Narcissus Plants: A Melting Pot of Potyviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Narcissus Plant Samples
2.2. Detection of NLSYV and NDV by RT-PCR and Sequencing
2.3. Sequencing of Full Genomics of NLSYV and NDV
2.4. Neighbor-Net Phylogenetic Network of CP Coding Regions
2.5. Maximum Likelihood Phylogenetic Tree of Polyprotein Coding Regions
2.6. Recombination Analysis
2.7. Genetic Diversity
2.8. Spatial Diffusion of NLSYV and NDV
2.9. Detection of Carlaviruses
3. Results and Discussion
3.1. Co- and Single-Infection of Narcissus Viruses
3.2. 3′ Terminal Region Sequences
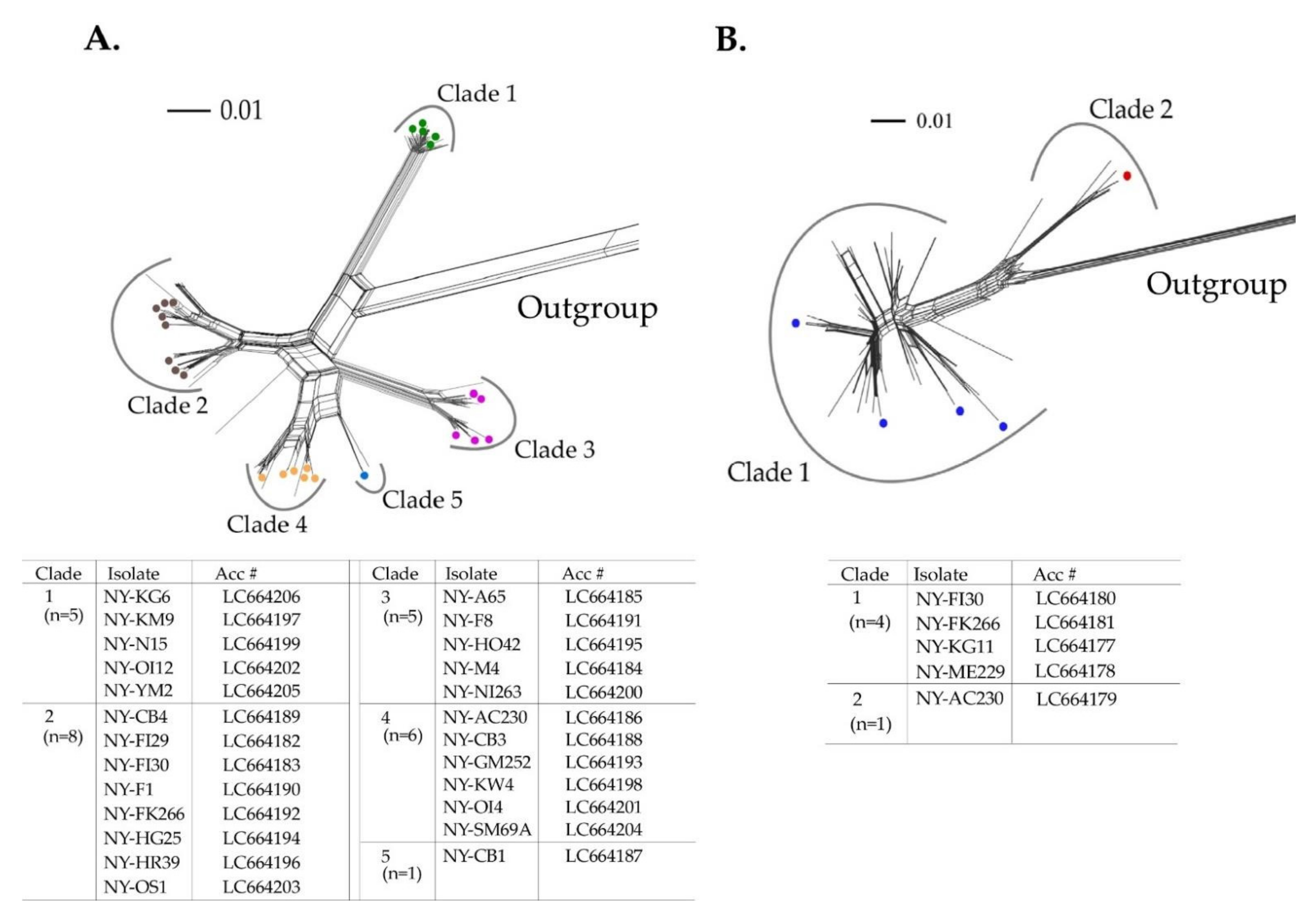
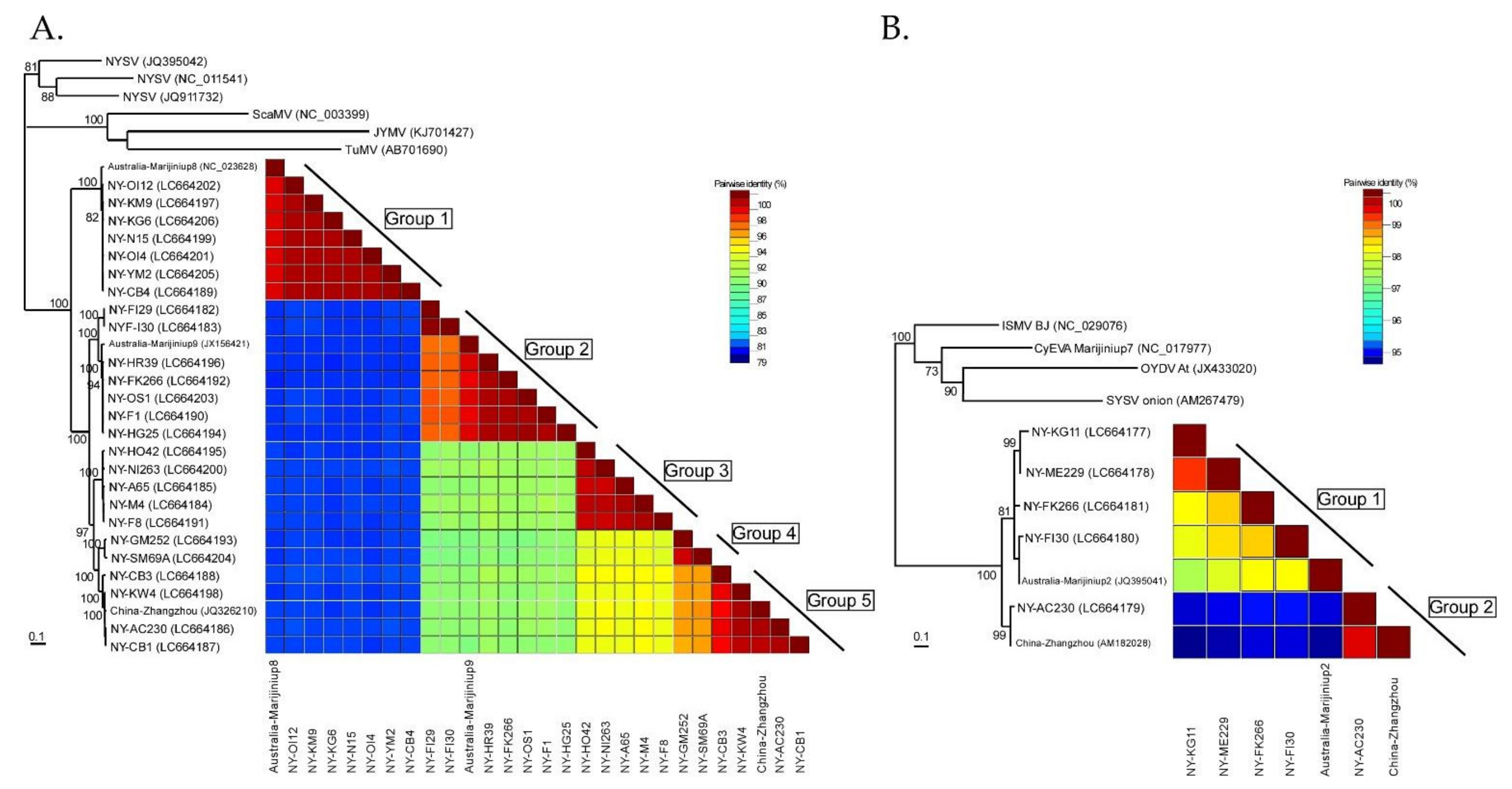
3.3. Phylogenetic Relationships and Nucleotide Identities Assessed by Coat Protein Coding Sequences
3.4. Co-Infection with Different Isolates and Quasispecies
3.5. Full Genomic Sequences
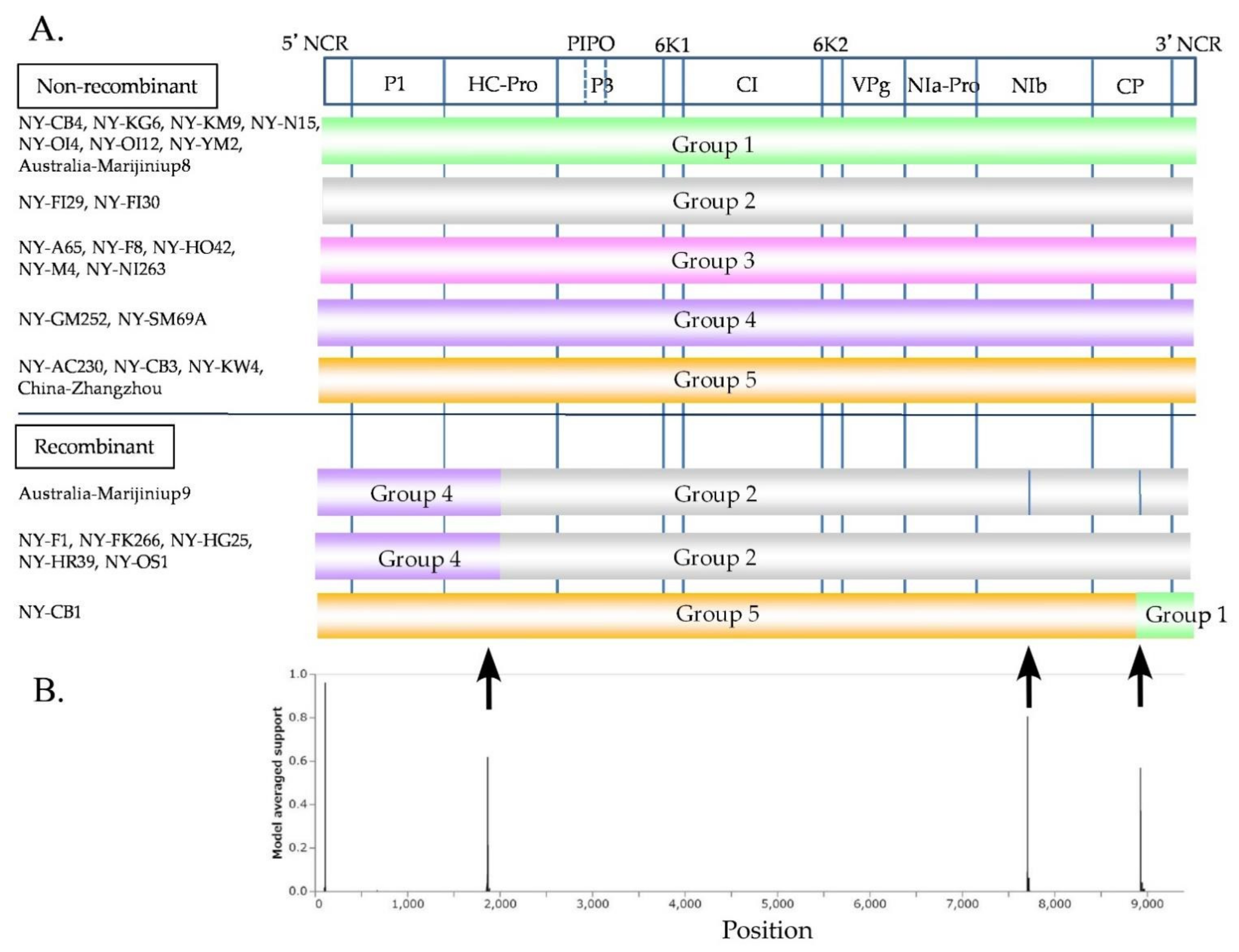
3.6. Inference of Recombination
3.7. Inference of Phylogenetic Relationships and Nucleotide Diversities Assessed by Polyprotein Coding Sequences
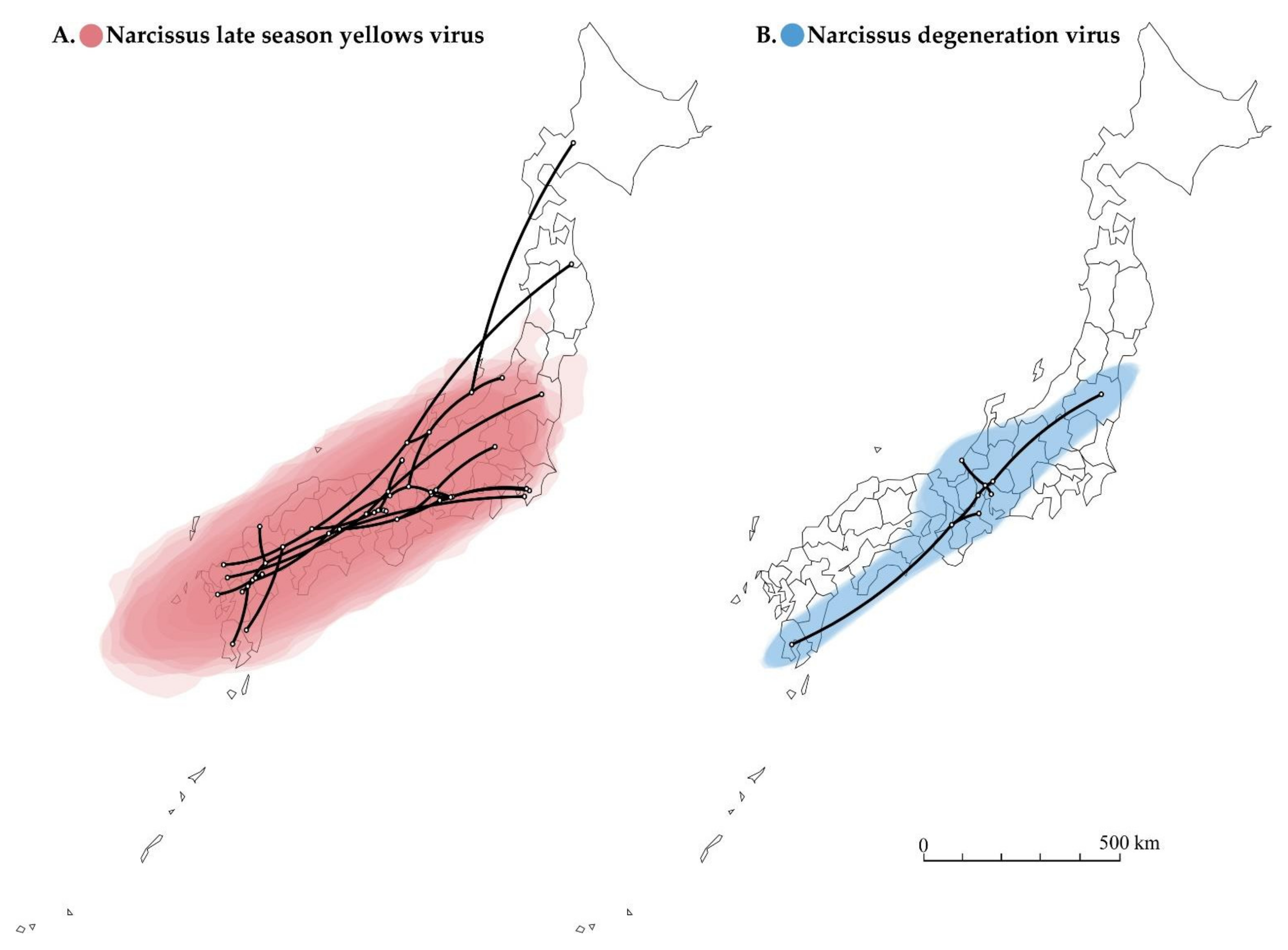
3.8. Timescale Analysis, Migration of Non-Recombinants and Recombinants
3.9. Co-Infection with Carlaviruses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Arenal, F.; Fraile, A.; Malpica, J.M. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001, 39, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Mackenzie, A.M.; Wei, K.J.; Gibbs, M.J. The potyviruses of Australia. Arch. Virol. 2008, 153, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Ohshima, K. Potyviruses and the digital revolution. Annu. Rev. Phytopathol. 2010, 48, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Hajizadeh, M.; Ohshima, K.; Jones, R.A.C. The potyviruses: An evolutionary synthesis is emerging. Viruses 2020, 12, 132. [Google Scholar] [CrossRef]
- Moreno Gonçalves, A.B.; López-Moya, J.J. When viruses play team sports: Mixed infections in plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Redinbaugh, M.G.; Stewart, L.R. Maize lethal necrosis: An emerging, synergistic viral disease. Annu. Rev. Virol. 2018, 5, 301–322. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato brown rugose fruit virus contributes to enhanced pepino mosaic virus titers in tomato plants. Viruses 2020, 12, 879. [Google Scholar] [CrossRef]
- Ontiveros, I.; Lopez-Moya, J.J.; Díaz-Pendón, J.A. Co-infection of tomato plants with Tomato yellow leaf curl virus and Tomato chlorosis virus affects the interaction with host and whiteflies. Phytopathology 2021, 26, 341. [Google Scholar] [CrossRef]
- Vinodhini, J.; Rajendran, L.; Abirami, R.; Karthikeyan, G. Co-existence of chlorosis inducing strain of Cucumber mosaic virus with tospoviruses on hot pepper (Capsicum annuum) in India. Sci. Rep. 2021, 11, 8796. [Google Scholar] [CrossRef]
- Stewart, L.R.; Willie, K. Maize yellow mosaic virus interacts with Maize chlorotic mottle virus and Sugarcane mosaic virus in mixed infections, but does not cause maize lethal necrosis. Plant Dis. 2021, 105, 3008–3014. [Google Scholar] [CrossRef]
- Kavalappara, S.R.; Milner, H.; Konakalla, N.C.; Morgan, K.; Sparks, A.N.; McGregor, C.; Culbreath, A.K.; Wintermantel, W.M.; Bag, S. High throughput sequencing-aided survey reveals widespread mixed infections of whitefly-transmitted viruses in cucurbits in Georgia, USA. Viruses 2021, 13, 988. [Google Scholar] [CrossRef]
- He, Z.; Yasaka, R.; Li, W.; Li, S.; Ohshima, K. Genetic structure of populations of sugarcane streak mosaic virus in China: Comparison with the populations in India. Virus Res. 2015, 211, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Nomiyama, R.; Mitoma, S.; Honda, Y.; Yasaka, R.; Tomimura, K. Evolutionary rates and genetic diversities of mixed potyviruses in Narcissus. Infect. Genet. Evol. 2016, 45, 213–223. [Google Scholar] [CrossRef] [PubMed]
- French, R.; Stenger, D.C. Population structure within lineages of Wheat streak mosaic virus derived from a common founding event exhibits stochastic variation inconsistent with the deterministic quasi-species model. Virology 2005, 343, 179–189. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohshima, K.; Akaishi, S.; Kajiyama, H.; Koga, R.; Gibbs, A.J. The evolutionary trajectory of turnip mosaic virus populations adapting to a new host. J. Gen. Virol. 2010, 91, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Roossinck, M.J. Analysis of quasispecies variation in single and mixed viral infection. Virus Evol. 2017, 3, vex037. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Kim, S.M.; Kim, S.L.; Lee, B.C.; Cho, W.K. The pepper virome: Natural co-infection of diverse viruses and their quasispecies. BMC Genom. 2017, 18, 453. [Google Scholar] [CrossRef]
- Syller, J. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef]
- McLeish, M.; Sacristán, S.; Fraile, A.; García-Arenal, F. Coinfection organizes epidemiological networks of viruses and hosts and reveals hubs of transmission. Phytopathology 2019, 109, 1003–1010. [Google Scholar] [CrossRef]
- Allen, L.J.S.; Bokil, V.A.; Cunniffe, N.J.; Hamelin, F.M.; Hilker, F.M.; Jeger, M.J. Modelling vector transmission and epidemiology of co-infecting plant viruses. Viruses 2019, 11, 1153. [Google Scholar] [CrossRef]
- Langeveld, S.A.; Derks, A.F.L.M.; Konicheva, V.; Muñoz, D.; Zhin-nan, C.; Denkova, S.T.; Lemmers, M.E.C.; Boonekamp, P.M. Molecular identification of potyviruses in Dutch stocks of Narcissus. Acta Hortic. 1997, 430, 641–648. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.P.; Langeveld, A.F.; Derks, A.F.L.M.; Adams, M.J. Molecular characterization of carla- and potyviruses from Narcissus in China. J. Phytopathol. 2003, 151, 26–29. [Google Scholar] [CrossRef]
- Chen, J.; Shi, Y.H.; Lu, Y.W.; Adams, M.J.; Chen, J.P. Narcissus symptomless virus: A new carlavirus of daffodils. Arch. Virol. 2006, 151, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, Y.H.; Adams, M.J.; Zheng, H.Y.; Qin, B.X.; Chen, J.P. Characterisation of an isolate of Narcissus degeneration virus from Chinese narcissus (Narcissus tazetta var. chinensis). Arch. Virol. 2007, 152, 441–448. [Google Scholar] [CrossRef]
- Monger, W.A.; Mumford, R.A. Vallota mosaic virus infecting nerine in the UK. Plant Pathol. 2008, 57, 768. [Google Scholar] [CrossRef]
- Lin, S.Q.; Shen, J.G.; Gao, F.L.; Cai, W.; Huang, Z.; Xie, L.Y.; Wu, Z.J. Complete genome sequence of Narcissus late season yellows virus infecting Chinese narcissus in China. Arch. Virol. 2012, 157, 1821–1824. [Google Scholar] [CrossRef]
- Wylie, S.J.; Jones, M.G.K. Complete genome sequences of seven carlavirus and potyvirus isolates from Narcissus and Hippeastrum plants in Australia, and proposals to clarify their naming. Arch. Virol. 2012, 157, 1471–1480. [Google Scholar] [CrossRef]
- Ohshima, K.; Mitoma, S.; Gibbs, A.J. The genetic diversity of narcissus viruses related to turnip mosaic virus blur arbitrary boundaries used to discriminate potyvirus species. PLoS ONE 2018, 13, e0190511. [Google Scholar] [CrossRef]
- Brunt, A.A. Narcissus. In Virus and Virus-Like Diseases of Bulb and Flower Crops; Loebenstein, G., Lawson, R.H., Brunt, A.A., Eds.; John Wiley and Sons: Chichester, UK, 1995; pp. 322–334. [Google Scholar]
- Wylie, S.J.; Li, H.; Sivasithamparam, K.; Jones, M.G.K. Complete genome analysis of three isolates of narcissus late season yellows virus and two of narcissus yellow stripe virus: Three species or one? Arch. Virol. 2014, 159, 1521–1525. [Google Scholar] [CrossRef]
- Raj, R.; Kaur, C.; Srivastava, A.; Kumar, S.; Raj, S.K. Sequence analyses of RT-PCR products obtained from seven infected leaf samples revealed existence of three potyvirus species in Indian narcissus (Narcissus tazetta L.). 3 Biotech 2020, 10, 428–430. [Google Scholar] [CrossRef]
- Valouzi, H.; Shahmohammadi, N.; Golnaraghi, A. Genetic diversity and evolutionary analyses of potyviruses infecting narcissus in Iran. J. Plant Pathol. 2021, 7, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.J.; Nguyen, H.D.; Ohshima, K. The emergence of turnip mosaic virus was probably a gene-forquasi-gene event. Curr. Opin. Virol. 2015, 10, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Fuji, S.; Mochizuki, T.; Okuda, M.; Tsuda, S.; Kagiwada, S.; Sekine, K.T.; Ugaki, M.; Natsuaki, K.T.; Isogai, M.; Maoka, T.; et al. Plant viruses and viroids in Japan. J. Gen. Plant Pathol. 2022, 88, 105–127. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Hunter, D.A.; Fletcher, J.D.; Davies, K.M.; Zhang, H. Colour break in reverse bicolour daffodils is associated with the presence of Narcissus mosaic virus. Virol. J. 2011, 8, 412. [Google Scholar] [CrossRef]
- Ward, L.I.; Veerakone, S.; Tang, J.; Clover, G.R.G. First report of Narcissus degeneration virus, Narcissus late season yellows virus, and Narcissus symptomless virus on Narcissus in New Zealand. Plant Dis. 2009, 93, 964. [Google Scholar] [CrossRef]
- Mowat, W.P.; Duncan, G.H.; Dawson, S. Narcissus late season yellows potyvirus: Symptoms, properties and serological detection. Ann. Appl. Biol. 1988, 113, 531–544. [Google Scholar] [CrossRef]
- Chandel, V.; Singh, M.K.; Hallan, V.Z.; Aijaz, A. Evidence for the occurrence of a distinct potyvirus on naturally growing Narcissus tazetta. Arch. Phytopathol. Plant Prot. 2010, 43, 209–214. [Google Scholar] [CrossRef]
- Sochacki, D.; Chojnowska, E. The frequency of viral infections on two narcissus plantations in central Poland. J. Hortic. Res. 2016, 24, 19–24. [Google Scholar] [CrossRef]
- Agoston, J.; Almasi, A.; Nemes, K.; Salanki, K.; Palkovics, L. First report of hippeastrum mosaic virus, narcissus late season yellows virus, narcissus latent virus and narcissus mosaic virus in daffodils from Hungary. J. Plant Pathol. 2020, 102, 1275–1276. [Google Scholar] [CrossRef]
- Wylie, S.J.; Nouri, S.; Coutts, B.A.; Jones, M.G.K. Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch. Virol. 2010, 155, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Lan, P.; Li, F.; Wang, M.; Li, R. Complete genome sequence of a divergent strain of Japanese yam mosaic virus from China. Arch. Virol. 2015, 160, 573–576. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Tomitaka, Y.; Ho, S.Y.W.; Duchêne, S.; Vetten, H.J.; Lesemann, D.; Walsh, J.A.; Gibbs, A.J.; Ohshima, K. Turnip mosaic potyvirus probably first spread to Eurasian brassica crops from wild orchids about 1000 years ago. PLoS ONE 2013, 8, e55336. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, H.; Chen, J.; Adams, M. Characterisation of a potyvirus and a potexvirus from Chinese scallion. Arch. Virol. 2002, 147, 683–693. [Google Scholar] [CrossRef]
- Li, Y.; Deng, C.; Shang, Q.; Zhao, X.; Liu, X.; Zhou, Q. The first complete genome sequence of iris severe mosaic virus. Arch. Virol. 2016, 161, 1069–1072. [Google Scholar] [CrossRef]
- Celli, M.G.; Torrico, A.K.; Kiehr, M.; Conci, V.C. Striking differences in the biological and molecular properties of onion and garlic isolates of onion yellow dwarf virus. Arch. Virol. 2013, 158, 1377–1382. [Google Scholar] [CrossRef]
- Chen, J.; Wei, C.B.; Zheng, H.Y.; Shi, Y.H.; Adams, M.J.; Lin, L.; Zhang, Q.Y.; Wang, S.J.; Chen, J.P. Characterisation of the welsh onion isolate of Shallot yellow stripe virus from China. Arch. Virol. 2005, 150, 2091–2099. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Weiller, G.F. TransAlign, Version 1.0; Research School of Biological Sciences: Canberra, Australia, 1999.
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.M. Treeview: An application to display phylogenetic trees on personal computer. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Sawyer, S.A. GENECONV: A Computer Package for the Statistical Detection of Gene Conversion; Department of Mathematics, Washington University: St. Louis, MO, USA, 1999. [Google Scholar]
- Salminen, M.O.; Carr, J.K.; Burke, D.S.; McCutchan, F.E. Identification of break points in intergenotypic recombinants of HIV type 1 by bootscanning. World J. Gastroenterol. 1995, 11, 1423–1425. [Google Scholar]
- Smith, M.J. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evolution of methods for detecting recombination from DNA sequence: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A monte calro procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2001, 16, 276–277. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Welch, J.J.; Suchard, M.A. Phylogeography takes a relaxed random walk-in continuous space and time. Mol. Biol. Evol. 2010, 27, 1877–1885. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Baele, G.; Li, W.L.; Drummond, A.J.; Suchard, M.A.; Lemey, P. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol. Biol. Evol. 2013, 30, 239–243. [Google Scholar] [CrossRef]
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive visualization of spatiotemporal history and trait evolutionary processes. Mol. Biol. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Chen, J.; Adams, M.J.; Chen, J.P. Complete nucleotide sequence and affinities of the genomic RNA of Narcissus common latent virus (genus Carlavirus). Arch. Virol. 2006, 151, 1667–1672. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, J.; Adams, M.J. Molecular characterisation of a complex mixture of viruses in garlic with mosaic symptoms in China. Arch. Virol. 2001, 146, 1841–1853. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Tomitaka, Y.; Wood, J.T.; Minematsu, Y.; Kajiyama, H.; Tomimura, K.; Gibbs, A.J. Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. J. Gen. Virol. 2007, 88, 298–315. [Google Scholar] [CrossRef]
- Ohshima, K.; Kawakubo, S.; Muraoka, S.; Gao, F.; Ishimaru, K.; Kayashima, T.; Fukuda, S. Genomic epidemiology and evolution of Scallion mosaic potyvirus from asymptomatic wild Japanese garlic. Front. Microbiol. 2021, 12, 789596. [Google Scholar] [CrossRef] [PubMed]
- Montarry, J.; Doumayrou, J.; Simon, V.; Moury, B. Genetic background matters: A plant-virus gene-for-gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 2011, 12, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Nigam, D.; LaTourrette, K.; Souza, P.F.N.; Garcia-Ruiz, H. Genome-wide variation in potyviruses. Front. Plant Sci. 2019, 12, 1439–1442. [Google Scholar] [CrossRef]
- Raj, R.; Kaur, C.; Agrawal, L.; Chauhan, P.S.; Kumar, S.; Raj, S.K. Full-length genome sequence of Cyrtanthus elatus virus—An isolated from Narcissus tazetta in India. 3 Biotech 2018, 8, 168–173. [Google Scholar] [CrossRef]
- Darlington, H.R. Yellow-stripe of daffodils. J. R. Hortic. Soc. 1908, 34, 161. [Google Scholar]
- Inouye, N.; Hakkaart, F.A. Preliminary description of a potyvirus from Vallota speciosa. Neth. J. Plant Pathol. 1980, 68, 265–275. [Google Scholar] [CrossRef]
- Kumar, S.; Raj, R.; Kaur, C.; Raj, S.K. First report of Cyrtanthus elatus virus A in Narcissus tazetta in India. Plant Dis. 2015, 99, 1658. [Google Scholar] [CrossRef]
- Brunt, A.A. Narcissus viruses. Rep. Glasshouse Crops Res. Inst. 1970, 134. [Google Scholar]
- Brunt, A.A.; Atkey, P.T. Rapid detection of Narcissus yellow stripe virus and two other filamentous viruses in crude negatively stained narcissus sap. Rep. Glasshouse Crops Res. Inst. 1967, 155–159. [Google Scholar]
- Ohshima, K.; Muraoka, S.; Yasaka, R.; Adachi, S.; Tokuda, M. First report of Scallion mosaic virus on wild Japanese garlic (Allium macrostemon) in Japan. J. Gen. Plant Pathol. 2016, 82, 61–64. [Google Scholar] [CrossRef]
- Santos-Gally, R.; Vargas, P.; Arroyo, J. Insights into neogene Mediterranean biogeography based on phylogenetic relationships of mountain and lowland lineages of Narcissus (Amaryllidaceae). J. Biogeogr. 2012, 39, 782–798. [Google Scholar] [CrossRef]
- Matsumoto, A.; Hori, D. Echizen Narcissus Mystery; Echizen-Board Education: Fukui, Japan, 2011; Available online: https://www.town.echizen.fukui.jp/otabunreki/panel/15.html (accessed on 1 March 2022).
- Park, C.H.; Song, E.G.; Ryu, K.H. Detection of co-infection of Notocactus leninghausii f. cristatus with six virus species in South Korea. Plant Pathol. J. 2018, 34, 65–70. [Google Scholar] [CrossRef]
- Gao, F.; Shen, J.; Liao, F.; Cai, W.; Lin, S.; Yang, H.; Chen, S. The first complete genome sequence of narcissus latent virus from Narcissus. Arch. Virol. 2018, 163, 1383–1386. [Google Scholar] [CrossRef]
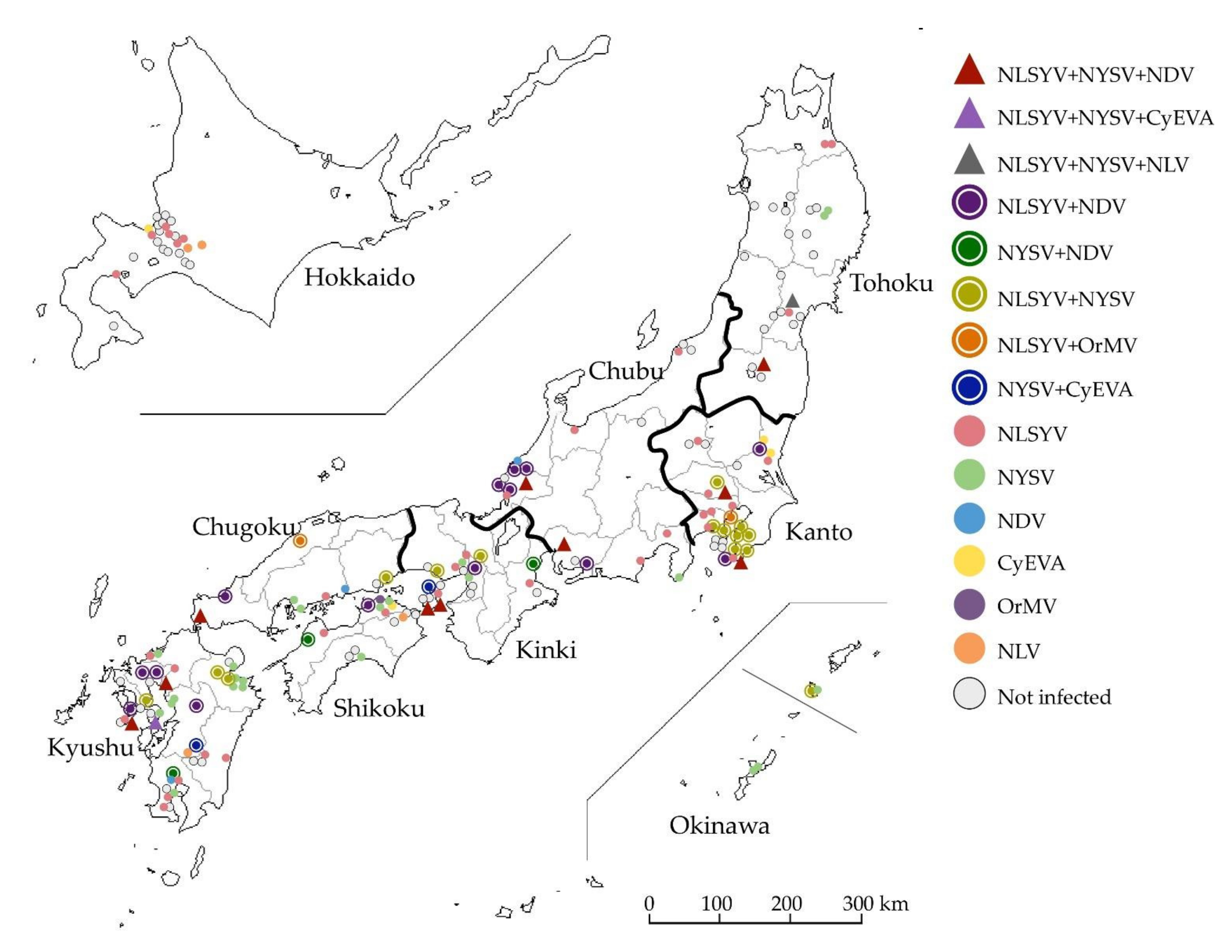
| Numbers of Plants (%, Number of Plants Detected out of 120 Detected Plants) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| District (Island) | Plants Examined (n) | Plants Detected (n) (%) 1 | Co-Infected with Three Viruses | Co-Infected with Two Viruses | Singly Infected with | |||||||||||
| Potyvirus 2 | Maclura-Virus | |||||||||||||||
| TuMV Group | OYDV Group | |||||||||||||||
| NLSYV NYSV NDV | NLSYV NYSV CyEVA | NLSYV NYSV NLV | NDV NLSYV | NDV NYSV | NLSYV NYSV | NLSYV OrMV | CyEVA NYSV | NLSYV | NYSV | NDV | CyEVA | OrMV | NLV | |||
| Hokkaido | 24 | 9 (37.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 1 | 0 | 2 |
| Tohoku | 25 | 7 (28.0) | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 |
| Kanto | 30 | 23 (76.7) | 2 | 0 | 0 | 2 | 0 | 8 | 1 | 0 | 8 | 0 | 0 | 2 | 0 | 0 |
| Chubu | 19 | 14 (73.7) | 2 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 5 | 1 | 1 | 0 | 0 | 0 |
| Kinki | 21 | 13 (61.9) | 2 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 4 | 2 | 0 | 0 | 0 | 0 |
| Chugoku | 9 | 8 (88.9) | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 0 |
| Shikoku | 15 | 10 (66.7) | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 3 | 0 | 1 | 1 | 1 |
| Kyushu and Okinawa | 46 | 36 (78.3) | 2 | 1 | 0 | 4 | 1 | 4 | 0 | 1 | 8 | 13 | 1 | 0 | 0 | 1 |
| Total | 189 | 120 (63.5) | 10 (8.3) | 1 (0.8) | 1 (0.8) | 14 (11.7) | 3 (2.5) | 15 (12.5) | 2 (1.7) | 2 (1.7) | 37 (30.8) | 23 (19.2) | 3 (2.5) | 4 (3.3) | 1 (0.8) | 4 (3.3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Probowati, W.; Kawakubo, S.; Ohshima, K. Narcissus Plants: A Melting Pot of Potyviruses. Viruses 2022, 14, 582. https://doi.org/10.3390/v14030582
Probowati W, Kawakubo S, Ohshima K. Narcissus Plants: A Melting Pot of Potyviruses. Viruses. 2022; 14(3):582. https://doi.org/10.3390/v14030582
Chicago/Turabian StyleProbowati, Wiwit, Shusuke Kawakubo, and Kazusato Ohshima. 2022. "Narcissus Plants: A Melting Pot of Potyviruses" Viruses 14, no. 3: 582. https://doi.org/10.3390/v14030582
APA StyleProbowati, W., Kawakubo, S., & Ohshima, K. (2022). Narcissus Plants: A Melting Pot of Potyviruses. Viruses, 14(3), 582. https://doi.org/10.3390/v14030582







