Characterization of Vaccine Breakthrough Cases during Measles Outbreaks in Milan and Surrounding Areas, Italy, 2017–2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Molecular and Serological Methods
2.3. Statistical Analysis
2.4. Phylogenetic Analysis of H Gene Sequences
3. Results
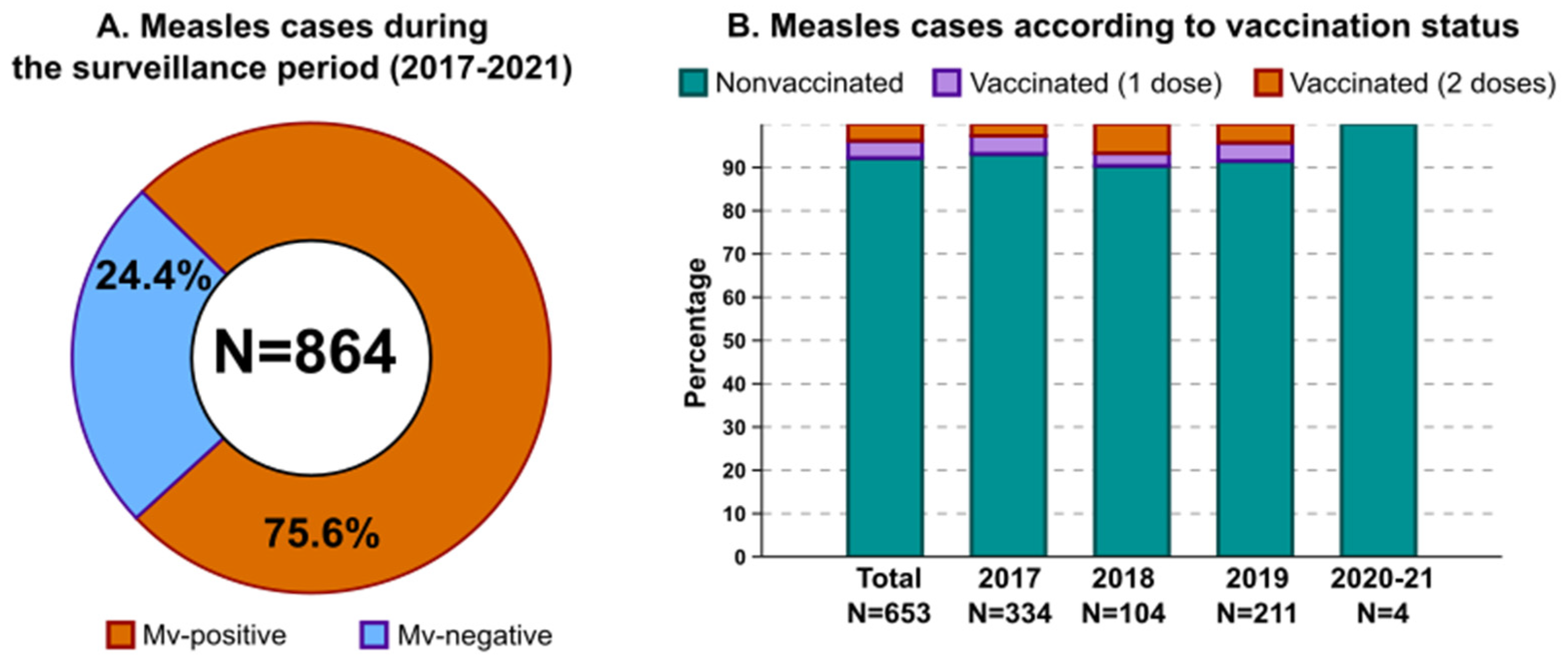
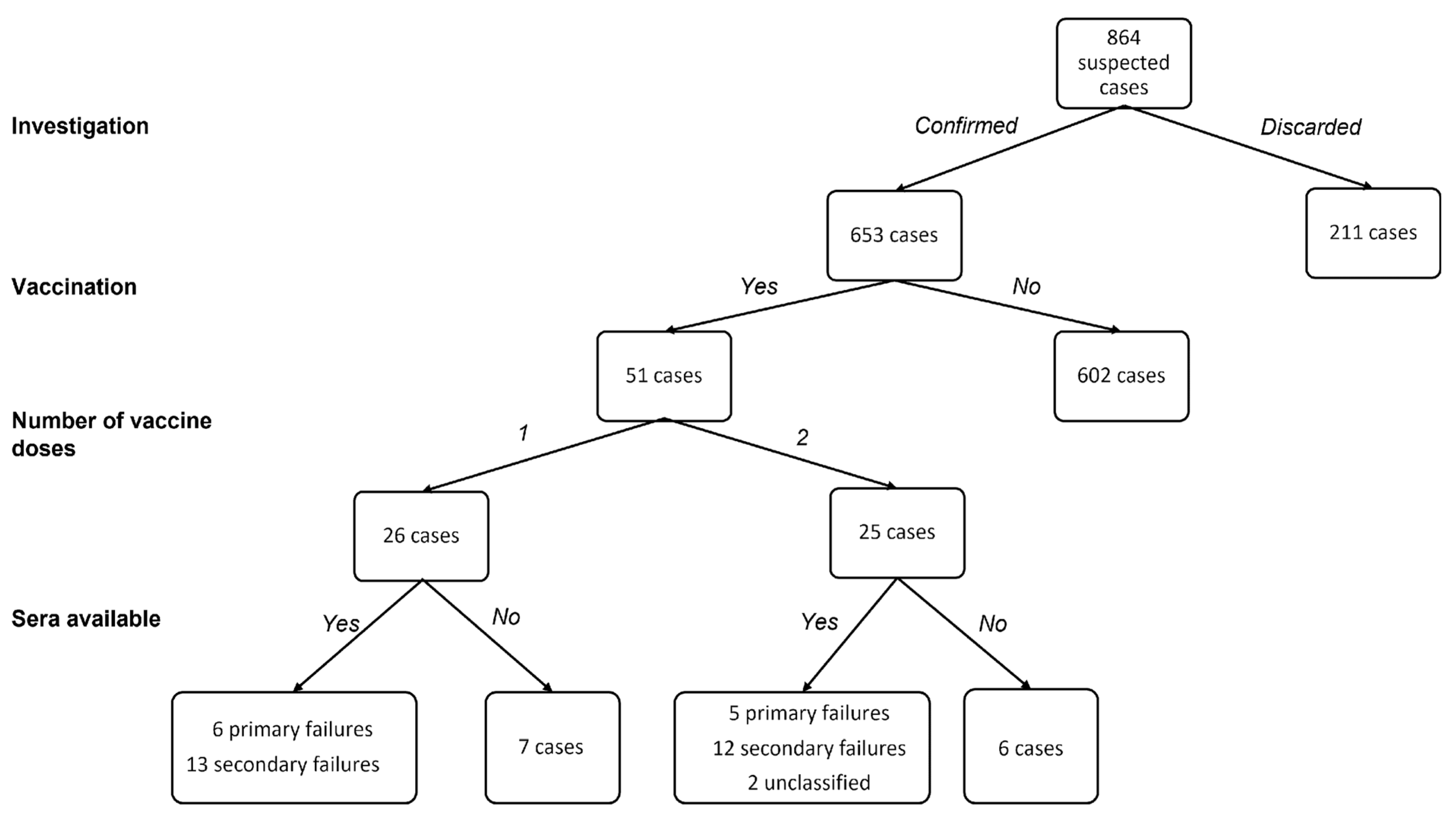
3.1. Measles Epidemiology in the Surveilled Area
3.2. Serological and Clinical Profiles of Breakthrough Cases
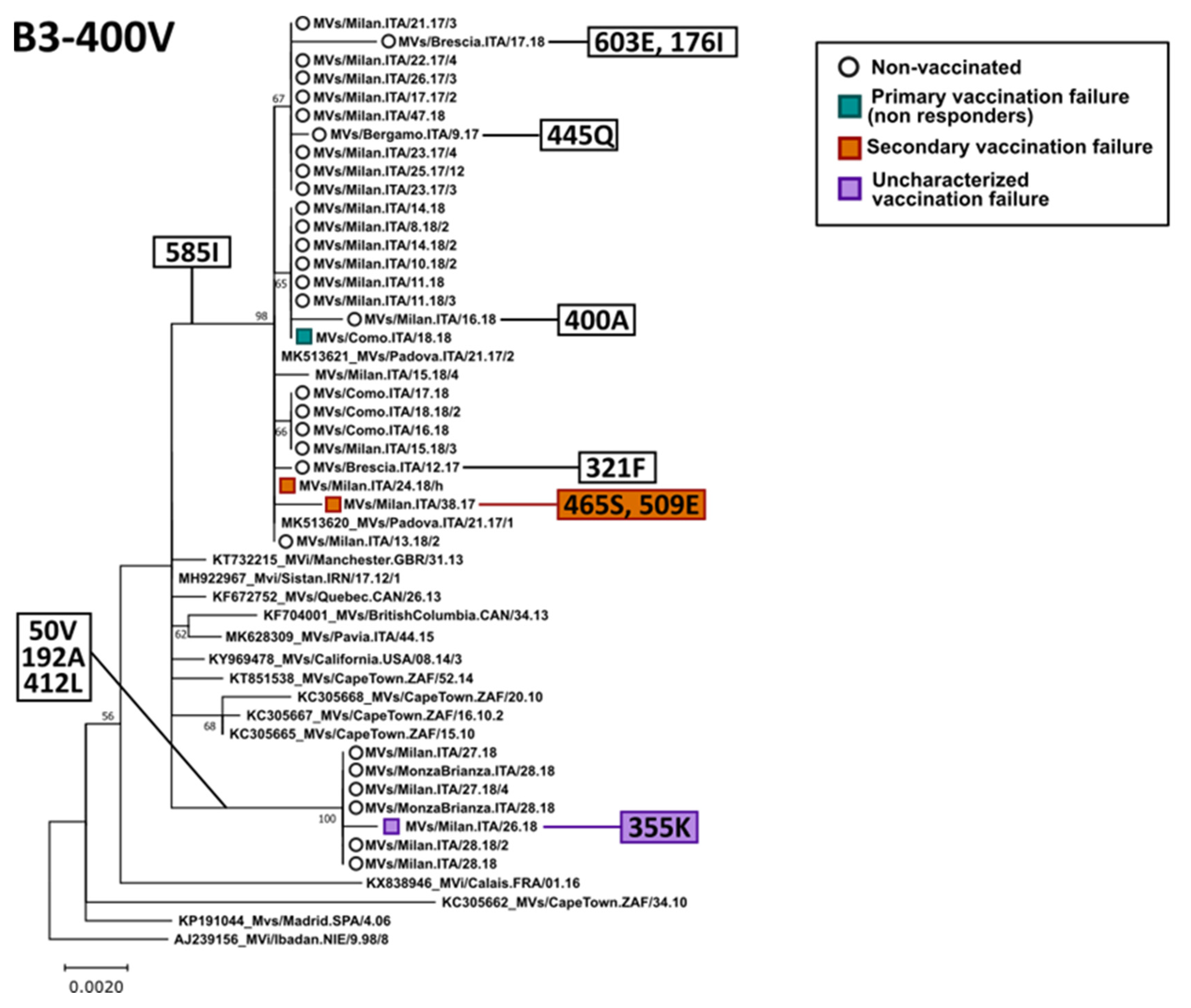
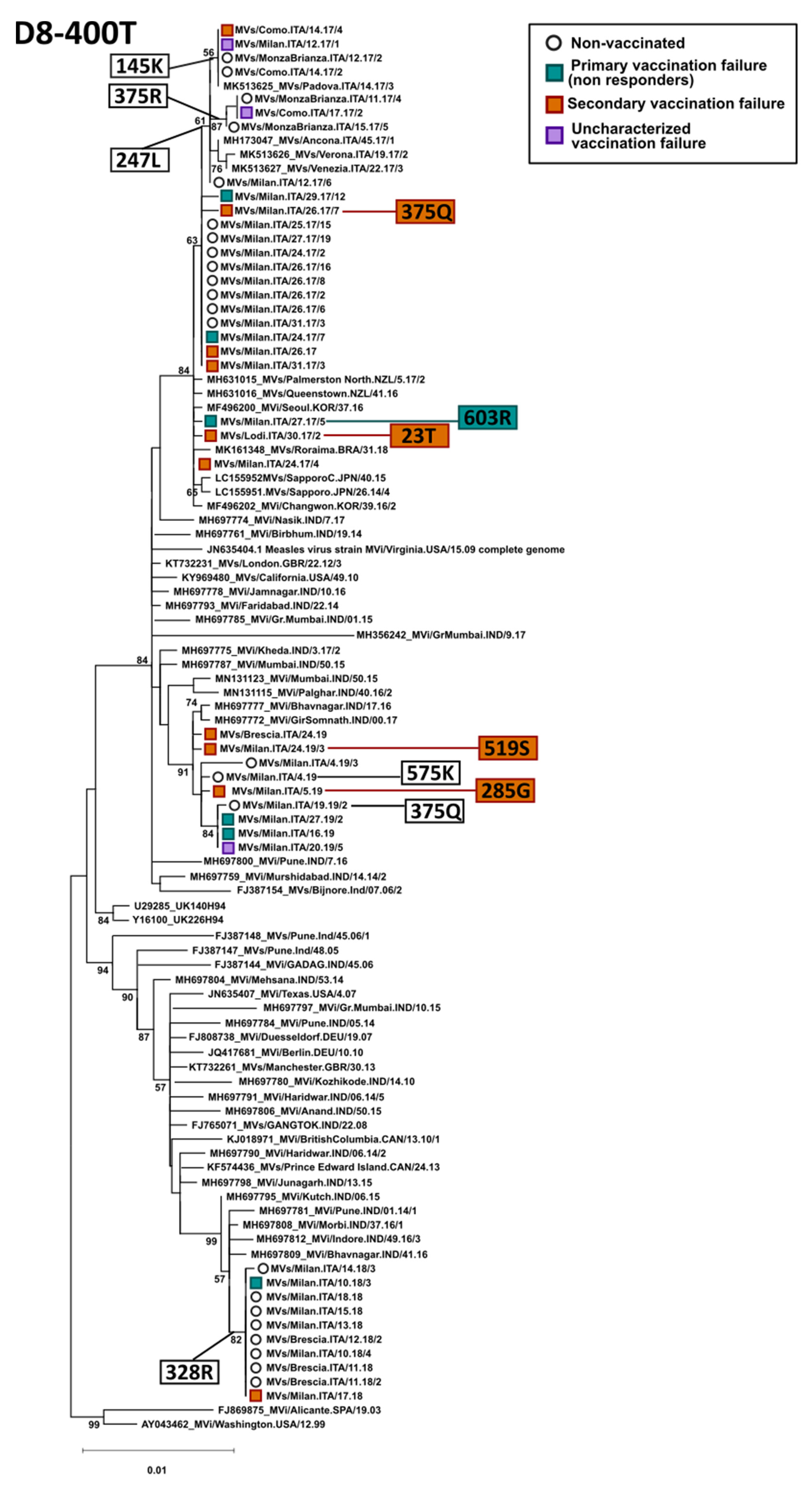
3.3. Phylogenetic Analysis of the H Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Newsroom. Fatcts in Pictures. Detail. Measles. Available online: https://www.who.int/news-room/facts-in-pictures/detail/measles (accessed on 26 January 2022).
- World Health Organization. Measles and Rubella Strategic Framework: 2021–2030. Available online: https://www.who.int/publications-detail-redirect/measles-and-rubella-strategic-framework-2021-2030 (accessed on 4 January 2022).
- Filia, A.; Bella, A.; Del Manso, M.; Rota, M.C. Who Is at Risk for Measles in Italy? Continued Measles Outbreaks in 2019 and Barriers to Elimination. Int. J. Infect. Dis. 2020, 101, 354. [Google Scholar] [CrossRef]
- Adamo, G.; Baccolini, V.; Massimi, A.; Barbato, D.; Cocchiara, R.; Di Paolo, C.; Mele, A.; Cianfanelli, S.; Angelozzi, A.; Castellani, F.; et al. Towards Elimination of Measles and Rubella in Italy: Progress and Challenges. PLoS ONE 2019, 14, e0226513. [Google Scholar] [CrossRef] [PubMed]
- Istituto Superiore di Sanità. EpiCentro. Copertura Vaccinale in Italia. Available online: https://www.epicentro.iss.it/vaccini/dati_Ita (accessed on 4 January 2022).
- European Centre for Diseases Prevention and Control. Vaccine Scheduler. Available online: https://vaccine-schedule.ecdc.europa.eu/Scheduler/ByCountry?SelectedCountryId=103&IncludeChildAgeGroup=true&IncludeAdultAgeGroup=true&SelectedVersionId=27 (accessed on 4 January 2022).
- Istituto Superiore di Sanità. EpiCentro. Vaccinazioni: Il Decreto Legge 73/2017. Available online: https://www.epicentro.iss.it/vaccini/DecretoVaccinazioni2017 (accessed on 26 February 2022).
- Centers for Disease Control and Prevention. Genetic Analysis of Measles Virus. Available online: https://www.cdc.gov/measles/lab-tools/genetic-analysis.html (accessed on 26 February 2022).
- Hashiguchi, T.; Maenaka, K.; Yanagi, Y. Measles Virus Hemagglutinin: Structural Insights into Cell Entry and Measles Vaccine. Front. Microbiol. 2011, 2, 247. [Google Scholar] [CrossRef]
- Baldo, A.; Galanis, E.; Tangy, F.; Herman, P. Biosafety Considerations for Attenuated Measles Virus Vectors Used in Virotherapy and Vaccination. Hum. Vaccin. Immunother. 2016, 12, 1102–1116. [Google Scholar] [CrossRef] [PubMed]
- Isituto Superiore di Sanità. EpiCentro. Morbillo Rosolia News: Il Bollettino della Sorveglianza Integrata Morbillo-Rosolia. Available online: https://www.epicentro.iss.it/morbillo/bollettino (accessed on 4 January 2022).
- Nicolay, N.; Mirinaviciute, G.; Mollet, T.; Celentano, L.P.; Bacci, S. Epidemiology of Measles during the COVID-19 Pandemic, a Description of the Surveillance Data, 29 EU/EEA Countries and the United Kingdom, January to May 2020. Euro Surveill. 2020, 25, 2001390. [Google Scholar] [CrossRef]
- Amendola, A.; Bianchi, S.; Frati, E.R.; Ciceri, G.; Faccini, M.; Senatore, S.; Colzani, D.; Lamberti, A.; Baggieri, M.; Cereda, D.; et al. Ongoing Large Measles Outbreak with Nosocomial Transmission in Milan, Northern Italy, March–August 2017. Euro Surveill. 2017, 22, 30596. [Google Scholar] [CrossRef]
- Javelle, E.; Colson, P.; Parola, P.; Raoult, D. Measles, the Need for a Paradigm Shift. Eur. J. Epidemiol. 2019, 34, 897–915. [Google Scholar] [CrossRef]
- Wiedermann, U.; Garner-Spitzer, E.; Wagner, A. Primary Vaccine Failure to Routine Vaccines: Why and What to Do? Hum. Vaccin. Immunother. 2016, 12, 239–243. [Google Scholar] [CrossRef]
- Sniadack, D.H.; Crowcroft, N.S.; Durrheim, D.N.; Rota, P.A. Roadmap to Elimination—Standard Measles and Rubella Surveillance. Wkly. Epidemiol. Rec. 2017, 92, 97105. [Google Scholar]
- Pannuti, C.S.; Morello, R.J.; de Moraes, J.C.; Curti, S.P.; Afonso, A.M.S.; Camargo, M.C.C.; de Souza, V.A.U.F. Identification of Primary and Secondary Measles Vaccine Failures by Measurement of Immunoglobulin G Avidity in Measles Cases during the 1997 São Paulo Epidemic. Clin. Diagn. Lab. Immunol. 2004, 11, 119–122. [Google Scholar] [CrossRef]
- Bouche, F.B.; Ertl, O.T.; Muller, C.P. Neutralizing B Cell Response in Measles. Viral Immunol. 2002, 15, 451–471. [Google Scholar] [CrossRef]
- Hübschen, J.M.; Kremer, J.R.; De Landtsheer, S.; Muller, C.P. A Multiplex TaqMan PCR Assay for the Detection of Measles and Rubella Virus. J. Virol. Methods 2008, 149, 246–250. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Manual for the Laboratory-Based Surveillance of Measles, Rubella, and Congenital Rubella Syndrome, 3rd ed.; WHO: Geneva, Switzerland, 2018; Available online: https://www.who.int/immunization/monitoring_surveillance/burden/laboratory/manual/en/ (accessed on 10 December 2021).
- Bianchi, S.; Frati, E.R.; Lai, A.; Colzani, D.; Ciceri, G.; Baggieri, M.; Lamberti, A.; Senatore, S.; Faccini, M.; Mazzilli, F.; et al. Genetic Characterisation of Measles Virus Variants Identified during a Large Epidemic in Milan, Italy, March–December 2017. Epidemiol. Infect. 2019, 147, e80. [Google Scholar] [CrossRef] [PubMed]
- Chibo, D.; Birch, C.J.; Rota, P.A.; Catton, M.G. Molecular Characterization of Measles Viruses Isolated in Victoria, Australia, between 1973 and 1998. J. Gen. Virol. 2000, 81, 2511–2518. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, S.; Canuti, M.; Ciceri, G.; Gori, M.; Colzani, D.; Dura, M.; Pennati, B.M.; Baggieri, M.; Magurano, F.; Tanzi, E.; et al. Molecular Epidemiology of B3 and D8 Measles Viruses through Hemagglutinin Phylogenetic History. Int. J. Mol. Sci. 2020, 21, 4435. [Google Scholar] [CrossRef] [PubMed]
- Bellini, W.J.; Icenogle, J.P. Measles and Rubella Virus. In Manual of Clinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R.H., Eds.; ASM Press: Washington, DC, USA, 2003; pp. 1389–1403. [Google Scholar]
- Dean, A.G.; Sullivan, K.M.; Soe, M.M. OpenEpi: Open Source Epidemiologic Statistics for Public Health. Available online: https://www.openepi.com (accessed on 16 July 2021).
- Ciceri, G.; Canuti, M.; Bianchi, S.; Gori, M.; Piralla, A.; Colzani, D.; Libretti, M.; Frati, E.R.; Baggieri, M.; Lai, A.; et al. Genetic Variability of the Measles Virus Hemagglutinin Gene in B3 Genotype Strains Circulating in Northern Italy. Infect. Genet. Evol. 2019, 75, 103943. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Rota, P.A.; Brown, K.; Mankertz, A.; Santibanez, S.; Shulga, S.; Muller, C.P.; Hübschen, J.M.; Siqueira, M.; Beirnes, J.; Ahmed, H.; et al. Global Distribution of Measles Genotypes and Measles Molecular Epidemiology. J. Infect. Dis. 2011, 204, S514–S523. [Google Scholar] [CrossRef]
- World Health Organization. Measles Vaccines: WHO Position Paper—April 2017. Available online: https://www.who.int/publications-detail-redirect/WER9217 (accessed on 26 February 2022).
- Felsenstein, J. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Kennedy, R.B.; Ovsyannikova, I.G.; Whitaker, J.A.; Poland, G.A. Variability in Humoral Immunity to Measles Vaccine: New Developments. Trends. Mol. Med. 2015, 21, 789–801. [Google Scholar] [CrossRef]
- Carazo Perez, S.; De Serres, G.; Bureau, A.; Skowronski, D.M. Reduced Antibody Response to Infant Measles Vaccination: Effects Based on Type and Timing of the First Vaccine Dose Persist After the Second Dose. Clin. Infect. Dis. 2017, 65, 1094–1102. [Google Scholar] [CrossRef]
- Sundell, N.; Dotevall, L.; Sansone, M.; Andersson, M.; Lindh, M.; Wahlberg, T.; Tyrberg, T.; Westin, J.; Liljeqvist, J.-Å.; Bergström, T.; et al. Measles Outbreak in Gothenburg Urban Area, Sweden, 2017 to 2018: Low Viral Load in Breakthrough Infections. Euro Surveill. 2019, 24, 1900114. [Google Scholar] [CrossRef] [PubMed]
- Arima, Y.; Oishi, K. Letter to the Editor: Measles Cases among Fully Vaccinated Persons. Euro Surveill. 2018, 23, 1800449. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.D.; Zahn, M. Clinical Characteristics of Measles in Previously Vaccinated and Unvaccinated Patients in California. Clin. Infect. Dis. 2018, 67, 1315–1319. [Google Scholar] [CrossRef]
- Muscat, M.; Marinova, L.; Mankertz, A.; Gatcheva, N.; Mihneva, Z.; Santibanez, S.; Kunchev, A.; Filipova, R.; Kojouharova, M. The Measles Outbreak in Bulgaria, 2009–2011: An Epidemiological Assessment and Lessons Learnt. Euro Surveill. 2016, 21, 30152. [Google Scholar] [CrossRef]
- Komitova, R.; Kevorkyan, A.; Boykinova, O.; Krumova, S.; Atanasova, M.; Raycheva, R.; Stoilova, Y.; Kunchev, A. Difficulties in Achieving and Maintaining the Goal of Measles Elimination in Bulgaria. Rev. Epidemiol. Sante Publique 2019, 67, 155–162. [Google Scholar] [CrossRef]
- Bianchi, S.; Faccini, M.; Lamberti, A.; Senatore, S.; Civeri, G.; Frati, E.R.; Colzani, D.; Gori, M.; Cereda, D.; Gramegna, M.; et al. Measles Surveillance Activities in the Metropolitan Area of Milan during 2017–2018. J. Prev. Med. Hyg. 2019, 60, E286–E292. [Google Scholar] [CrossRef]
- World Health Organization = Organisation Mondiale de la Santé. Weekly Epidemiological Record = Relevé Épidémiologique Hebdomadaire, 2020, 95, 48, 585–608. Available online: https://apps.who.int/iris/handle/10665/337100 (accessed on 31 January 2022).
- Mulholland, K.; Kretsinger, K.; Wondwossen, L.; Crowcroft, N. Action Needed Now to Prevent Further Increases in Measles and Measles Deaths in the Coming Years. Lancet 2020, 396, 1782–1784. [Google Scholar] [CrossRef]
- Durrheim, D.N.; Andrus, J.K.; Tabassum, S.; Bashour, H.; Githanga, D.; Pfaff, G. A Dangerous Measles Future Looms beyond the COVID-19 Pandemic. Nat. Med. 2021, 27, 360–361. [Google Scholar] [CrossRef] [PubMed]
| Breakthrough Cases | |||||||
|---|---|---|---|---|---|---|---|
| Genotype | Total Cases | Cases | Outbreaks | ||||
| Vaccinated (%) | Sporadic Vaccinated | N Outbreaks | Index Cases | Subjects Involved (Vaccinated; Not Vaccinated) | Mean Outbreak Size | ||
| D8 | 438 | 28 (6.4) | 8 | 15 | 5 | 20; 31 | 3.4 |
| B3 | 160 | 8 (5.0) | 3 | 5 | 3 | 5; 12 | 3.4 |
| NT 1 | 55 | 15 (27.3) | 15 | 0 | 0 | 0 | 0 |
| Total | 653 | 51 (7.8) | 26 | 20 | 8 | 25; 43 | 3.4 |
| Median Age, Years (Range) | Time from Rash Onset to Last Vaccine Dose, Average Years (Range) | Mean Age at Last Vaccine Dose, Years (Range) | Evidence of Onward Transmission, N (%) | ||
|---|---|---|---|---|---|
| Primary failure (N = 11) 1 | Two vaccine doses (N = 5) | 27 (6–34) | 17 (5–31) | 9.2 (5–12) | 2 (40.0) |
| One vaccine dose (N = 6) | 29 (2–43) | 18 (1–27) | 8.7 (1–33) | 2 (33.3) | |
| Secondary failure (N = 25) 2 | Two vaccine doses (N = 12) | 18 (10–27) | 12.3 (5–22) | 7.6 (5–11) | 1 (8.3) |
| One vaccine dose (N = 13) | 26 (2–38) | 20.2 (1–37) | 2.6 (1–8) | 3 (23.1) | |
| Unclassified (N = 15) 3 | Two vaccine doses (N = 8) | 19 (9–31) | 12 (1–31) | 7.8 (5–16) | 1 (12.5) |
| One vaccine dose (N = 7) | 26 (3–35) | 18 (0–33) | 6.9 (1–37) | 1 (14.3) |
| Oropharyngeal Swab | Urine | |||||
|---|---|---|---|---|---|---|
| N | Ct Value (Mean) | p Value | N | Ct Value (Mean) | p Value | |
| Patients with vaccination history | 42 | 30.71 | 0.0001774 | 46 | 31.96 | 0.000000150 |
| Patients with no vaccination history | 331 | 27.34 | 518 | 27.21 | ||
| Patients with primary vaccination failure | 9 | 26.38 | 0.01068 | 11 | 28.34 | 0.00004798 |
| Patients with secondary vaccination failure | 25 | 32.58 | 25 | 34.36 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchi, S.; Gori, M.; Fappani, C.; Ciceri, G.; Canuti, M.; Colzani, D.; Dura, M.; Terraneo, M.; Lamberti, A.; Baggieri, M.; et al. Characterization of Vaccine Breakthrough Cases during Measles Outbreaks in Milan and Surrounding Areas, Italy, 2017–2021. Viruses 2022, 14, 1068. https://doi.org/10.3390/v14051068
Bianchi S, Gori M, Fappani C, Ciceri G, Canuti M, Colzani D, Dura M, Terraneo M, Lamberti A, Baggieri M, et al. Characterization of Vaccine Breakthrough Cases during Measles Outbreaks in Milan and Surrounding Areas, Italy, 2017–2021. Viruses. 2022; 14(5):1068. https://doi.org/10.3390/v14051068
Chicago/Turabian StyleBianchi, Silvia, Maria Gori, Clara Fappani, Giulia Ciceri, Marta Canuti, Daniela Colzani, Marco Dura, Mara Terraneo, Anna Lamberti, Melissa Baggieri, and et al. 2022. "Characterization of Vaccine Breakthrough Cases during Measles Outbreaks in Milan and Surrounding Areas, Italy, 2017–2021" Viruses 14, no. 5: 1068. https://doi.org/10.3390/v14051068
APA StyleBianchi, S., Gori, M., Fappani, C., Ciceri, G., Canuti, M., Colzani, D., Dura, M., Terraneo, M., Lamberti, A., Baggieri, M., Senatore, S., Faccini, M., Magurano, F., Tanzi, E., & Amendola, A. (2022). Characterization of Vaccine Breakthrough Cases during Measles Outbreaks in Milan and Surrounding Areas, Italy, 2017–2021. Viruses, 14(5), 1068. https://doi.org/10.3390/v14051068








