Pivoting Novel Exosome-Based Technologies for the Detection of SARS-CoV-2
Abstract
:1. Introduction
2. NIH RADx Initiative
3. RADx-Rad
4. Exosomes
5. Novel Exosome-Based Technologies
6. RADx-Rax Exosome Technologies
7. Regulatory Considerations
8. Commercialization Considerations
9. Scale-Up and Manufacturing Considerations
10. Post-Acute Sequelae of SARS-CoV-2 Infection (PASC)
11. Future Pandemics
Author Contributions
Funding
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.; Mehra, M.R.; Scholkmann, F.; Schupbach, R.; Ruschitzka, F.; et al. Electron microscopy of SARS-CoV-2: A challenging task—Authors’ reply. Lancet 2020, 395, e100. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharm. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8. [Google Scholar] [CrossRef]
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W.V.; Gerlo, S.; Vandekerckhove, L. On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLoS Pathog. 2020, 16, e1009037. [Google Scholar] [CrossRef]
- Peng, L.; Liu, J.; Xu, W.; Luo, Q.; Chen, D.; Lei, Z.; Huang, Z.; Li, X.; Deng, K.; Lin, B.; et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J. Med. Virol. 2020, 92, 1676–1680. [Google Scholar] [CrossRef]
- Zapor, M. Persistent detection and infectious potential of SARS-CoV-2 virus in clinical specimens from COVID-19 patients. Viruses 2020, 12, 1384. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID Data Tracker. Available online: https://covid.cdc.gov/covid-data-tracker (accessed on 10 May 2022).
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 14 April 2022).
- Food and Drug Administration. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicron (accessed on 16 January 2022).
- Tromberg, B.J.; Schwetz, T.A.; Perez-Stable, E.J.; Hodes, R.J.; Woychik, R.P.; Bright, R.A.; Fleurence, R.L.; Collins, F.S. Rapid scaling up of COVID-19 diagnostic testing in the United States—The NIH RADx initiative. N. Engl. J. Med. 2020, 383, 1071–1077. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. [Google Scholar] [CrossRef]
- Scohy, A.; Anantharajah, A.; Bodeus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef]
- Brownstein, N.C.; Chen, Y.A. Predictive values, uncertainty, and interpretation of serology tests for the novel coronavirus. Sci. Rep. 2021, 11, 5491. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for COVID-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar Khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Liu, G.; Rusling, J.F. COVID-19 antibody tests and their limitations. ACS Sens. 2021, 6, 593–612. [Google Scholar] [CrossRef]
- Brooks, Z.C.; Das, S. COVID-19 testing. Am. J. Clin. Pathol. 2020, 154, 575–584. [Google Scholar] [CrossRef]
- Benzigar, M.R.; Bhattacharjee, R.; Baharfar, M.; Liu, G. Current methods for diagnosis of human coronaviruses: Pros and cons. Anal. Bioanal. Chem. 2021, 413, 2311–2330. [Google Scholar] [CrossRef]
- Ong, D.S.Y.; Fragkou, P.C.; Schweitzer, V.A.; Chemaly, R.F.; Moschopoulos, C.D.; Skevaki, C.; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Respiratory Viruses (ESGREV). How to interpret and use COVID-19 serology and immunology tests. Clin. Microbiol. Infect. 2021, 27, 981–986. [Google Scholar] [CrossRef]
- Muller, L.; Hong, C.S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef] [Green Version]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Torregrosa Paredes, P.; Esser, J.; Admyre, C.; Nord, M.; Rahman, Q.K.; Lukic, A.; Radmark, O.; Gronneberg, R.; Grunewald, J.; Eklund, A.; et al. Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy 2012, 67, 911–919. [Google Scholar] [CrossRef]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum exosome MicroRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef] [PubMed]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Asea, A.; Jean-Pierre, C.; Kaur, P.; Rao, P.; Linhares, I.M.; Skupski, D.; Witkin, S.S. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J. Reprod. Immunol. 2008, 79, 12–17. [Google Scholar] [CrossRef]
- Domenis, R.; Zanutel, R.; Caponnetto, F.; Toffoletto, B.; Cifu, A.; Pistis, C.; Di Benedetto, P.; Causero, A.; Pozzi, M.; Bassini, F.; et al. Characterization of the proinflammatory profile of synovial fluid-derived exosomes of patients with osteoarthritis. Mediat. Inflamm. 2017, 2017, 4814987. [Google Scholar] [CrossRef]
- Klingeborn, M.; Dismuke, W.M.; Bowes Rickman, C.; Stamer, W.D. Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye Res. 2017, 59, 158–177. [Google Scholar] [CrossRef]
- Srinivasan, S.; Vannberg, F.O.; Dixon, J.B. Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci. Rep. 2016, 6, 24436. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Wang, Y.; Nie, J.; Li, Q.; Tang, L.; Deng, X.; Wang, F.; Xu, B.; Wu, X.; Zhang, X.; et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget 2017, 8, 69995–70005. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Happel, C.; Ganguly, A.; Tagle, D.A. Extracellular RNAs as potential biomarkers for cancer. J. Cancer Metastasis Treat. 2020, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New molecular targets of diseases. Acta Pharm. Sin. 2018, 39, 501–513. [Google Scholar] [CrossRef]
- Femmino, S.; Penna, C.; Margarita, S.; Comita, S.; Brizzi, M.F.; Pagliaro, P. Extracellular vesicles and cardiovascular system: Biomarkers and cardioprotective effectors. Vasc. Pharm. 2020, 135, 106790. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating extracellular vesicles in human disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Crenshaw, B.J.; Gu, L.; Sims, B.; Matthews, Q.L. Exosome biogenesis and biological function in response to viral infections. Open Virol. J. 2018, 12, 134–148. [Google Scholar] [CrossRef] [Green Version]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Thery, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef]
- Wang, J.; Chen, S.; Bihl, J. Exosome-mediated transfer of ACE2 (angiotensin-converting enzyme 2) from endothelial progenitor cells promotes survival and function of endothelial cell. Oxid. Med. Cell Longev. 2020, 2020, 4213541. [Google Scholar] [CrossRef] [Green Version]
- Hassanpour, M.; Rezaie, J.; Nouri, M.; Panahi, Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 2020, 85, 104422. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, Content, release, and uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Liangsupree, T.; Multia, E.; Riekkola, M.L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef] [PubMed]
- Mizenko, R.R.; Brostoff, T.; Rojalin, T.; Koster, H.J.; Swindell, H.S.; Leiserowitz, G.S.; Wang, A.; Carney, R.P. Tetraspanins are unevenly distributed across single extracellular vesicles and bias sensitivity to multiplexed cancer biomarkers. J. Nanobiotechnol. 2021, 19, 250. [Google Scholar] [CrossRef]
- Han, C.; Kang, H.; Yi, J.; Kang, M.; Lee, H.; Kwon, Y.; Jung, J.; Lee, J.; Park, J. Single-vesicle imaging and co-localization analysis for tetraspanin profiling of individual extracellular vesicles. J. Extracell. Vesicles. 2021, 10, e12047. [Google Scholar] [CrossRef]
- Food and Drug Administration. In Vitro Diagnostics EUAs. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas#covid19ivdTemplates (accessed on 10 May 2022).
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Huang, N.; Perez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef]
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; Williamson, D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00776-20. [Google Scholar] [CrossRef] [Green Version]
- Abasiyanik, M.F.; Flood, B.; Lin, J.; Ozcan, S.; Rouhani, S.J.; Pyzer, A.; Trujillo, J.; Zhen, C.; Wu, P.; Jumic, S.; et al. Sensitive detection and quantification of SARS-CoV-2 in saliva. Sci. Rep. 2021, 11, 12425. [Google Scholar] [CrossRef]
- Tan, S.H.; Allicock, O.; Armstrong-Hough, M.; Wyllie, A.L. Saliva as a gold-standard sample for SARS-CoV-2 detection. Lancet Respir. Med. 2021, 9, 562–564. [Google Scholar] [CrossRef]
- Teo, A.K.J.; Choudhury, Y.; Tan, I.B.; Cher, C.Y.; Chew, S.H.; Wan, Z.Y.; Cheng, L.T.E.; Oon, L.L.E.; Tan, M.H.; Chan, K.S.; et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci. Rep. 2021, 11, 3134. [Google Scholar] [CrossRef] [PubMed]
- Reategui, E.; van der Vos, K.E.; Lai, C.P.; Zeinali, M.; Atai, N.A.; Aldikacti, B.; Floyd, F.P., Jr.; Khankhel, A.H.; Thapar, V.; Hochberg, F.H.; et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Chen, C.; Mao, Z.; Bachman, H.; Becker, R.; Rufo, J.; Wang, Z.; Zhang, P.; Mai, J.; Yang, S.; et al. Acoustofluidic centrifuge for nanoparticle enrichment and separation. Sci. Adv. 2021, 7, eabc0467. [Google Scholar] [CrossRef]
- Food and Drug Administration. Policy for Evaluating Impact of Viral Mutations on COVID-19 Tests; Food and Drug Administration: White Oak, MD, USA, 2021. [Google Scholar]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Mina, M.J.; Peto, T.E.; Garcia-Finana, M.; Semple, M.G.; Buchan, I.E. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet 2021, 397, 1425–1427. [Google Scholar] [CrossRef]
- Magleby, R.; Westblade, L.F.; Trzebucki, A.; Simon, M.S.; Rajan, M.; Park, J.; Goyal, P.; Safford, M.M.; Satlin, M.J. Impact of severe acute respiratory syndrome coronavirus 2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin. Infect. Dis. 2021, 73, e4197–e4205. [Google Scholar] [CrossRef]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration. Determination of Public Health Emergency; Department of Health and Human Services: Washington, DC, USA, 2020; pp. 7316–7317.
- Bianchi, D.W.B.P.F.; Gibbons, G.H.; Gordon, J.; Hodes, R.J.; Lorsch, J.R.; Mensah, G.A.; Perez-Stable, E.J.; Riley, W.; Schwetz, T.A.; Volkow, N.D. Why Testing is the Key to Getting Back to Normal. 2020. Available online: https://nlmdirector.nlm.nih.gov/2020/09/04/why-testing-is-the-key-to-getting-back-to-normal/ (accessed on 6 July 2021).
- United States Congress. United States Code: Federal Food, Drug, and Cosmetic Act; 21 U.S.C.; U.S. Government Publishing Office: Washington, DC, USA, 1934.
- Roback, J.D.; Tyburski, E.A.; Alter, D.; Asakrah, S.; Chahroudi, A.; Esper, A.; Farmer, S.; Figueroa, J.; Frediani, J.K.; Gonzalez, M.D.; et al. The need for new test verification and regulatory support for innovative diagnostics. Nat. Biotechnol. 2021, 39, 1060–1062. [Google Scholar] [CrossRef] [PubMed]
- Shuren, J.; Stenzel, T. COVID-19 molecular diagnostic testing—Lessons learned. N. Engl. J. Med. 2020, 383, e97. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, M.; Tessier, P.; Collins, J.; Caro, E. The RADx tech deep dive and work package 1 process. IEEE Open J. Eng. Med. Biol. 2021, 2, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, S.; Davis, R.W.; Saha, A.K. Microfluidic point-of-care testing: Commercial landscape and future directions. Front. Bioeng. Biotechnol. 2020, 8, 602659. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Gilligan, K.E.; Dwyer, R.M. Role of extracellular vesicles (EVs) in cell stress response and resistance to cancer therapy. Cancers 2019, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Kim, J.H. Diverse effects of exosomes on COVID-19: A perspective of progress from transmission to therapeutic developments. Front. Immunol. 2021, 12, 716407. [Google Scholar] [CrossRef]
- Mosquera-Heredia, M.I.; Morales, L.C.; Vidal, O.M.; Barcelo, E.; Silvera-Redondo, C.; Velez, J.I.; Garavito-Galofre, P. Exosomes: Potential disease biomarkers and new therapeutic targets. Biomedicines 2021, 9, 1061. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, T.; Deng, R.; Xia, X.; Li, B.; Wang, X. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci. Rep. 2021, 11, 7991. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtsev, I.; Kalinina, O.; Bezrukikh, V.; Melnik, O.; Golovkin, A. The significance of phenotyping and quantification of plasma extracellular vesicles levels using high-sensitivity flow cytometry during COVID-19 treatment. Viruses 2021, 13, 767. [Google Scholar] [CrossRef] [PubMed]
- Rosell, A.; Havervall, S.; von Meijenfeldt, F.; Hisada, Y.; Aguilera, K.; Grover, S.P.; Lisman, T.; Mackman, N.; Thalin, C. Patients with COVID-19 Have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality-brief report. Arter. Thromb Vasc. Biol. 2021, 41, 878–882. [Google Scholar] [CrossRef] [PubMed]
- Campello, E.; Radu, C.M.; Simion, C.; Spiezia, L.; Bulato, C.; Gavasso, S.; Tormene, D.; Perin, N.; Turatti, G.; Simioni, P. Longitudinal trend of plasma concentrations of extracellular vesicles in patients hospitalized for COVID-19. Front. Cell Dev. Biol. 2021, 9, 770463. [Google Scholar] [CrossRef]
- HHS Launches First Venture Capital Partnership to Develop Transformative Technologies to Combat Future Pandemics, Other Health Emergencies. 2021. Available online: https://www.hhs.gov/about/news/2021/06/01/hhs-launches-first-venture-capital-partnership-develop-transformative-technologies-combat-future-pandemic.html (accessed on 1 June 2021).
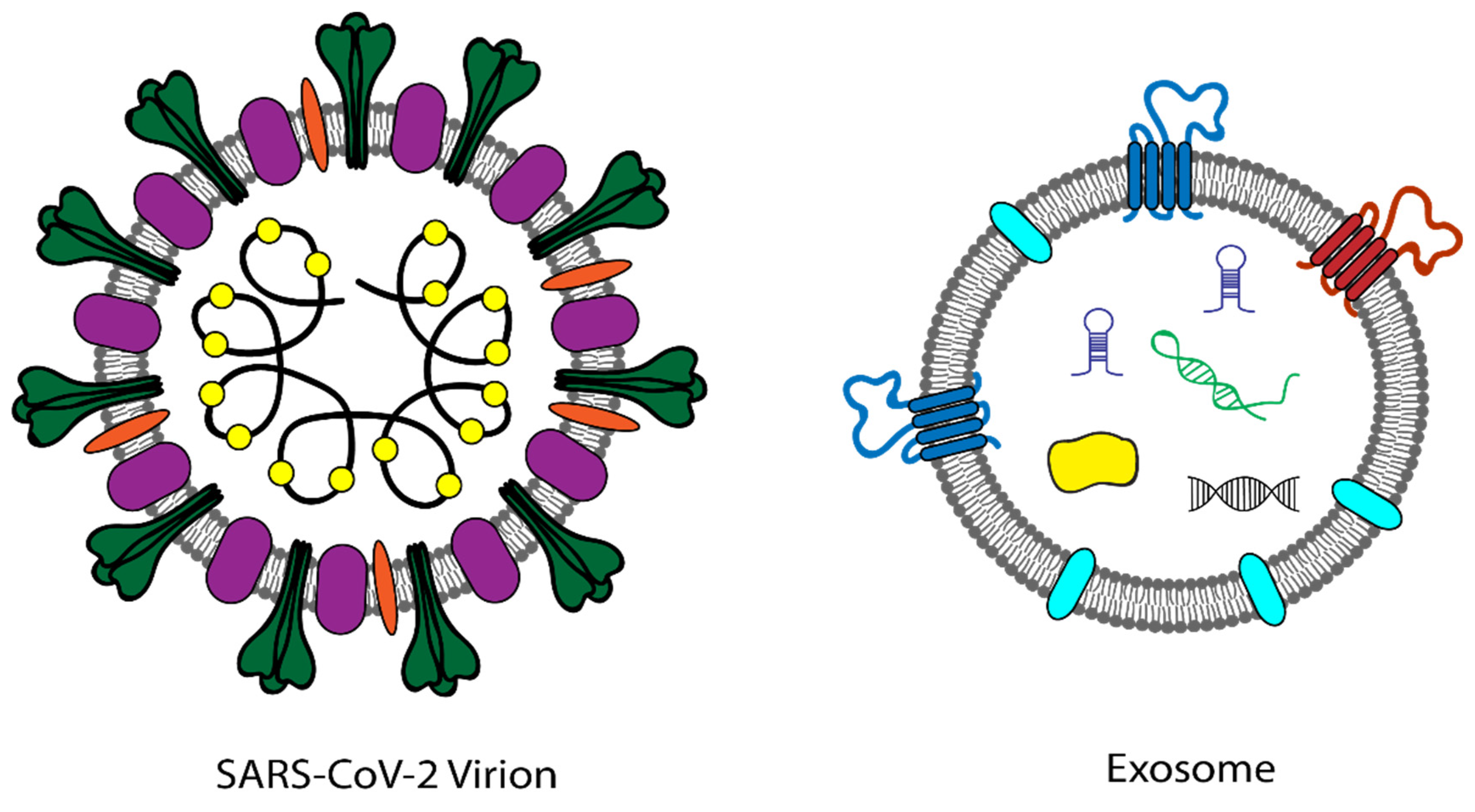
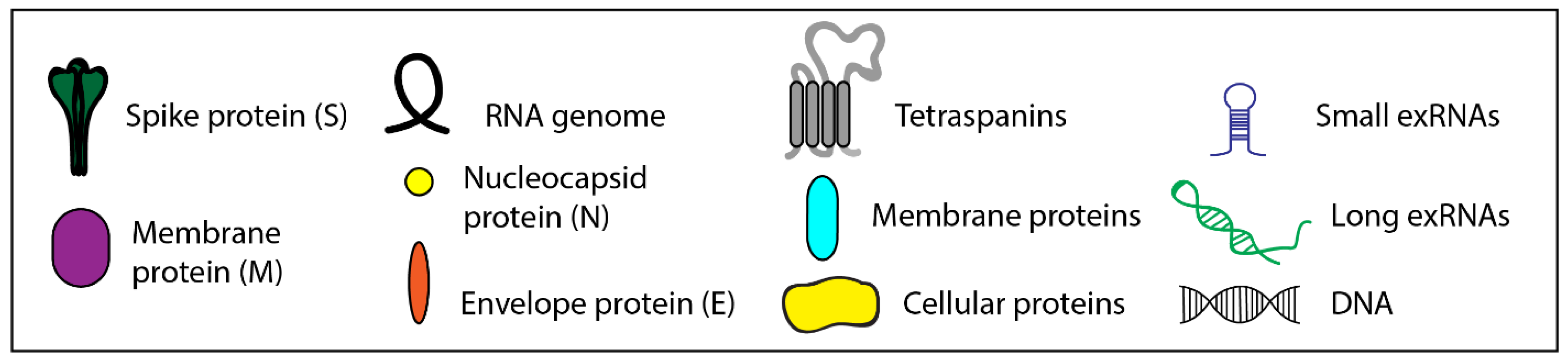
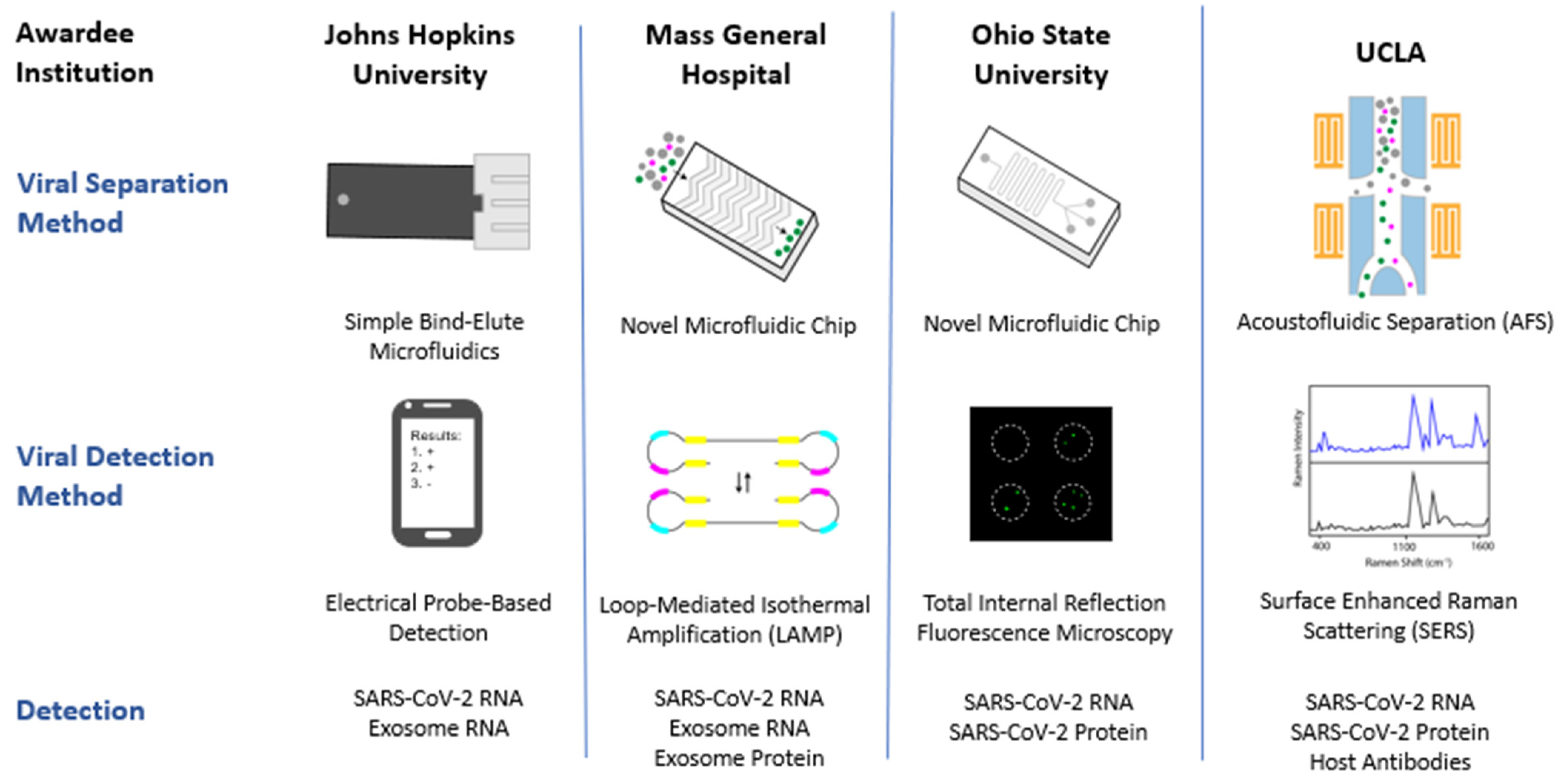
| RADx-Rad Awardee | Collaborators | Project Title |
|---|---|---|
| Samarjit Das— Johns Hopkins University (JHU) | Anubhav Dubey—Sognef Kenneth Witwer—JHU | Exosome-based Non-traditional Technologies Towards Multi-Parametric and Integrated Approaches for SARS-CoV-2 |
| Shannon L. Stott—Massachusetts General Hospital (MGH) | Genevieve Boland—MGH Xu Yu—MGH Seyed Rabi—MGH Jochen Lennerz—MGH | Multi-parametric Integrated Molecular Detection of SARS-CoV-2 from Biofluids by Adapting Single Extracellular Vesicle Characterization Technologies |
| Eduardo Reategui—Ohio State University (OSU) | L. James Lee—OSU Preeti Pancholi—OSU Shan-Lu Liu—OSU Kai Wang—Institute for Systems Biology Xioakui Mo—OSU | Microfluidics Array Based Sorting, Isolation, and RNA Analysis in Single Extracellular Vesicles |
| David T. Wong—University of California Los Angeles (UCLA) | Yong Kim—UCLA Ya-Hong Xie—UCLA Samantha Chiang—UCLA David Elashoff—UCLA Jennifer Fulcher—UCLA Wayne Grody—UCLA Feng Li—UCLA Fang Wei—UCLA Otto Yang—UCLA Tony Jun Huang—Duke University | AFS/SERS Saliva-based SARS-CoV-2 Earliest Infection and Antibodies Detection |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Happel, C.; Peñalber-Johnstone, C.; Tagle, D.A. Pivoting Novel Exosome-Based Technologies for the Detection of SARS-CoV-2. Viruses 2022, 14, 1083. https://doi.org/10.3390/v14051083
Happel C, Peñalber-Johnstone C, Tagle DA. Pivoting Novel Exosome-Based Technologies for the Detection of SARS-CoV-2. Viruses. 2022; 14(5):1083. https://doi.org/10.3390/v14051083
Chicago/Turabian StyleHappel, Christine, Chariz Peñalber-Johnstone, and Danilo A. Tagle. 2022. "Pivoting Novel Exosome-Based Technologies for the Detection of SARS-CoV-2" Viruses 14, no. 5: 1083. https://doi.org/10.3390/v14051083
APA StyleHappel, C., Peñalber-Johnstone, C., & Tagle, D. A. (2022). Pivoting Novel Exosome-Based Technologies for the Detection of SARS-CoV-2. Viruses, 14(5), 1083. https://doi.org/10.3390/v14051083







