Structural Basis of the Avian Influenza NS1 Protein Interactions with the Cell Polarity Regulator Scribble
Abstract
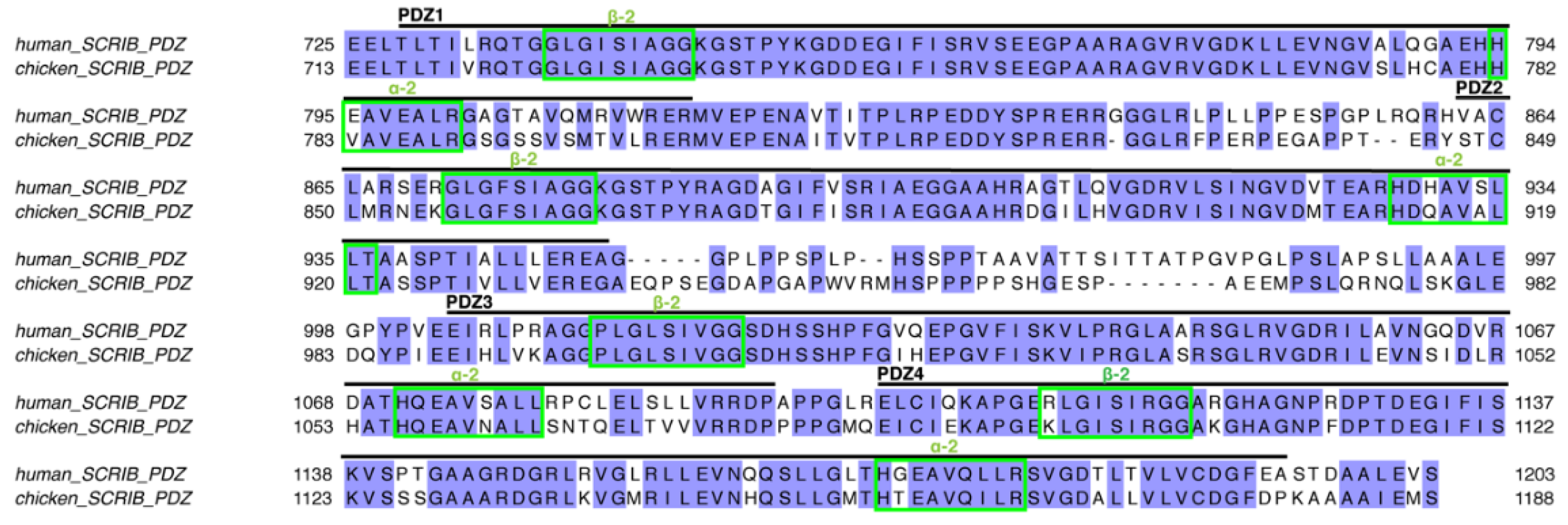
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
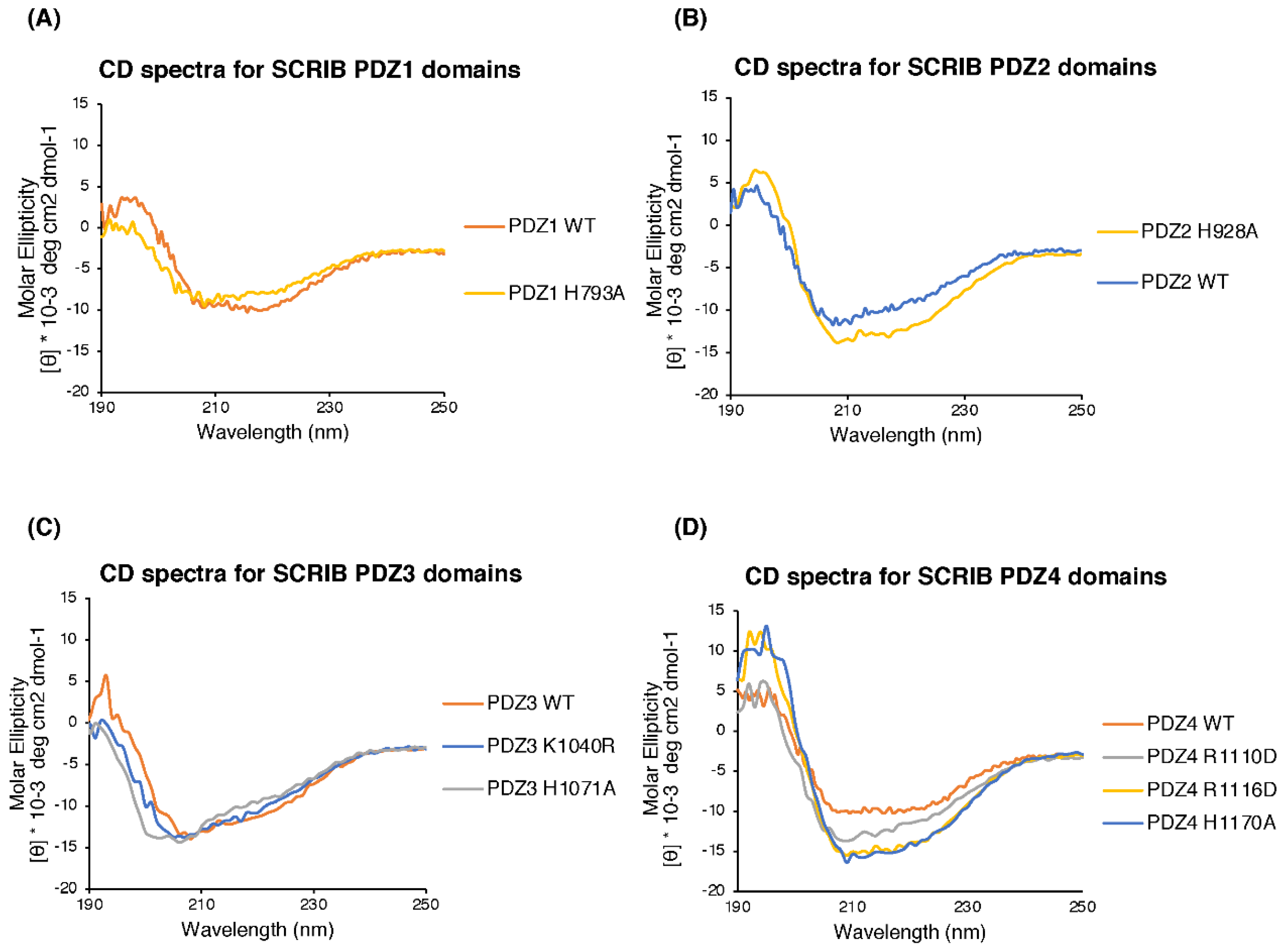
2.2. Circular Dichroism (CD) Spectroscopy
2.3. Isothermal Titration Calorimetry (ITC)
2.4. Crystallisation and Structure Determination
3. Results
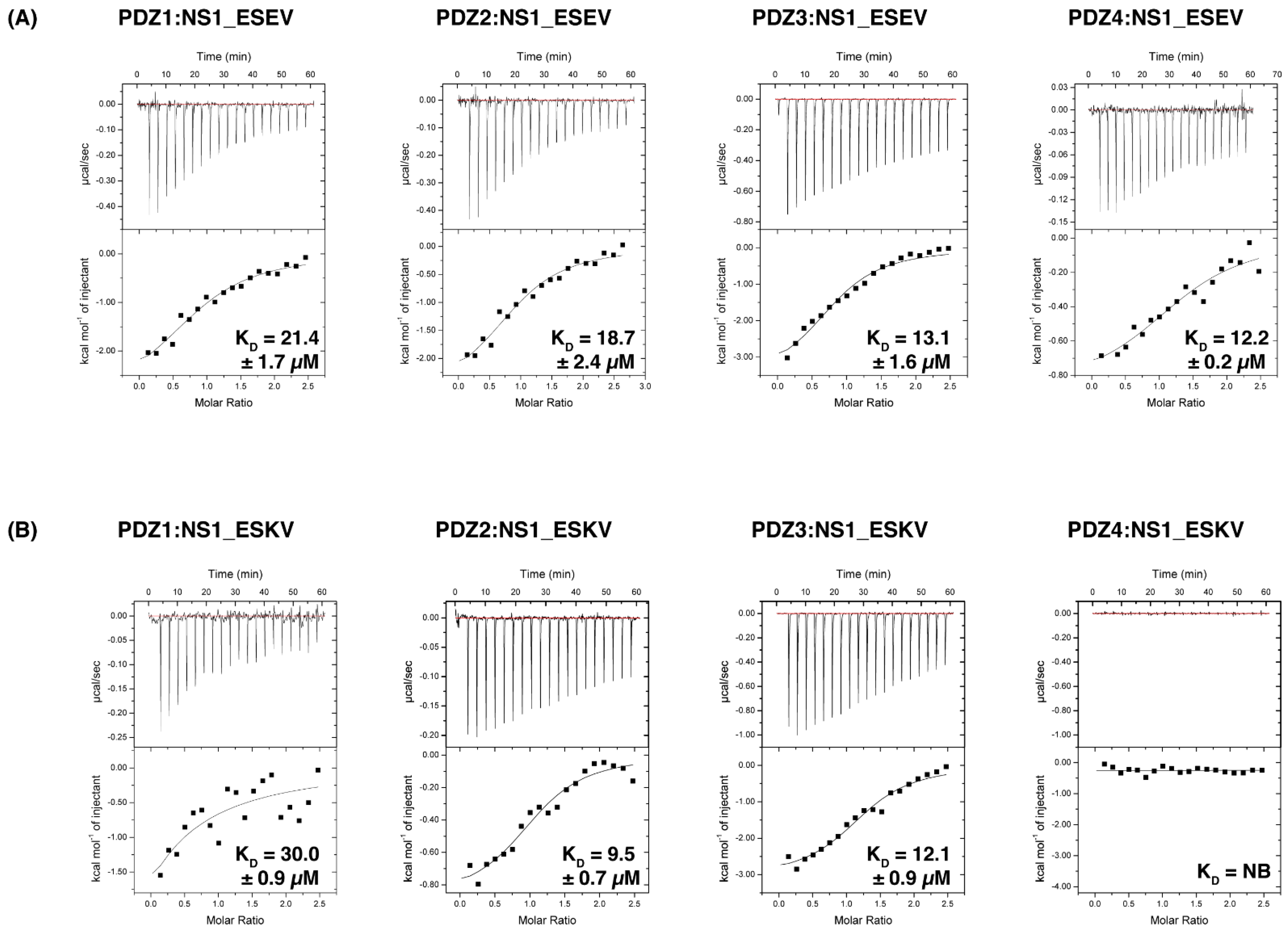
3.1. Biochemical Characterisation of SCRIB PDZ Domains and NS1 Peptide Interactions
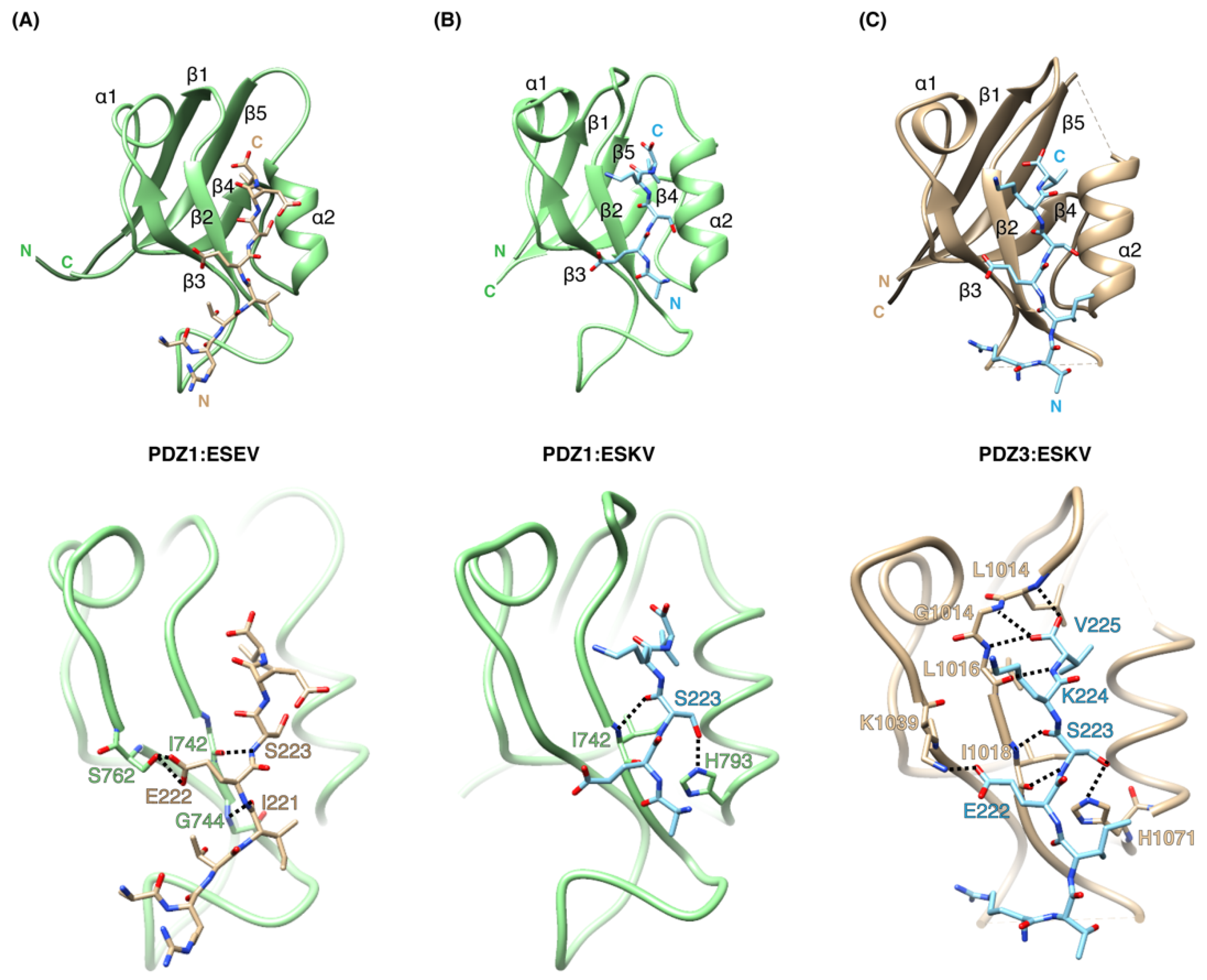
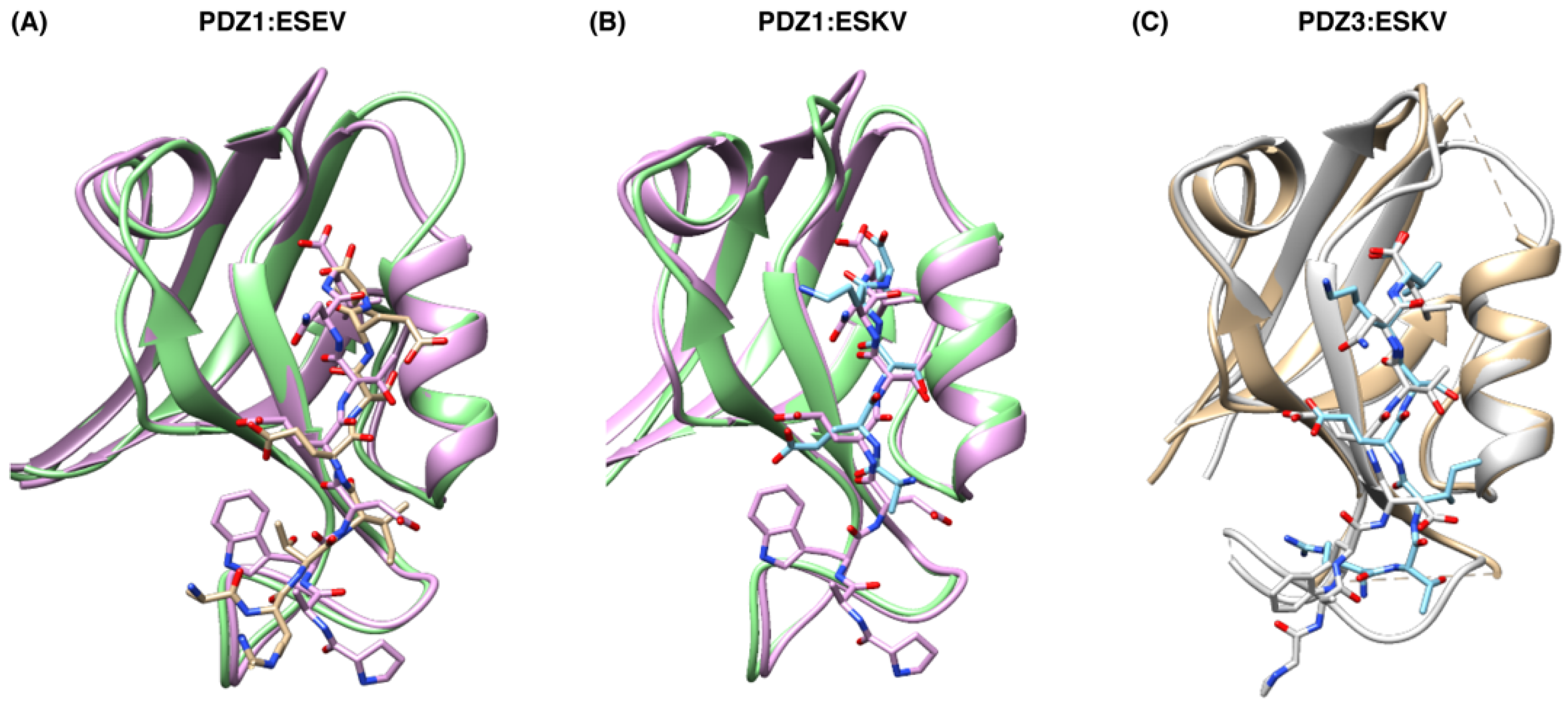
3.2. Structural Analysis of SCRIB PDZ-NS1 Interactions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, W.J. Adaptation of core mechanisms to generate cell polarity. Nature 2003, 422, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Allam, A.H.; Charnley, M.; Russell, S.M. Context-Specific Mechanisms of Cell Polarity Regulation. J. Mol. Biol. 2018, 430, 3457–3471. [Google Scholar] [CrossRef] [PubMed]
- Jarjour, A.A.; Boyd, A.; Dow, L.E.; Holloway, R.K.; Goebbels, S.; Humbert, P.O.; Williams, A.; Ffrench-Constant, C. The Polarity Protein Scribble Regulates Myelination and Remyelination in the Central Nervous System. PLoS Biol. 2015, 13, e1002107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krummel, M.F.; Macara, I. Maintenance and modulation of T cell polarity. Nat. Immunol. 2006, 7, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Royer, C.; Lu, X. Epithelial cell polarity: A major gatekeeper against cancer? Cell Death Differ. 2011, 18, 1470–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahirovic, S.; Bradke, F. Neuronal polarity. Cold Spring Harb. Perspect. Biol. 2009, 1, a001644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.; Banks, L. Upsetting the Balance: When Viruses Manipulate Cell Polarity Control. J. Mol. Biol. 2018, 430, 3481–3503. [Google Scholar] [CrossRef] [PubMed]
- Assemat, E.; Bazellieres, E.; Pallesi-Pocachard, E.; Le Bivic, A.; Massey-Harroche, D. Polarity complex proteins. Biochim. Biophys. Acta 2008, 1778, 614–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoni, M.J.; Kashyap, R.; Camoin, L.; Borg, J.P. The Scribble family in cancer: Twentieth anniversary. Oncogene 2020, 39, 7019–7033. [Google Scholar] [CrossRef] [PubMed]
- Bonello, T.T.; Peifer, M. Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell Biol. 2019, 218, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Stephens, R.; Lim, K.; Portela, M.; Kvansakul, M.; Humbert, P.O.; Richardson, H.E. The Scribble Cell Polarity Module in the Regulation of Cell Signaling in Tissue Development and Tumorigenesis. J. Mol. Biol. 2018, 430, 3585–3612. [Google Scholar] [CrossRef]
- Bryant, P.J.; Huwe, A. LAP proteins: What’s up with epithelia? Nat. Cell Biol. 2000, 2, E141–E143. [Google Scholar] [CrossRef] [PubMed]
- Javier, R.T.; Rice, A.P. Emerging theme: Cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 2011, 85, 11544–11556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takizawa, S.; Nagasaka, K.; Nakagawa, S.; Yano, T.; Nakagawa, K.; Yasugi, T.; Takeuchi, T.; Kanda, T.; Huibregtse, J.M.; Akiyama, T.; et al. Human scribble, a novel tumor suppressor identified as a target of high-risk HPV E6 for ubiquitin-mediated degradation, interacts with adenomatous polyposis coli. Genes Cells 2006, 11, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Okajima, M.; Takahashi, M.; Higuchi, M.; Ohsawa, T.; Yoshida, S.; Yoshida, Y.; Oie, M.; Tanaka, Y.; Gejyo, F.; Fujii, M. Human T-cell leukemia virus type 1 Tax induces an aberrant clustering of the tumor suppressor Scribble through the PDZ domain-binding motif dependent and independent interaction. Virus Genes 2008, 37, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Wigerius, M.; Melik, W.; Elväng, A.; Johansson, M. Rac1 and Scribble are targets for the arrest of neurite outgrowth by TBE virus NS5. Mol. Cell. Neurosci. 2010, 44, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Golebiewski, L.; Liu, H.; Javier, R.T.; Rice, A.P. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J. Virol. 2011, 85, 10639–10648. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal. 2010, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Giallourakis, C.; Cao, Z.; Green, T.; Wachtel, H.; Xie, X.; Lopez-Illasaca, M.; Daly, M.; Rioux, J.; Xavier, R. A molecular-properties-based approach to understanding PDZ domain proteins and PDZ ligands. Genome Res. 2006, 16, 1056–1072. [Google Scholar] [CrossRef] [Green Version]
- Nourry, C.; Grant, S.G.; Borg, J.P. PDZ domain proteins: Plug and play! Sci. STKE 2003, 2003, RE7. [Google Scholar] [CrossRef]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.-H.; Reva, B.; Held, H.A.; Appleton, B.A.; Evangelista, M.; Wu, Y. A specificity map for the PDZ domain family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Golebiewski, L.; Dow, E.C.; Krug, R.M.; Javier, R.T.; Rice, A.P. The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting Scribble. J. Virol. 2010, 84, 11164–11174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hale, B.G.; Randall, R.E.; Ortin, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Soubies, S.M.; Volmer, C.; Croville, G.; Loupias, J.; Peralta, B.; Costes, P.; Lacroux, C.; Guerin, J.L.; Volmer, R. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J. Virol. 2010, 84, 6733–6747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obenauer, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wang, J.; Ma, J.; Fan, Y.; et al. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Krug, R.M.; Garcia-Sastre, A. The NS1 protein: A master regulator of host and viral functions. In Textbook of Influenza, 2nd ed.; Wiley: Bridgewater, NJ, USA, 2013; pp. 114–132. [Google Scholar] [CrossRef]
- Uiprasertkul, M.; Puthavathana, P.; Sangsiriwut, K.; Pooruk, P.; Srisook, K.; Peiris, M.; Nicholls, J.M.; Chokephaibulkit, K.; Vanprapar, N.; Auewarakul, P. Influenza A H5N1 replication sites in humans. Emerg. Infect. Dis. 2005, 11, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.B.; Gödde, N.J.; Humbert, P.O.; Kvansakul, M. Structural basis for the differential interaction of Scribble PDZ domains with the guanine nucleotide exchange factor β-PIX. J. Biol. Chem. 2017, 292, 20425–20436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caria, S.; Stewart, B.Z.; Jin, R.; Smith, B.J.; Humbert, P.O.; Kvansakul, M. Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains. FEBS J. 2019, 286, 4910–4925. [Google Scholar] [CrossRef] [PubMed]
- How, J.Y.; Stephens, R.K.; Lim, K.Y.B.; Humbert, P.O.; Kvansakul, M. Structural basis of the human Scribble-Vangl2 association in health and disease. Biochem. J. 2021, 478, 1321–1332. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Macken, C.A.; Li, C.; Ozawa, M.; Goto, H.; Iswahyudi, N.F.; Nidom, C.A.; Chen, H.; Neumann, G.; Kawaoka, Y. Synergistic effect of the PDZ and p85beta-binding domains of the NS1 protein on virulence of an avian H5N1 influenza A virus. J. Virol. 2013, 87, 4861–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wang, W. Organization of signaling complexes by PDZ-domain scaffold proteins. ACC Chem. Res. 2003, 36, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Javorsky, A.; Humbert, P.O.; Kvansakul, M. Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain. Commun. Biol. 2021, 4, 724. [Google Scholar] [CrossRef] [PubMed]
- Maddumage, J.C.; Stewart, B.Z.; Humbert, P.O.; Kvansakul, M. Crystallographic Studies of PDZ Domain-Peptide Interactions of the Scribble Polarity Module. Methods Mol. Biol. 2021, 2256, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Cryst. D Biol. Cryst. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Cryst. D Biol. Cryst. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Cryst. D Biol. Cryst. 2007, 63, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werme, K.; Wigerius, M.; Johansson, M. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell. Microbiol. 2008, 10, 696–712. [Google Scholar] [CrossRef]
- Audebert, S.; Navarro, C.; Nourry, C.; Chasserot-Golaz, S.; Lécine, P.; Bellaiche, Y.; Dupont, J.-L.; Premont, R.T.; Sempéré, C.; Strub, J.-M.; et al. Mammalian Scribble Forms a Tight Complex with the βPIX Exchange Factor. Curr. Biol. 2004, 14, 987–995. [Google Scholar] [CrossRef] [Green Version]
- Kallay, L.M.; McNickle, A.; Brennwald, P.J.; Hubbard, A.L.; Braiterman, L.T. Scribble associates with two polarity proteins, lgl2 and vangl2, via distinct molecular domains. J. Cell. Biochem. 2006, 99, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Dow, L.E.; Kauffman, J.S.; Caddy, J.; Peterson, A.S.; Jane, S.M.; Russell, S.M.; Humbert, P.O. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: Regulation of Rho GTPase recruitment to the leading edge. Oncogene 2007, 26, 2272–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaelis, U.R.; Chavakis, E.; Kruse, C.; Jungblut, B.; Kaluza, D.; Wandzioch, K.; Manavski, Y.; Heide, H.; Santoni, M.J.; Potente, M.; et al. The polarity protein Scrib is essential for directed endothelial cell migration. Circ. Res. 2013, 112, 924–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, P.A.; Socias, S.; Key, J.; Ransey, E.; Tjon, E.C.; Buschiazzo, A.; Lei, M.; Botka, C.; Withrow, J.; Neau, D.; et al. Data publication with the structural biology data grid supports live analysis. Nat. Commun. 2016, 7, 10882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
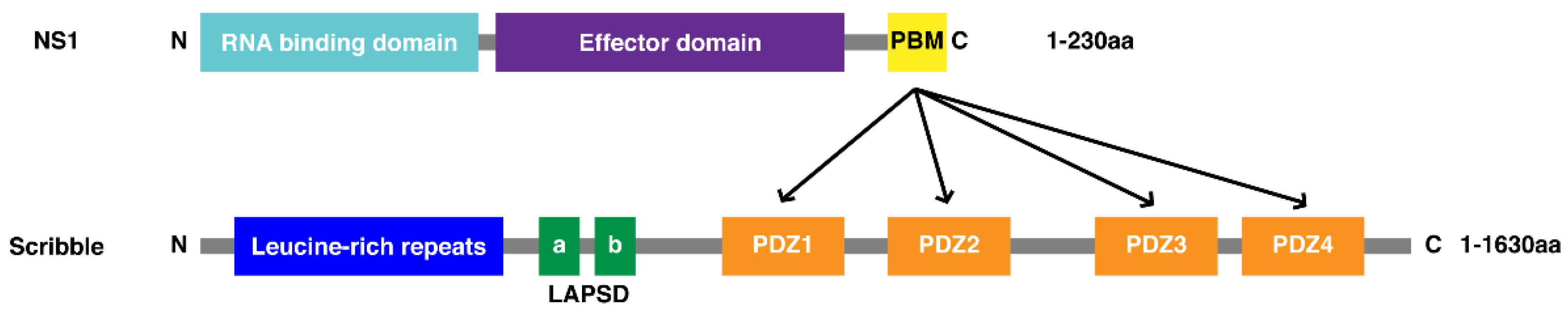
| NS1 Peptide Origin | Peptide Sequence | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vietnam_H5N1 | K | M | A | R | T | I | E | S | E | V |
| Hubei_H5N1 | K | M | A | R | T | I | E | S | K | V |
| Brevig_H1N1 | K | M | A | R | T | I | K | S | E | V |
| Memphis_H3N2 | K | M | A | R | T | A | R | S | K | V |
| Puerto Rico_H1N1 | E | M | A | G | T | I | R | S | E | V |
| HongKong_H5N1 | K | M | E | R | T | I | E | P | E | V |
| : | * | * | . | : | * | |||||
| PBM Peptides | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCRIB PDZ Constructs | Vietnam (ESEV) | Hubei (ESKV) | Brevig (KSEV) | Memphis (RSKV) | Puerto Rico (RSEV) | Hong Kong $ (EPEV) | ||||||
| N | KD (µM) | N | KD (µM) | N | KD (µM) | N | KD (µM) | N | KD (µM) | N | KD (µM) | |
| PDZ1 | 1.1 ± 0.1 | 21.4 ± 1.7 | 1.2 | 30.0 ± 0.9 | NB | NB | NB | NB | NB | NB | NB | NB |
| PDZ2 | 1.0 ± 0.03 | 18.7 ± 2.4 | 1.1 | 9.5 ± 0.7 | NB | NB | NB | NB | NB | NB | NB | NB |
| PDZ3 | 1.1 ± 0.1 | 13.1 ± 1.6 | 1.1 | 12.1 ± 0.9 | NB | NB | NB | NB | NB | NB | NB | NB |
| PDZ4 | 1.0 ± 0.1 | 12.2 ± 0.2 | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
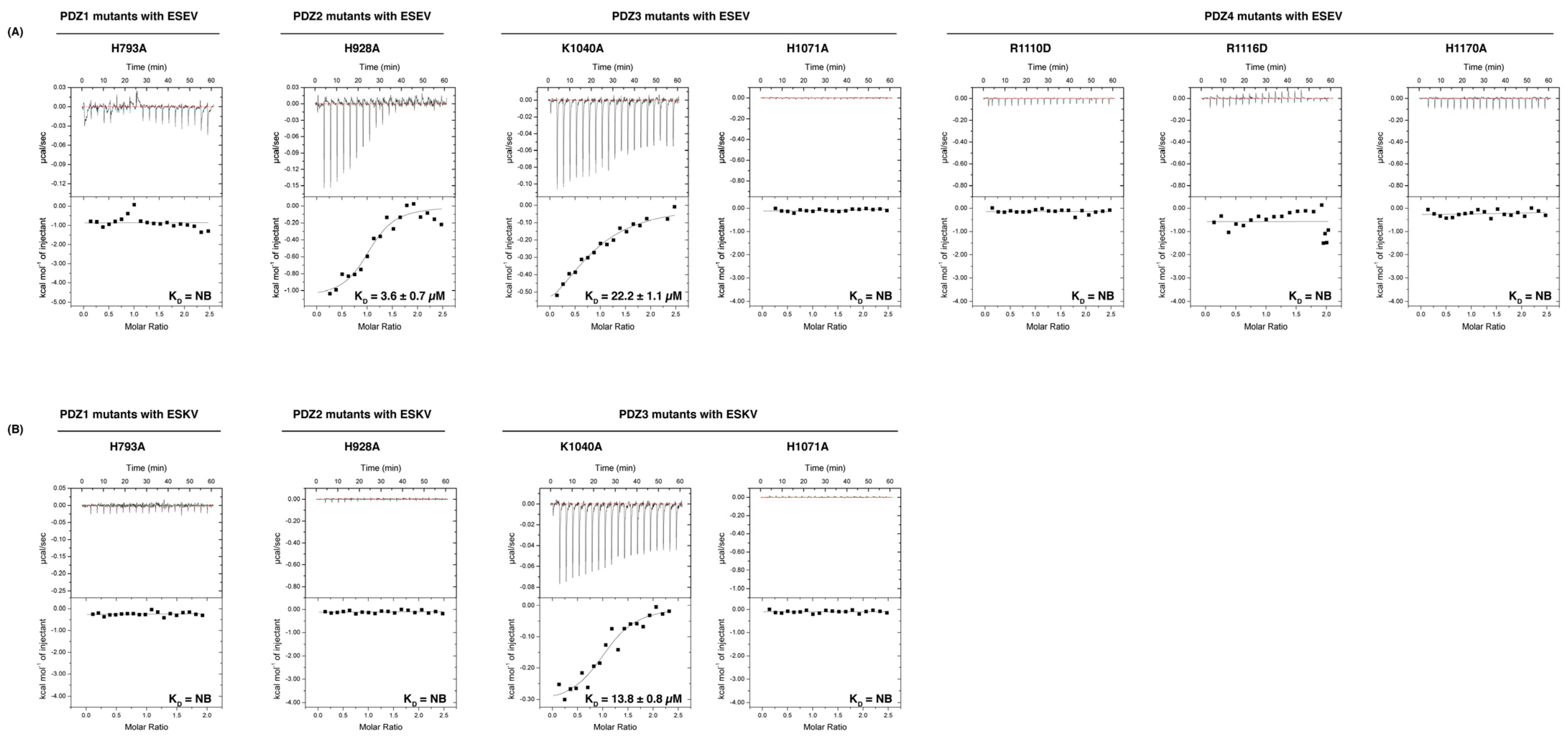
| PDZ1 H793A | NB | NB | NB | NB | ||||||||
| PDZ2 H928A | 1.1 ± 0.1 | 3.6 ± 0.7 | NB | NB | ||||||||
| PDZ3 K1040A | 1.1 ± 0.1 | 22.2 ± 1.1 | 1.1 | 13.8 ± 0.8 | ||||||||
| PDZ3 H1071A | NB | NB | NB | NB | ||||||||
| PDZ4 R1110D | NB | NB | ||||||||||
| PDZ4 R1116D | NB | NB | ||||||||||
| PDZ4 H1170A | NB | NB | ||||||||||
| Data Collection | SCRIB PDZ1: ESEV | SCRIB PDZ1: ESKV | SCRIB PDZ3: ESKV |
|---|---|---|---|
| Space group | I2 | C2 | C2 |
| No. of molecules in AU | 2 | 1 | 4 |
| Cell dimensions | |||
| a, b, c (Å) | 56.91; 57.01; 61.60 | 58.54; 51.20; 27.87 | 77.84; 77.72; 64.75 |
| α, β, γ (°) | 90.00; 117.372; 90.00 | 90.00; 90.55; 90.00 | 90.00; 94.16; 90.00 |
| Wavelength (Å) | 0.95 | 0.95 | 0.95 |
| Resolution (Å) | 39.74 –3.50 (3.63–3.50 | 29.26–2.06 (2.13–2.06) | 42.94–2.83 (2.93–2.83) |
| Rsym or Rmerge | 0.08177 (0.1554) | 0.0176 (0.068) | 0.05437 (0.5045) |
| I/σI | 6.63 (4.20) | 13.58 (6.80) | 9.79 (1.94) |
| CC (1/2) | 0.982 (0.94) | 0.999 (0.987) | 0.999 (0.866) |
| Completeness (%) | 97.52 (99.10) | 99.90 (100.0) | 96.63 (94.74) |
| Multiplicity | 1.9 (1.9) | 2.0 (2.0) | 1.9 (1.8) |
| Wilson B-factor | 43.63 | 30.93 | 56.7 |
| Refinement | |||
| Resolution (Å) | 30.47–3.50 (3.63–3.50) | 29.26–2.06 (2.13–2.06) | 42.94–2.84 (2.94–2.84) |
| No. reflections | 2199 (220) | 10215 (1022) | 16752 (1584) |
| Rwork/Rfree | 0.242/0.266 | 0.277/0.279 | 0.289/0.295 |
| No. atoms | |||
| Protein | 1423 | 689 | 2620 |
| Ligand/ion | 0 | 0 | 0 |
| Water | 0 | 43 | 0 |
| B-factors | |||
| Protein | 26.32 | 48.46 | 48.84 |
| Ligand/ion | 0 | 0 | 0 |
| Water | 0 | 42.46 | 0 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.006 | 0.003 |
| Bond angles (°) | 0.55 | 0.9 | 0.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javorsky, A.; Humbert, P.O.; Kvansakul, M. Structural Basis of the Avian Influenza NS1 Protein Interactions with the Cell Polarity Regulator Scribble. Viruses 2022, 14, 583. https://doi.org/10.3390/v14030583
Javorsky A, Humbert PO, Kvansakul M. Structural Basis of the Avian Influenza NS1 Protein Interactions with the Cell Polarity Regulator Scribble. Viruses. 2022; 14(3):583. https://doi.org/10.3390/v14030583
Chicago/Turabian StyleJavorsky, Airah, Patrick O. Humbert, and Marc Kvansakul. 2022. "Structural Basis of the Avian Influenza NS1 Protein Interactions with the Cell Polarity Regulator Scribble" Viruses 14, no. 3: 583. https://doi.org/10.3390/v14030583
APA StyleJavorsky, A., Humbert, P. O., & Kvansakul, M. (2022). Structural Basis of the Avian Influenza NS1 Protein Interactions with the Cell Polarity Regulator Scribble. Viruses, 14(3), 583. https://doi.org/10.3390/v14030583







