The Role of the Flavivirus Replicase in Viral Diversity and Adaptation
Abstract
1. Introduction
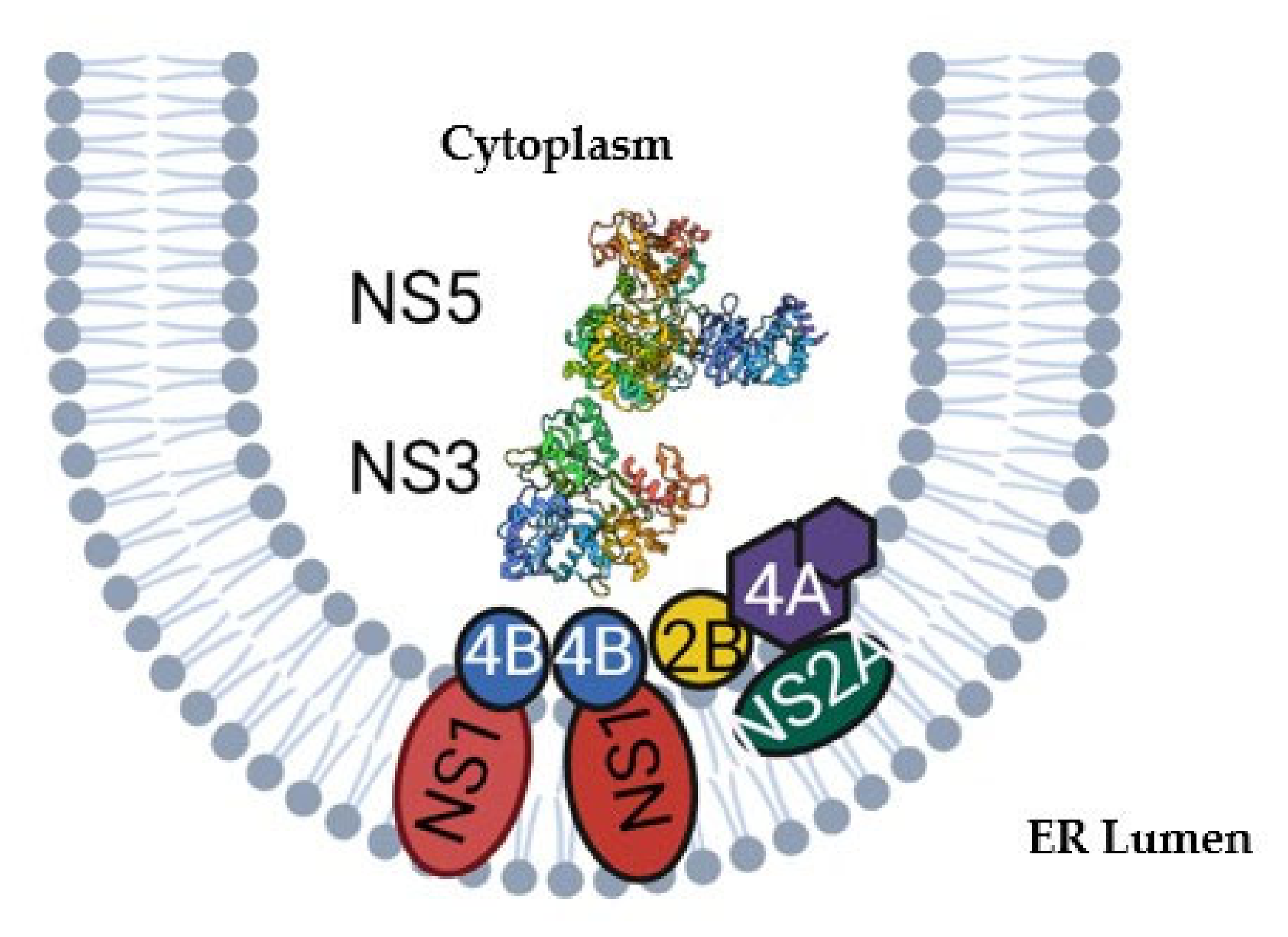
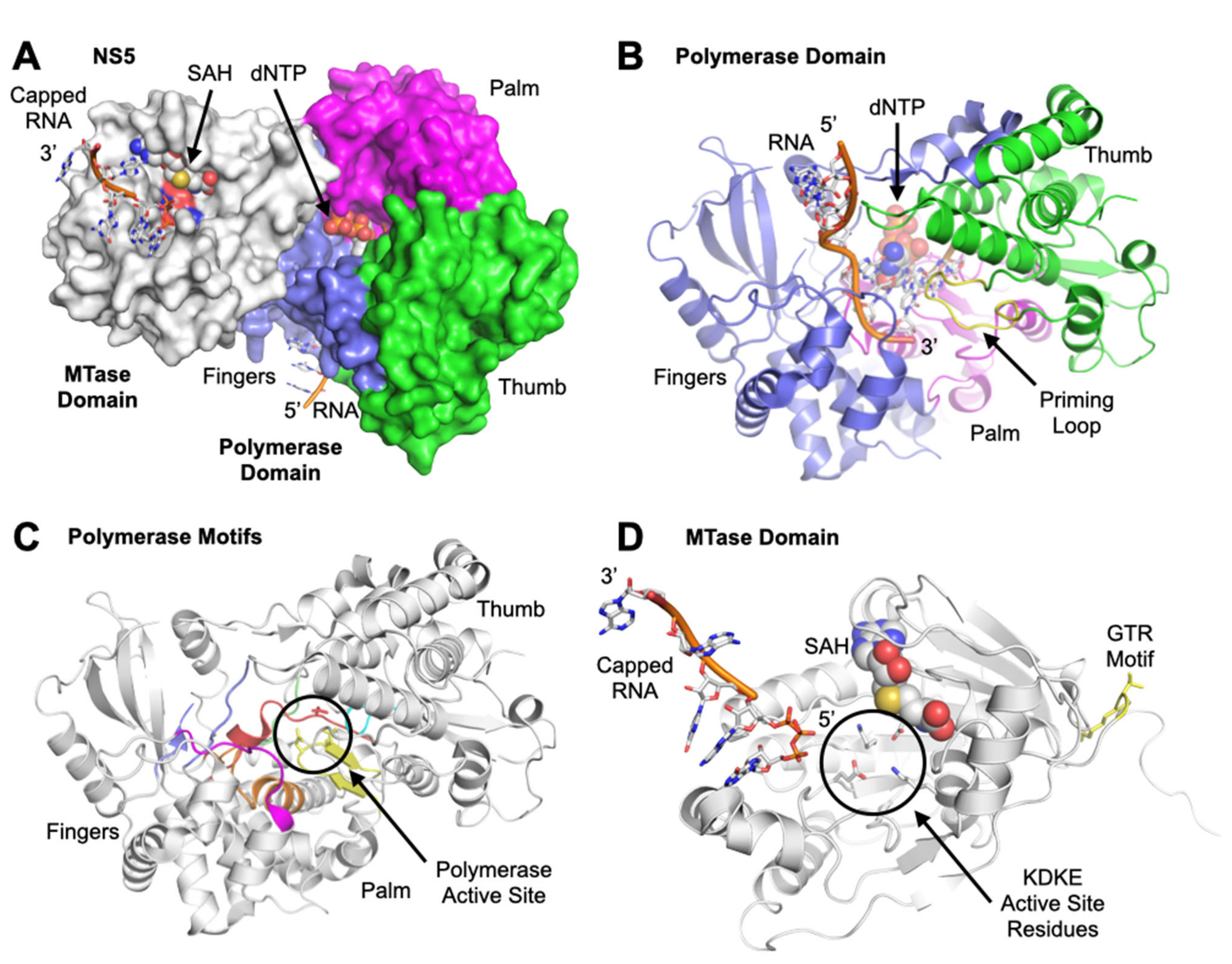
2. Structure and Function of NS3 and NS5
3. Fidelity
4. Quasispecies and Host Adaptation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kilpatrick, A.M. Globalization, land use, and the invasion of West Nile virus. Science 2011, 334, 323–327. [Google Scholar] [CrossRef]
- Kramer, L.D.; Styer, L.M.; Ebel, G.D. A Global Perspective on the Epidemiology of West Nile Virus. Annu. Rev. Entomol. 2007, 53, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Ronca, S.E.; Ruff, J.C.; Murray, K.O. A 20-year historical review of West Nile virus since its initial emergence in North America: Has West Nile virus become a neglected tropical disease? PLoS Negl. Trop. Dis. 2021, 15, e0009190. [Google Scholar] [CrossRef] [PubMed]
- Didier, M.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC); Brunette, G.W.; Nemhauser, J.B. CDC Yellow Book 2020 Health Information for International Travel: Health Information for International Travel; Oxford University Press: Oxford, UK, 2019. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Nwaiwu, A.U.; Musekiwa, A.; Tamuzi, J.L.; Sambala, E.Z.; Nyasulu, P.S. The incidence and mortality of yellow fever in Africa: A systematic review and meta-analysis. BMC Infect. Dis. 2021, 21, 1089. [Google Scholar] [CrossRef]
- Gardner, C.L.; Ryman, K.D. Yellow Fever: A Reemerging Threat. Clin. Lab. Med. 2010, 30, 237–260. [Google Scholar] [CrossRef] [PubMed]
- Erlanger, T.E.; Weiss, S.; Keiser, J.; Utzinger, J.; Wiedenmayer, K. Past, present, and future of Japanese encephalitis. Emerg. Infect. Dis. 2009, 15, 1–7. [Google Scholar] [CrossRef]
- Therkelsen, M.D.; Klose, T.; Vago, F.; Jiang, W.; Rossmann, M.G.; Kuhn, R.J. Flaviviruses have imperfect icosahedral symmetry. Proc. Natl. Acad. Sci. USA 2018, 115, 11608. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.; Soto-Acosta, R.; Yeh, S.; Schott-Lerner, G.; Pompon, J.; Sessions, O.; Bradrick, S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef]
- Chen, Y.; Maguire, T.; Hileman, R.E.; Fromm, J.; Esko, J.; Lindhardt, R.; Marks, R. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 1997, 3, 866–871. [Google Scholar] [CrossRef]
- Pujhari, S.; Brustolin, M.; Macias, V.M.; Nissly, R.; Nomura, M.; Kuchipudi, S.; Rasgon, J. Heat shock protein 70 (Hsp70) mediates Zika virus entry, replication, and egress from host cells. Emerg. Microbes Infect. 2019, 8, 8–16. [Google Scholar] [CrossRef]
- Reyes-Del Valle, J.; Chávez-Salinas, S.; Medina, F.; del Angel, R.M. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 2005, 79, 4557–4567. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Webb, A.I.; Chan, V.; Jumainsong, A.; Davidson, A.; Mongolsapaya, J.; Screaton, G. Lectin Switching During Dengue Virus Infection. J. Infect. Dis. 2011, 203, 1775–1783. [Google Scholar] [CrossRef]
- Meertens, L.; Carnec, X.; Lecoin, M.P.; Ramdasi, R.; Guivel-Benhassine, F.; Lew, E.; Lemke, G.; Schwartz, O.; Amara, A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012, 12, 544–557. [Google Scholar] [CrossRef]
- Navarro-Sanchez, E.; Altmeyer, R.; Amara, A.; Schwartz, O.; Fieschi, F.; Virelizier, J.; Arenzana-Seisdedos, F.; Desprès, P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003, 4, 723–728. [Google Scholar] [CrossRef]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.; et al. DC-SIGN (CD209) Mediates Dengue Virus Infection of Human Dendritic Cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef]
- Chu, J.J.H.; Ng, M.L. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 2004, 78, 10543–10555. [Google Scholar] [CrossRef]
- Hackett, B.A.; Cherry, S. Flavivirus internalization is regulated by a size-dependent endocytic pathway. Proc. Natl. Acad. Sci. USA 2018, 115, 4246–4251. [Google Scholar] [CrossRef]
- Acosta, E.G.; Castilla, V.; Damonte, E.B. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J. Gen. Virol. 2008, 89, 474–484. [Google Scholar] [CrossRef]
- Chiu, W.W.; Kinney, R.M.; Dreher, T.W. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J. Virol. 2005, 79, 8303–8315. [Google Scholar] [CrossRef]
- Sanford, T.J.; Mears, H.; Fajardo, T.; Locker, N.; Sweeney, T.R. Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res. 2019, 47, 9789–9802. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Linares, A.; Cahour, A.; Després, P.; Girard, M.; Bouloy, M. Processing of yellow fever virus polyprotein: Role of cellular proteases in maturation of the structural proteins. J. Virol. 1989, 63, 4199–4209. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Blanco, M.A.; Vasudevan, S.G.; Bradrick, S.S.; Nicchitta, C. Flavivirus RNA transactions from viral entry to genome replication. Antivir. Res. 2016, 134, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.W.; Campos, R.K.; Child, J.R.; Zheng, T.; Chan, K.; Bradrick, S.; Vasudevan, S.; Garcia-Blanco, M.; Nicchitta, C. Dengue Virus Selectively Annexes Endoplasmic Reticulum-Associated Translation Machinery as a Strategy for Co-opting Host Cell Protein Synthesis. J. Virol. 2018, 92, e01766-17. [Google Scholar] [CrossRef] [PubMed]
- Den Boon, J.A.; Ahlquist, P. Organelle-Like Membrane Compartmentalization of Positive-Strand RNA Virus Replication Factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.; Walther, P.; Fuller, S.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009, 5, 365–375. [Google Scholar] [CrossRef]
- Oliveira, E.R.A.; de Alencastro, R.B.; Horta, B.A.C. New insights into flavivirus biology: The influence of pH over interactions between prM and E proteins. J. Comput. Aided Mol. Des. 2017, 31, 1009–1019. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [CrossRef]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular Localization and Membrane Topology of the Dengue Virus Type 2 Non-structural Protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular Localization and Some Biochemical Properties of the Flavivirus Kunjin Nonstructural Proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.Y. Membrane topology and function of dengue virus NS2A protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the Dengue Virus Nonstructural Glycoprotein NS1 Suggests a Role in Viral RNA Replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Cyr, M.; Longenecker, K.; Tripathi, R.; Sun, C.; Kempf, D.J. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr. Sect. F 2017, 73, 116–122. [Google Scholar] [CrossRef]
- Tian, H.; Ji, X.; Yang, X.; Xie, W.; Yang, K.; Chen, C.; Wu, C.; Chi, H.; Mu, Z.; Wang, Z.; et al. The crystal structure of Zika virus helicase: Basis for antiviral drug design. Protein Cell 2016, 7, 450–454. [Google Scholar] [CrossRef]
- Koonin, E. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991, 72, 2197–2206. [Google Scholar] [CrossRef]
- Koonin, E. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and 2 protein of reovirus. J. Gen. Virol. 1993, 74, 733–740. [Google Scholar] [CrossRef]
- Issur, M.; Geiss, B.J.; Bougie, I.; Picard-Jean, F.; Despins, S.; Mayette, J.; Hobdey, S.; Bisaillon, M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 2009, 15, 2340–2350. [Google Scholar] [CrossRef]
- Egloff, M.P.; Benarroch, D.; Selisko, B.; Romette, J.L.; Canard, B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002, 21, 2757–2768. [Google Scholar] [CrossRef]
- Ray, D.; Shah, A.; Tilgner, M.; Guo, Y.; Zhao, Y.; Dong, H.; Deas, T.; Zhou, Y.; Li, H.; Shi, P. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006, 80, 8362–8370. [Google Scholar] [CrossRef]
- Tan, B.H.; Fu, J.; Sugrue, R.J.; Yap, E.H.; Chan, Y.C.; Tan, Y.H. Recombinant Dengue Type 1 Virus NS5 Protein Expressed inEscherichia coliExhibits RNA-Dependent RNA Polymerase Activity. Virology 1996, 216, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Ahola, T.; Kääriäinen, L. Reaction in alphavirus mRNA capping: Formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 1995, 92, 507–511. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Falgout, B.; Markoff, L.; Padmanabhan, R. In vitro RNA Synthesis from Exogenous Dengue Viral RNA Templates Requires Long Range Interactions between 5′- and 3′-Terminal Regions That Influence RNA Structure. J. Biol. Chem. 2001, 276, 15581–15591. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Khromykh, A.A. Kunjin RNA replication and applications of Kunjin replicons. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2003; Volume 59, pp. 99–140. [Google Scholar] [CrossRef]
- Bhatia, S.; Narayanan, N.; Nagpal, S.; Nair, D.T. Antiviral therapeutics directed against RNA dependent RNA polymerases from positive-sense viruses. Mol. Asp. Med. 2021, 81, 101005. [Google Scholar] [CrossRef]
- Ng, K.K.S.; Arnold, J.J.; Cameron, C.E. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top. Microbiol. Immunol. 2008, 320, 137–156. [Google Scholar] [CrossRef]
- Klema, V.J.; Ye, M.; Hindupur, A.; Teramoto, T.; Gottipati, K.; Padmanabhan, R.; Choi, K. Dengue Virus Nonstructural Protein 5 (NS5) Assembles into a Dimer with a Unique Methyltransferase and Polymerase Interface. PLoS Pathog. 2016, 12, e1005451. [Google Scholar] [CrossRef]
- Peng, G.; Peersen, O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 22505–22510. [Google Scholar] [CrossRef]
- Yap, L.J.; Luo, D.; Chung, K.Y.; Lim, S.P.; Bodenreider, C.; Noble, C.; Shi, P.Y.; Lescar, J.; Lescar, J. Crystal Structure of the Dengue Virus Methyltransferase Bound to a 5′-Capped Octameric RNA. PLoS ONE 2010, 5, e12836. [Google Scholar] [CrossRef]
- Lu, G.; Gong, P. Crystal Structure of the Full-Length Japanese Encephalitis Virus NS5 Reveals a Conserved Methyltransferase-Polymerase Interface. PLoS Pathog. 2013, 9, e1003549. [Google Scholar] [CrossRef]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.; Vaughan, R.; Sankaran, B.; Kao, C.; Li, P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef]
- Hansen, J.L.; Long, A.M.; Schultz, S.C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 1997, 5, 1109–1122. [Google Scholar] [CrossRef]
- Bruenn, J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003, 31, 1821–1829. [Google Scholar] [CrossRef]
- Selisko, B.; Papageorgiou, N.; Ferron, F.; Canard, B. Structural and Functional Basis of the Fidelity of Nucleotide Selection by Flavivirus RNA-Dependent RNA Polymerases. Viruses 2018, 10, 59. [Google Scholar] [CrossRef]
- Lohmann, V.; Körner, F.; Herian, U.; Bartenschlager, R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 1997, 71, 8416–8428. [Google Scholar] [CrossRef]
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Luo, D.; Xu, T.; Watson, R.P.; Scherer-Becker, D.; Sampath, A.; Jahnke, W.; Yeong, S.; Wang, C.; Lim, S.; Strongin, A.; et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008, 27, 3209–3219. [Google Scholar] [CrossRef]
- Kawaoka, J.; Jankowsky, E.; Pyle, A.M. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 2004, 11, 526–530. [Google Scholar] [CrossRef]
- Benarroch, D.; Egloff, M.P.; Mulard, L.; Guerreiro, C.; Romette, J.L.; Canard, B. A Structural Basis for the Inhibition of the NS5 Dengue Virus mRNA 2′-O-Methyltransferase Domain by Ribavirin 5′-Triphosphate. J. Biol. Chem. 2004, 279, 35638–35643. [Google Scholar] [CrossRef]
- Tackett, A.J.; Wei, L.; Cameron, C.E.; Raney, K.D. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001, 29, 565–572. [Google Scholar] [CrossRef]
- Taylor, S.D.; Solem, A.; Kawaoka, J.; Pyle, A.M. The NPH-II Helicase Displays Efficient DNA˙RNA Helicase Activity and a Pronounced Purine Sequence Bias. J. Biol. Chem. 2010, 285, 11692–11703. [Google Scholar] [CrossRef] [PubMed]
- Guenther, U.P.; Handoko, L.; Laggerbauer, B.; Jablonka, S.; Chari, A.; Alzheimer, M.; Ohmer, J.; Plöttner, O.; Gehring, N.; Sickmann, A.; et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1). Hum. Mol. Genet. 2009, 18, 1288–1300. [Google Scholar] [CrossRef] [PubMed]
- Chernov, A.; Shiryaev, S.A.; Aleshin, A.E.; Ratnikov, B.; Smith, J.; Liddington, R.; Strongin, A. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J. Biol. Chem. 2008, 283, 17270–17278. [Google Scholar] [CrossRef] [PubMed]
- Amberg, S.M.; Rice, C.M. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 1999, 73, 8083–8094. [Google Scholar] [CrossRef]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.; et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir. Res. 2010, 87, 125–148. [Google Scholar] [CrossRef]
- Wu, J.; Bera, A.K.; Kuhn, R.J.; Smith, J.L. Structure of the Flavivirus helicase: Implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 2005, 79, 10268–10277. [Google Scholar] [CrossRef]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.; Lescar, J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 2005, 79, 10278–10288. [Google Scholar] [CrossRef]
- Klema, V.J.; Padmanabhan, R.; Choi, K.H. Flaviviral replication complex: Coordination between RNA synthesis and 5′-RNA capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef]
- Martin, J.L.; McMillan, F.M. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011, 10, 51–65. [Google Scholar] [CrossRef]
- Cleaves, G.R.; Dubin, D.T. Methylation status of intracellular dengue type 2 40 S RNA. Virology 1979, 96, 159–165. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A. Host Factors Involved in West Nile Virus Replication. Ann. N. Y. Acad. Sci. 2001, 951, 207–219. [Google Scholar] [CrossRef]
- Brault, A.C.; Huang, C.Y.H.; Langevin, S.A.; Kinney, R.; Bowen, R.; Ramey, W.; Panella, N.; Holmes, E.; Powers, A.; Miller, B. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat. Genet. 2007, 39, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.A.; Bowen, R.A.; Reisen, W.K.; Andrade, C.; Ramey, W.; Maharaj, P.; Anishchenko, M.; Kenney, J.; Duggal, N.; Romo, H.; et al. Host Competence and Helicase Activity Differences Exhibited by West Nile Viral Variants Expressing NS3-249 Amino Acid Polymorphisms. PLoS ONE 2014, 9, e100802. [Google Scholar] [CrossRef] [PubMed]
- Du Pont, K.E.; Davidson, R.B.; McCullagh, M.; Geiss, B.J. Motif V regulates energy transduction between the flavivirus NS3 ATPase and RNA-binding cleft. J. Biol. Chem. 2020, 295, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Zhang, L.; Ramachandra, M.; Kusukawa, J.; Ebner, K.E.; Padmanabhan, R. Association between NS3 and NS5 Proteins of Dengue Virus Type 2 in the Putative RNA Replicase Is Linked to Differential Phosphorylation of NS5. J. Biol. Chem. 1995, 270, 19100–19106. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, G.; Zhang, B.; Gong, P. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J. Virol. 2015, 89, 249–261. [Google Scholar] [CrossRef]
- Zhao, Y.; Soh, T.S.; Zheng, J.; Chan, K.; Phoo, W.; Lee, C.; Tay, M.; Swaminathan, K.; Cornvik, T.; Lim, S.; et al. A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication. PLoS Pathog. 2015, 11, e1004682. [Google Scholar] [CrossRef]
- Li, X.D.; Shan, C.; Deng, C.L.; Ye, H.; Shi, P.; Yuan, Z.; Gong, P.; Zhang, B. The Interface between Methyltransferase and Polymerase of NS5 Is Essential for Flavivirus Replication. PLoS Negl. Trop. Dis. 2014, 8, e2891. [Google Scholar] [CrossRef]
- El Sahili, A.; Soh, T.S.; Schiltz, J.; Gharbi-Ayachi, A.; Seh, C.; Shi, P.; Lim, S.; Lescar, J. NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues. J. Virol. 2019, 94, e01294-19. [Google Scholar] [CrossRef]
- Drake, J.W. The Distribution of Rates of Spontaneous Mutation over Viruses, Prokaryotes, and Eukaryotes. Ann. N. Y. Acad. Sci. 1999, 870, 100–107. [Google Scholar] [CrossRef]
- Drake, J.W.; Holland, J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910. [Google Scholar] [CrossRef]
- John, H.; Katherine, S.; Frank, H.; Elizabeth, G.; Stuart, N.; Scott, V. Rapid Evolution of RNA Genomes. Science 1982, 215, 1577–1585. [Google Scholar] [CrossRef]
- Pfeiffer, J.K.; Kirkegaard, K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 2003, 100, 7289. [Google Scholar] [CrossRef]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006, 439, 344–348. [Google Scholar] [CrossRef]
- Van Slyke, G.A.; Ciota, A.T.; Willsey, G.G.; Jaeger, J.; Shi, P.Y.; Kramer, L.D. Point mutations in the West Nile virus (Flaviviridae; Flavivirus) RNA-dependent RNA polymerase alter viral fitness in a host-dependent manner in vitro and in vivo. Virology 2012, 427, 18–24. [Google Scholar] [CrossRef]
- Ciota, A.T.; Ehrbar, D.J.; van Slyke, G.A.; Willsey, G.G.; Kramer, L.D. Cooperative interactions in the West Nile virus mutant swarm. BMC Evol. Biol. 2012, 12, 58. [Google Scholar] [CrossRef]
- Griesemer, S.B.; Kramer, L.D.; van Slyke, G.A.; Pata, J.; Gohara, D.; Cameron, C.; Ciota, A. Mutagen resistance and mutation restriction of St. Louis encephalitis virus. J. Gen. Virol. 2017, 98, 201–211. [Google Scholar] [CrossRef]
- Caldwell, H.S.; Ngo, K.; Pata, J.D.; Kramer, L.D.; Ciota, A.T. West Nile Virus fidelity modulates the capacity for host cycling and adaptation. J. Gen. Virol. 2020, 101, 410–419. [Google Scholar] [CrossRef]
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J.J.; Ahlquist, P. RNA Genetics; CRC Press Inc.: Boca Raton, FL, USA, 1988. [Google Scholar]
- Qiu, L.; Patterson, S.E.; Bonnac, L.F.; Geraghty, R.J. Nucleobases and corresponding nucleosides display potent antiviral activities against dengue virus possibly through viral lethal mutagenesis. PLoS Negl. Trop. Dis. 2018, 12, e0006421. [Google Scholar] [CrossRef]
- Agudo, R.; Arias, A.; Domingo, E. 5-fluorouracil in lethal mutagenesis of foot-and-mouth disease virus. Future Med. Chem. 2009, 1, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S.; Maag, D.; Arnold, J.J.; Zhong, W.; Lau, J.; Hong, Z.; Andino, R.; Cameron, C. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000, 6, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Escribano-Romero, E.; Jiménez de Oya, N.; Domingo, E.; Saiz, J.C. Extinction of West Nile Virus by Favipiravir through Lethal Mutagenesis. Antimicrob. Agents Chemother. 2017, 61, e01400-17. [Google Scholar] [CrossRef] [PubMed]
- Deval, J.; Symons, J.A.; Beigelman, L. Inhibition of viral RNA polymerases by nucleoside and nucleotide analogs: Therapeutic applications against positive-strand RNA viruses beyond hepatitis C virus. Curr. Opin. Virol. 2014, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Beeharry, Y.; Bordería, A.; Blanc, H.; Vignuzzi, M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. USA 2011, 108, 16038. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Vignuzzi, M.; Stone, J.K.; Andino, R.; Cameron, C.E. Remote Site Control of an Active Site Fidelity Checkpoint in a ViralRNA-dependent RNAPolymerase. J. Biol. Chem. 2005, 280, 25706–25716. [Google Scholar] [CrossRef]
- Gnädig, N.F.; Beaucourt, S.; Campagnola, G.; Borderia, A.; Sanz-Ramos, M.; Gong, P.; Blanc, H.; Peerson, O.; Vignuzzi, M. Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E2294. [Google Scholar] [CrossRef]
- McDonald, S.; Block, A.; Beaucourt, S.; Moratorio, G.; Vignuzzi, M.; Peersen, O.B. Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in vivo. J. Biol. Chem. 2016, 291, 13999–14011. [Google Scholar] [CrossRef]
- Van Slyke, G.A.; Arnold, J.J.; Lugo, A.J.; Griesemer, S.B.; Moustafa, I.M.; Kramer, L.D.; Cameron, C.E.; Ciota, A.T. Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes. PLoS Pathog. 2015, 11, e1005009. [Google Scholar] [CrossRef]
- Castro, C.; Arnold, J.J.; Cameron, C.E. Incorporation fidelity of the viral RNA-dependent RNA polymerase: A kinetic, thermodynamic and structural perspective. Virus Res. 2005, 107, 141–149. [Google Scholar] [CrossRef]
- Xie, X.; Wang, H.; Zeng, J.; Li, C.; Zhou, G.; Yang, D.; Yu, L. Foot-and-mouth disease virus low-fidelity polymerase mutants are attenuated. Arch. Virol. 2014, 159, 2641–2650. [Google Scholar] [CrossRef]
- Rai, D.K.; Diaz-San Segundo, F.; Campagnola, G.; Keith, A.; Schafer, E.A.; Campagnola, G.; Kloc, A.; de Los Santos, T.; Peersen, O.; Rieder, E. Attenuation of Foot-and-Mouth Disease Virus by Engineered Viral Polymerase Fidelity. J. Virol. 2017, 91, e00081-17. [Google Scholar] [CrossRef]
- Rozen-Gagnon, K.; Stapleford, K.A.; Mongelli, V.; Blanc, H.; Failloux, A.; Saleh, M.; Vignuzzi, M. Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models. PLoS Pathog. 2014, 10, e1003877. [Google Scholar] [CrossRef]
- Blair, C.D. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011, 6, 265–277. [Google Scholar] [CrossRef]
- Kautz, T.F.; Forrester, N.L. RNA virus fidelity mutants: A useful tool for evolutionary biology or a complex challenge? Viruses 2018, 10, 600. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies Theory and the Behavior of RNA Viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Viral quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef]
- Wilke, C.O.; Wang, J.L.; Ofria, C.; Lenski, R.E.; Adami, C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 2001, 412, 331–333. [Google Scholar] [CrossRef]
- Duarte, E.; Clarke, D.; Moya, A.; Domingo, E.; Holland, J. Rapid fitness losses in mammalian RNA virus clones due to Mullers ratchet. Proc. Natl. Acad. Sci. USA 1992, 89, 6015. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.L.; Roosinck, M.J. Genetic Diversity in RNA Virus Quasispecies Is Controlled by Host-Virus Interactions. J. Virol. 2001, 75, 6566–6571. [Google Scholar] [CrossRef]
- Novella, I.; Quer, J.; Domingo, E.; Holland, J.J. Exponential Fitness Gains of RNA Virus Populations Are Limited by Bottleneck Effects. J. Virol. 1999, 73, 1668–1671. [Google Scholar] [CrossRef]
- Borrow, P.; Shaw, G.M. Cytotoxic T-lymphocyte escape viral variants: How important are they in viral evasion of immune clearance in vivo? Immunol Rev. 1998, 164, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Escarmís, C.; Lázaro, E.; Arias, A.; Domingo, E. Repeated Bottleneck Transfers Can Lead to Non-cytocidal Forms of a Cytopathic Virus: Implications for Viral Extinction. J. Mol. Biol. 2008, 376, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; McCaskill, J.; Schuster, P. Molecular quasi-species. J. Phys. Chem. 1988, 92, 6881–6891. [Google Scholar] [CrossRef]
- Escarmis, C.; Perales, C.; Esteban, D. Biological Effect of Muller’s Ratchet: Distant Capsid Site Can Affect Picornavirus Protein Processing. J. Virol. 2009, 83, 6748–6756. [Google Scholar] [CrossRef] [PubMed]
- De Ávila, A.I.; Gallego, I.; Soria, M.E.; Gregori, J.; Quer, J.; Esteban, J.; Rice, C.; Domingo, E.; Perales, C. Lethal Mutagenesis of Hepatitis C Virus Induced by Favipiravir. PLoS ONE 2016, 11, e0164691. [Google Scholar] [CrossRef]
- Dapp, M.J.; Patterson, S.E.; Mansky, L.M. Back to the future: Revisiting HIV-1 lethal mutagenesis. Trends Microbiol. 2013, 21, 56–62. [Google Scholar] [CrossRef]
- Bassi, M.R.; Sempere, R.N.; Meyn, P.; Polacek, C.; Arias, A. Extinction of Zika Virus and Usutu Virus by Lethal Mutagenesis Reveals Different Patterns of Sensitivity to Three Mutagenic Drugs. Antimicrob. Agents Chemother. 2018, 62, e00380-18. [Google Scholar] [CrossRef]
- Borrow, P.; Lewicki, H.; Wei, X.; Horwitz, M.S.; Peffer, N.; Mayers, H.; Nelson, J.A.; Gairin, J.E.; Hahn, B.H.; Oldstone, M.; et al. Antiviral pressure exerted by HIV-l-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 1997, 3, 205–211. [Google Scholar] [CrossRef]
- Weaver, S.C. Evolutionary influences in arboviral disease. Curr. Top. Microbiol. Immunol. 2006, 299, 285–314. [Google Scholar] [CrossRef]
- Ciota, A.T.; Lovelace, A.O.; Jia, Y.; Davis, L.J.; Young, D.S.; Kramer, L.D. Characterization of mosquito-adapted West Nile virus. J. Gen. Virol. 2008, 89 (Pt 7), 1633–1642. [Google Scholar] [CrossRef]
- Ciota, A.T.; Lovelace, A.O.; Jones, S.A.; Payne, A.; Kramer, L.D. Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. J. Gen. Virol. 2007, 88, 2398–2406. [Google Scholar] [CrossRef]
- Deardorff, E.R.; Fitzpatrick, K.A.; Jerzak, G.V.S.; Shi, P.Y.; Kramer, L.D.; Ebel, G.D. West Nile Virus Experimental Evolution in vivo and the Trade-off Hypothesis. PLoS Pathog. 2011, 7, e1002335. [Google Scholar] [CrossRef]
- Jerzak, G.; Bernard, K.A.; Kramer, L.D.; Ebel, G.D. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J. Gen. Virol. 2005, 86, 2175–2183. [Google Scholar] [CrossRef]
- Ciota, A.T.; Jia, Y.; Payne, A.F.; Jerzak, G.; Davis, L.; Young, S.D.; Ehrbar, D.; Kramer, L.D. Experimental Passage of St. Louis Encephalitis Virus In vivo in Mosquitoes and Chickens Reveals Evolutionarily Significant Virus Characteristics. PLoS ONE 2009, 4, e7876. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Smith, D.R.; Brackney, D.E.; Bosco-Lauth, A.; Fauver, J.; Campbell, C.; Felix, T.; Romo, H.; Duggal, N.; Dietrich, E.; et al. Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness. PLoS Pathog. 2015, 11, e1004874. [Google Scholar] [CrossRef]
- Caldwell, H.S.; Lasek-Nesselquist, E.; Follano, P.; Kramer, L.D.; Ciota, A.T. Divergent mutational landscapes of consensus and minority genotypes of west nile virus demonstrate host and gene-specific evolutionary pressures. Genes 2020, 11, 1299. [Google Scholar] [CrossRef]
- Brackney, D.E.; Schirtzinger, E.E.; Harrison, T.D.; Ebel, G.D.; Hanley, K.A. Modulation of flavivirus population diversity by RNA interference. J. Virol. 2015, 89, 4035–4039. [Google Scholar] [CrossRef]
- Brackney, D.E.; Beane, J.E.; Ebel, G.D. RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification. PLoS Pathog. 2009, 5, e1000502. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Rückert, C.; Armstrong, P.M.; Bransfield, A.; Anderson, J.; Ebel, G.; Brackney, D. Transmission bottlenecks and RNAi collectively influence tick-borne flavivirus evolution. Virus Evol. 2016, 2, vew033. [Google Scholar] [CrossRef] [PubMed]
- Moudy, R.M.; Meola, M.A.; Morin, L.L.L.; Ebel, G.D.; Kramer, L.D. A Newly Emergent Genotype of West Nile Virus Is Transmitted Earlier and More Efficiently by Culex Mosquitoes. Am. J. Trop. Med. Hyg. 2007, 77, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Bialosuknia, S.M.; Tan, Y.; Zink, S.D.; Koetzner, C.; Maffei, J.; Halpin, R.; Mueller, E.; Novotny, M.; Shilts, M.; Fedorova, N.; et al. Evolutionary dynamics and molecular epidemiology of West Nile virus in New York State: 1999–2015. Virus Evol. 2019, 5, vez020. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Xing-Yao, H.; Zhong-Yu, L.; Feng, Z.; Xing-Liang, Z.; Jiu-Yang, Y.; Xue, J.; Yan-Peng, X.; Guanghui, L.; Cui, L.; et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 2017, 358, 933–936. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Du, S.; Shan, C.; Nie, K.; Zhang, R.; Li, X.; Zhang, R.; Wang, T.; Qin, C.; et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature 2017, 545, 482–486. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldwell, H.S.; Pata, J.D.; Ciota, A.T. The Role of the Flavivirus Replicase in Viral Diversity and Adaptation. Viruses 2022, 14, 1076. https://doi.org/10.3390/v14051076
Caldwell HS, Pata JD, Ciota AT. The Role of the Flavivirus Replicase in Viral Diversity and Adaptation. Viruses. 2022; 14(5):1076. https://doi.org/10.3390/v14051076
Chicago/Turabian StyleCaldwell, Haley S., Janice D. Pata, and Alexander T. Ciota. 2022. "The Role of the Flavivirus Replicase in Viral Diversity and Adaptation" Viruses 14, no. 5: 1076. https://doi.org/10.3390/v14051076
APA StyleCaldwell, H. S., Pata, J. D., & Ciota, A. T. (2022). The Role of the Flavivirus Replicase in Viral Diversity and Adaptation. Viruses, 14(5), 1076. https://doi.org/10.3390/v14051076




