miniature Poodle | M | A | 8 m | N/A | N/A |
| DIAPD-57861/2 | 2017 | Friuli Venezia Giulia | English Pointer | F | A | 5.5 m | N/A | N/A |
| DIAPD-766/5 | 2018 | Veneto | Chihuahua | F | A | 9 m | Y | N/A |
| DIAPD-1100/3 | 2018 | Friuli Venezia Giulia | Mix breed | M | P | 4 m | N/A | N/A |
| DIAPD-1399/7 | 2018 | Veneto | Maltese | M | P | 3.5 m | N/A | N/A |
| DIAPD-150/3 | 2018 | Friuli Venezia Giulia | Poodle | M | P | 2 m | Y | N/A |
| DIAPD-2874/6 | 2018 | Veneto | Pincher | M | A | 4 y | Y | N/A |
| DIAPD-3417/6 | 2018 | Puglia | Mix breed | F | P | 3 m | Y | N/A |
| DIAPD-3617/4 | 2018 | Veneto | Mix breed | F | P | 3 m | Y | N/A |
| DIAPD-50765/13 | 2018 | Trentino Alto Adige | Shiba Inu | M | P | 3 m | N/A | N/A |
| DIAPD-51020/3 | 2018 | Friuli Venezia Giulia | Mix breed | F | P | 2 m | N/A | N/A |
| DIAPD-51301/13 | 2018 | Trentino Alto Adige | N/A | N/A | N/A | N/A | N/A | N/A |
| DIAPD-52097/6 | 2018 | Marche | N/A | N/A | N/A | N/A | N | N/A |
| DIAPD-52386/3 | 2018 | Veneto | American Staffordshire Terrier | M | A | 3 y | Y | Y * |
| DIAPD-52568/4 | 2018 | Veneto | N/A | F | A | 4 y | N/A | N/A |
| DIAPD-52707/2 | 2018 | Friuli Venezia Giulia | Mix breed | M | P | 3 m | Y | N/A |
| DIAPD-52984/2 | 2018 | Friuli Venezia Giulia | Mix breed | F | P | 5 m | N/A | N/A |
| DIAPD-53807/2 | 2018 | Friuli Venezia Giulia | N/A | F | A | 8 m | Y | N/A |
| DIAPD-54123/2 | 2018 | Veneto | N/A | N/A | A | 2 y | N/A | Y * |
| DIAPD-55181/3 | 2018 | Friuli Venezia Giulia | Italian Hound | M | P | 3 m | N/A | N/A |
| DIAPD-55182/3 | 2018 | Friuli Venezia Giulia | Italian Hound | M | P | 3 m | N/A | N/A |
| DIAPD-55509/2 | 2018 | Veneto | N/A | F | P | 3 m | N/A | N/A |
| DIAPD-56178/6 | 2018 | Veneto | N/A | N/A | N/A | N/A | N/A | N/A |
| DIAPD-56237/5 | 2018 | Veneto | Poodle Toy | F | P | 2 m | N/A | N/A |
| DIAPD-56531/4 | 2018 | Friuli Venezia Giulia | Golden Retriever | M | P | 2 m | N/A | N/A |
| DIAPD-56557/6 | 2018 | Veneto | Alaskan Malamute | F | A | 9 m | N/A | N/A |
| DIAPD-58450/8 | 2018 | Trentino Alto Adige | N/A | N/A | N/A | N/A | N/A | N/A |
| DIAPD-855/18 | 2018 | Veneto | Chihuahua | M | P | 2 m | Y | N/A |
| DIAPD-855/19 | 2018 | Veneto | Chihuahua | F | P | 2 m | Y | N/A |
| DIAPD-855/42 | 2018 | Veneto | Chihuahua | M | P | 2 m | Y | N/A |
| DIAPD-855/6 | 2018 | Veneto | Chihuahua | M | P | 2 m | Y | N/A |
| DIAPD-883/6 | 2018 | Veneto | Maltese | F | N/A | N/A | N/A | N/A |
| DIAPD-200/5 | 2019 | Friuli Venezia Giulia | Boxer | M | A | 6 m | Y | N/A |
| DIAPD-2836/7 | 2019 | Veneto | Boxer | M | A | N/A | Y | N/A |
| DIAPD-3366/4 | 2019 | Veneto | Yorkshire Terrier | F | P | 2 m | N | N/A |
| DIAPD-3485/3 | 2019 | Veneto | Bloodhound | M | P | 2 m | Y | N/A |
| DIAPD-50725/5 | 2019 | Veneto | N/A | N/A | P | 2 m | Y | N/A |
| DIAPD-51808/4 | 2019 | Veneto | French Bulldog | M | P | 2 m | N/A | N/A |
| DIAPD-52182/8 | 2019 | Trentino Alto Adige | Cavalier King Charles Spaniel | M | P | 2 m | N/A | N/A |
| DIAPD-55450/9 | 2019 | Veneto | German Shepherd | M | P | 4 m | N/A | N/A |
| Sequence Presenting the Mutation in the VP2 Gene | Phylogenetic Group | VP2 Nucleotidic Residue | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 156 | 268 | 459 | 633 | 812 | 882 | 923 | 936 | 946 | 1002 | 1008 | 1020 | 1024 | 1039 | 1043 | 1053 | 1054 | 1057 | 1079 | 1091 | 1126 | 1174 | 1182 | 1297 | 1380 | 1485 | ||
| MT996031 CPV2-Canis-Italy-TrentinoAltoAdige-strain 18DIAPD-50765-13-2018-new2a | 2A.3 | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - |
| MT996032 CPV2-Canis-Italy-TrentinoAltoAdige-strain 18DIAPD-51301-13-2018-new2a | 2A.3 | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996030 CPV2-Canis-Italy-Puglia-strain DIAPD 18DIAPD-3417-6-2018-new2b | 2B | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996020 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-1100-3-2018-new2b | 2B | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996025 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-53807-2-2018-new2b | 2B | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996036 CPV2-Canis-Italy-Veneto-strain 17DIAPD-52386-3-2017-new2b | 2B | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996022 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-51020-3-2018-OR | OR | - | - | - | C | G | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996057 CPV2-Canis-Italy-Veneto-strain 19DIAPD-3485-3-2019-OR | OR | - | - | - | - | G | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996019 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 17DIAPD-57861-2-2017-new2a | / | - | - | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| OM525670 CPV2-Canis-Italy-Veneto-13VIR2697-8-2013-new2b | 2B | - | - | - | - | - | - | G | G | - | A | - | T | - | T | T | - | - | - | A | - | - | - | - | - | - | - |
| MT996047 CPV2-Canis-Italy-Veneto-strain 18DIAPD-55509-2-2018-new2b | 2B | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| OM525673 CPV2-Canis-Italy-TrentinoAltoAdige-14VIR3493-3-2014-new2b | 2B | - | - | - | - | - | - | - | - | - | - | G | - | - | - | - | - | - | - | - | G | - | - | - | - | - | - |
| MT996023 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-52707-2-2018-new2a | / | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - |
| MT996044 CPV2-Canis-Italy-Veneto-strain 18DIAPD-3617-4-2018-new2a | 2A.3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | - | - | - | - |
| MT996058 CPV2-Canis-Italy-Veneto-strain 19DIAPD-50725-5-2019-2c | 2C.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | T | - | - | - |
| OM525667 CPV2-Canis-Italy-Veneto-15VIR1684-6-2015-2c | 2C.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | - | - | - | - | - |
| OM525674 CPV2-Canis-Italy-TrentinoAltoAdige-14VIR3058-2-2014-new2a | 2A.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| MT996035 CPV2-Canis-Italy-Veneto-strain 17DIAPD-2046-16-2017-2c | 2C.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - |
| OM525666 CPV2-Canis-Italy-Veneto-15VIR1838-10-2015-new2a | 2A.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - |
| MT996026 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-55181-3-2018-new2a | 2A.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C |
| MT996027 CPV2-Canis-Italy-FriuliVeneziaGiulia-strain 18DIAPD-55182-3-2018-new2a | 2A.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C |
| MT996050 CPV2-Canis-Italy-Veneto-strain 18DIAPD-855-18-2018-new2a | 2A.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C |
| Nucleotidic mutation | A156G | A268C | T459C | T633C | A812G | G882A | T923G | A936G | G946A | T1002A | T1008G | A1020T | T1024C | G1039T | C1043T | G1053A | C1054T | T1057A | G1079A | C1091G | C1126A | G1174A | A1182T | A1297T | A1380C | T1485C | |
| Frequency in the Italian population (%) | 0.4 | 0.2 | 0.6 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 0.3 | 0.3 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.6 | |
| Silent/missense/non sense | sil | mis | sil | sil | mis | sil | mis | sil | mis | sil | sil | sil | mis | mis | mis | sil | mis | mis | mis | mis | mis | mis | sil | mis | sil | sil | |
| AA change | T90P | K271R | V308G | V316I | Y342H | A347S | S348F | P352S | F353I | G360E | A364G | P376T | G392R | T433S | |||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrino, M.; Tassoni, L.; Campalto, M.; Cavicchio, L.; Mion, M.; Corrò, M.; Natale, A.; Beato, M.S. Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering. Viruses 2022, 14, 917. https://doi.org/10.3390/v14050917
Carrino M, Tassoni L, Campalto M, Cavicchio L, Mion M, Corrò M, Natale A, Beato MS. Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering. Viruses. 2022; 14(5):917. https://doi.org/10.3390/v14050917
Chicago/Turabian StyleCarrino, Marilena, Luca Tassoni, Mery Campalto, Lara Cavicchio, Monica Mion, Michela Corrò, Alda Natale, and Maria Serena Beato. 2022. "Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering" Viruses 14, no. 5: 917. https://doi.org/10.3390/v14050917
APA StyleCarrino, M., Tassoni, L., Campalto, M., Cavicchio, L., Mion, M., Corrò, M., Natale, A., & Beato, M. S. (2022). Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering. Viruses, 14(5), 917. https://doi.org/10.3390/v14050917








