Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank
Abstract
1. Introduction
2. Materials and Methods
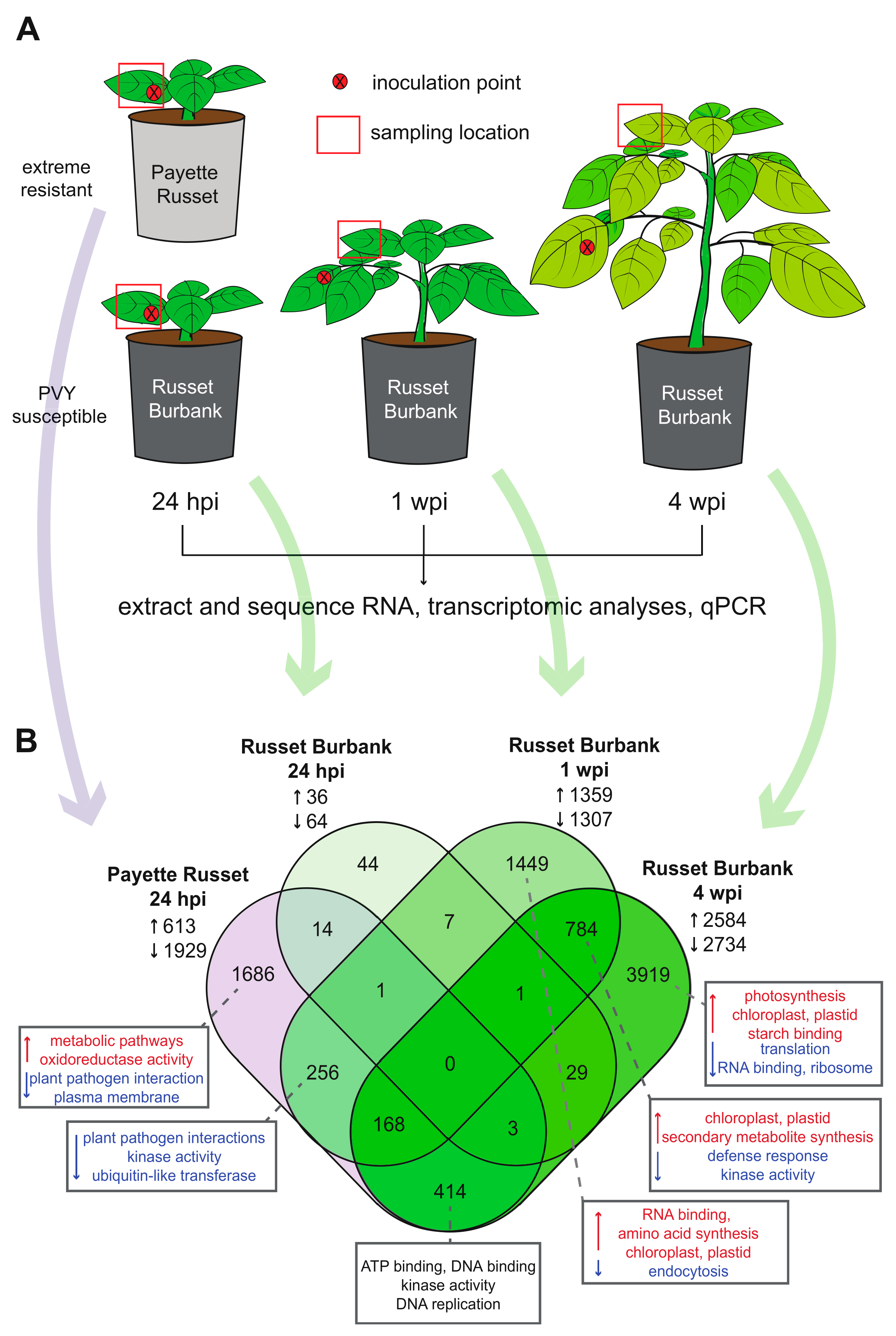
2.1. Potato Sources and Growing Conditions
2.2. PVY Inoculum Preparation and Inoculation Protocol
2.3. RNA Isolation and cDNA Synthesis
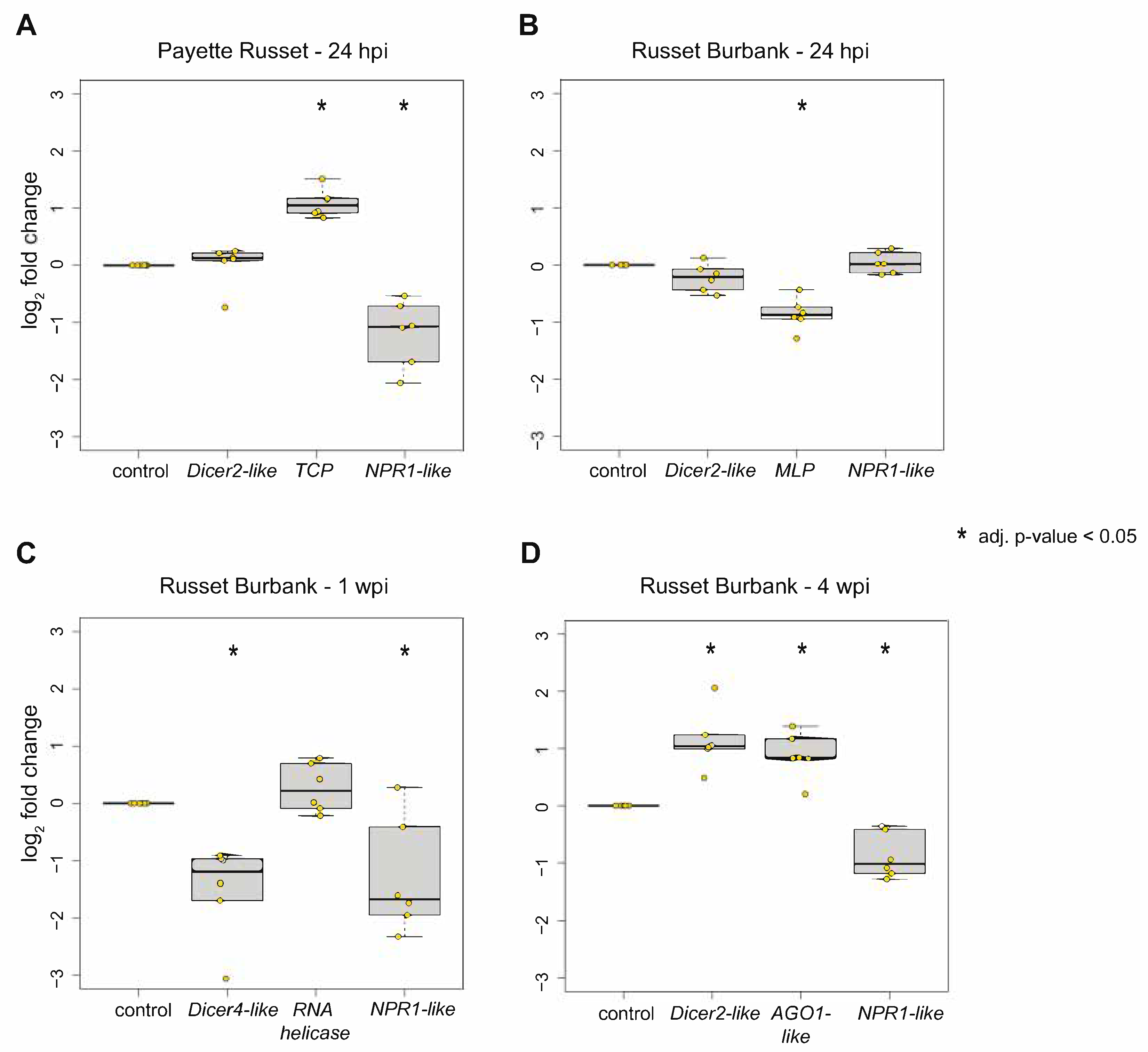
2.4. Quantitative PCR (qPCR)
2.5. Transcriptome Library Preparation and Sequencing
2.6. Differential Gene Expression Analysis
2.7. Blast Search of Potato Transcript Sequences against Potato Genome, Wild Potato Species, and All Organisms
3. Results and Discussion
3.1. Transcriptome Level Assessment of Potato Virus Y (PVY)-Infected Potato Cultivars
3.1.1. Transcriptomic Differences between PVY-Resistant Payette Russet and Susceptible Russet Burbank Potato Cultivars 24-hours Post PVY-Inoculation
3.1.2. Upper, Uninoculated Russet Burbank Leaves Exhibit Different Transcriptional Responses before and after Systemic Infection
3.2. Differences and Similarities in Differentially Expressed Genes between Potato Cultivars and Time Points Reveal Patterns of Infection and Possible Antiviral Genes
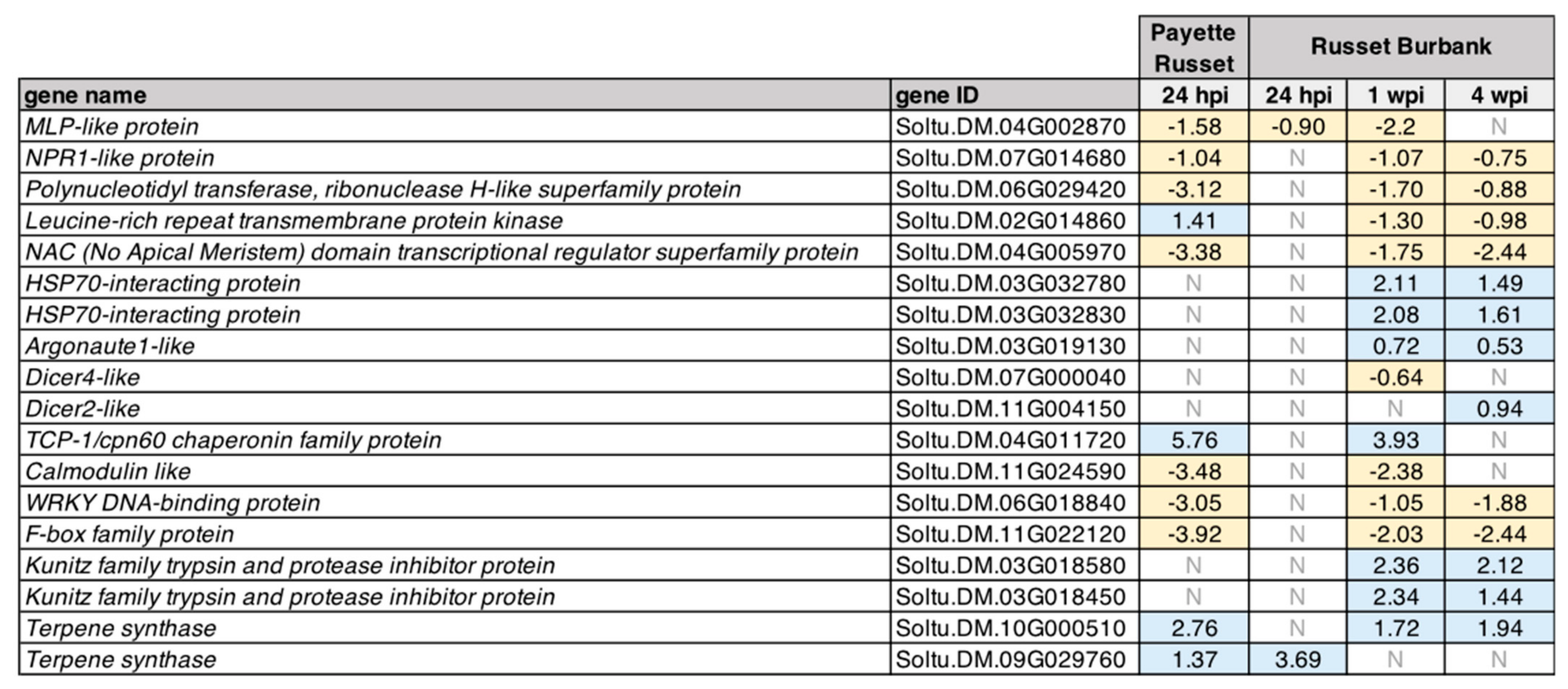
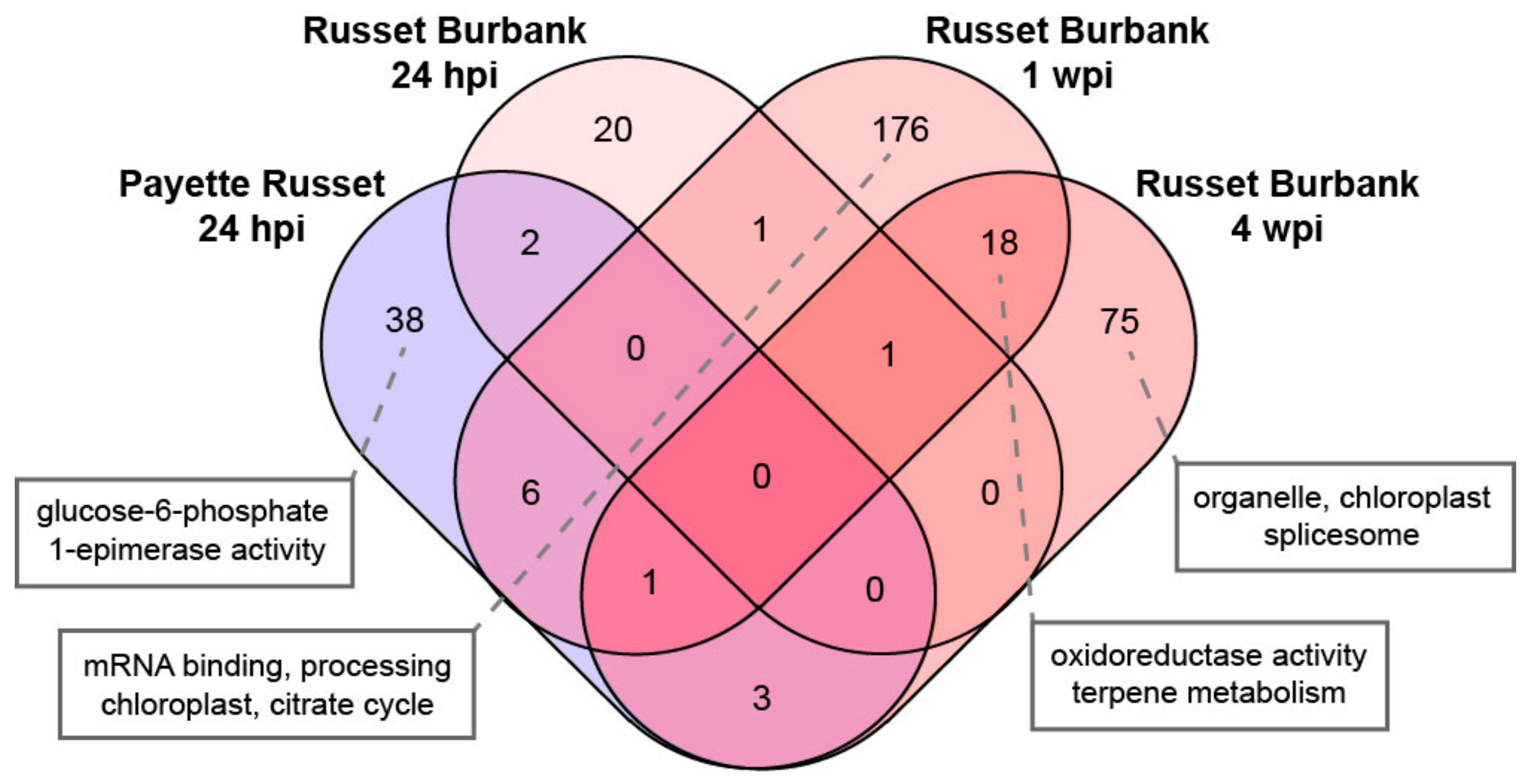
3.2.1. DEGs Shared between PVY-Infected Payette Russet and Russet Burbank May Indicate Antiviral Roles
3.2.2. DEGs Shared between Russet Burbank Time Points May Be Important to Successfully Establishing PVY Infection or Involved in Antiviral Defense
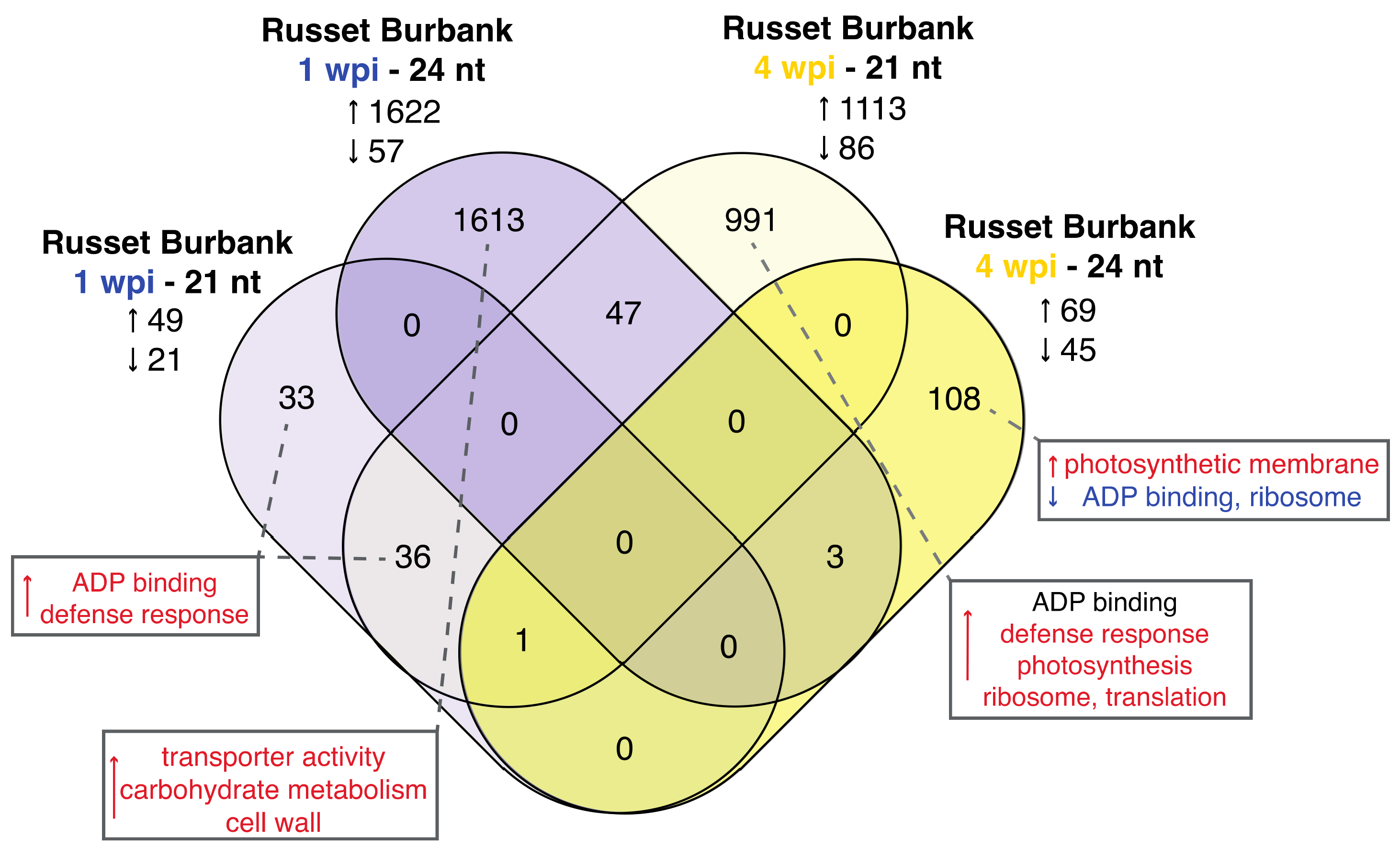
3.3. Expression and Type of Small RNA Varies by Time Point and Cultivar
3.3.1. The miRNA Profiles of PVY-Infected Russet Burbank Plants Mirrors mRNA Results and Is Similar to PVY-Tolerant Potato Cultivars
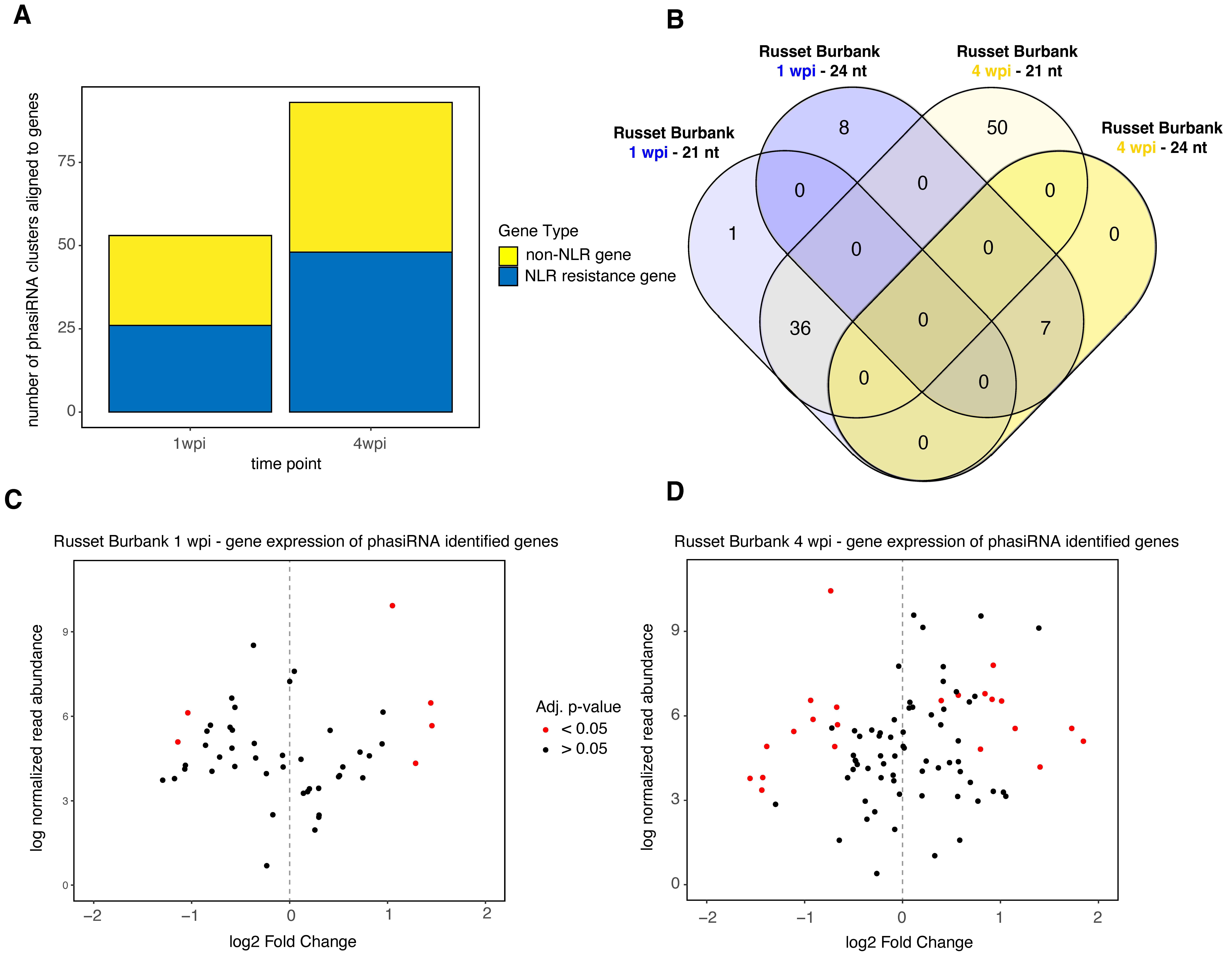
3.3.2. Analysis of phasiRNAs in PVY-Infected Potato Plants Reveals Enrichment of NLR Resistance Gene Targets and Similarities between Time Points
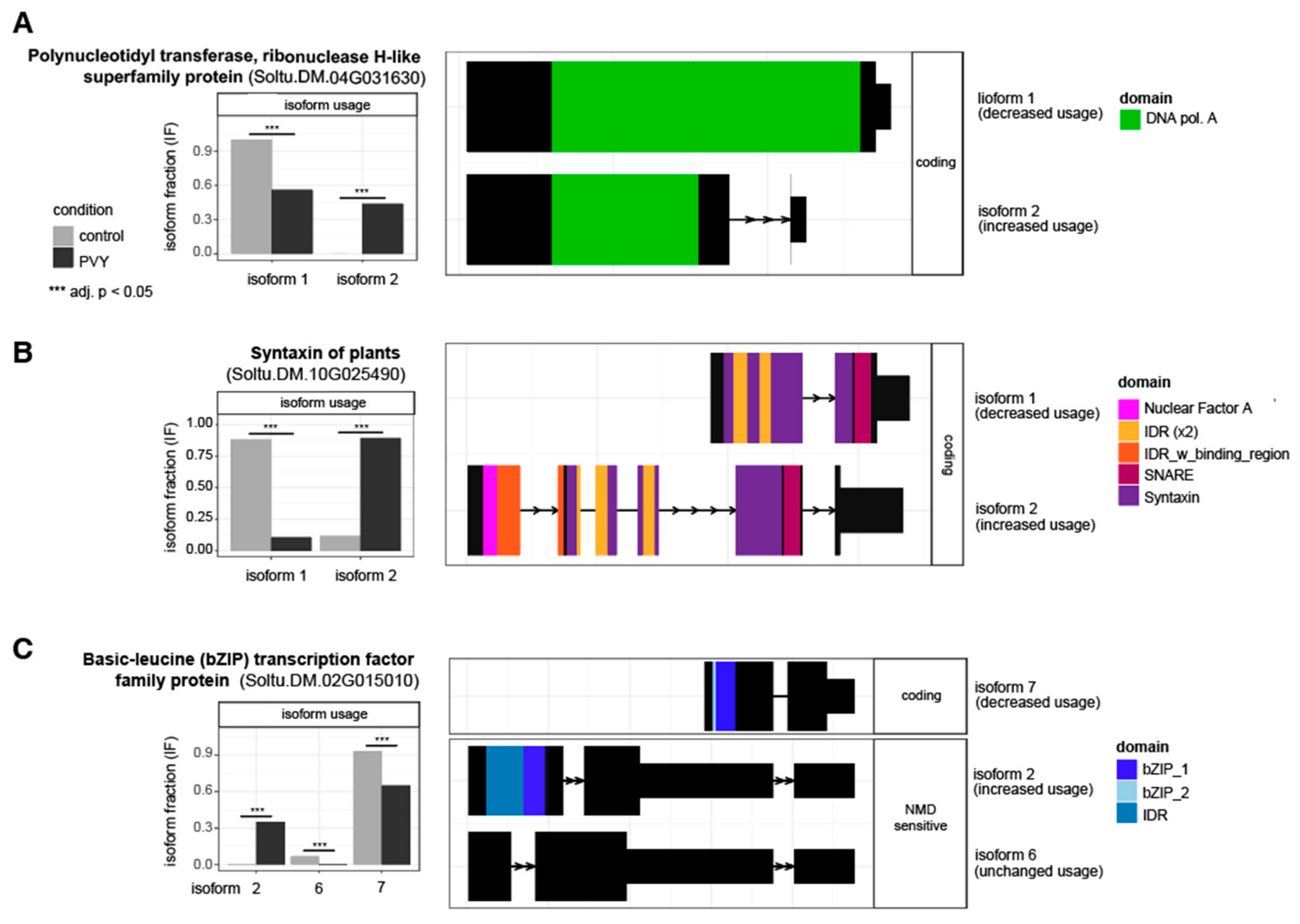
3.4. Alternative Splicing Analysis Reveals Genes outside of Differential Expression Analysis with Potential Impacts on PVY Infection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Devaux, A.; Kromann, P.; Ortiz, O. Potatoes for Sustainable Global Food Security. Potato Res. 2014, 57, 185–199. [Google Scholar] [CrossRef]
- Steinwand, M.A.; Ronald, P.C. Crop Biotechnology and the Future of Food. Nat. Food 2020, 1, 273–283. [Google Scholar] [CrossRef]
- Lal, M.; Yadav, S.; Pant, R.P.; Dua, V.; Singh, B.; Kaushik, S. Impact of Global Climate Change on Potato Diseases and Strategies for Their Mitigation. In Sustainable Potato Production and the Impact of Climate Change; IGI Global: Hershey, PA, USA, 2017; pp. 205–227. [Google Scholar]
- Karasev, A.V.; Gray, S.M. Continuous and Emerging Challenges of Potato virus Y in Potato. Annu. Rev. Phytopathol. 2013, 51, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Talianksy, M.E. Potato virus Y Emergence and Evolution from the Andes of South America to Become a Major Destructive Pathogen of Potato and Other Solanaceous Crops Worldwide. Viruses 2020, 12, 1430. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; De Boer, S.; Lorenzen, J.; Karasev, A.; Whitworth, J.; Nolte, P.; Singh, R.; Boucher, A.; Xu, H. Potato virus Y: An Evolving Concern for Potato Crops in the United States and Canada. Plant Dis. 2010, 94, 1384–1397. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Durand-Tardif, M.; Tronchet, M.; Boudazin, G.; Astier-Manifacier, S.; Casse-Delbart, F. Nucleotide Sequence of Potato virus Y (N Strain) Genomic RNA. J. Gen. Virol. 1989, 70, 935–947. [Google Scholar] [CrossRef]
- Fageria, M.; Nie, X.; Gallagher, A.; Singh, M. Mechanical Transmission of Potato virus Y (PVY) through Seed Cutting and Plant Wounding. Am. J. Potato Res. 2014, 92, 143–147. [Google Scholar] [CrossRef]
- Ng, J.C.K.; Falk, B.W. Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annu. Rev. Phytopathol. 2006, 44, 183–212. [Google Scholar] [CrossRef]
- Nie, B.; Singh, M.; Murphy, A.; Sullivan, A.; Xie, C.; Nie, X. Response of Potato Cultivars to Five Isolates Belonging to Four Strains of Potato virus Y. Plant Dis. 2012, 96, 1422–1429. [Google Scholar] [CrossRef]
- Funke, C.N.; Nikolaeva, O.V.; Green, K.J.; Tran, L.T.; Chikh-Ali, M.; Quintero-Ferrer, A.; Cating, R.A.; Frost, K.E.; Hamm, P.B.; Olsen, N.; et al. Strain-Specific Resistance to Potato Virus Y (PVY) in Potato and Its Effect on the Relative Abundance of PVY Strains in Commercial Potato Fields. Plant Dis. 2017, 101, 20–28. [Google Scholar] [CrossRef]
- Goyer, A.; Hamlin, L.; Crosslin, J.M.; Buchanan, A.; Chang, J.H. Rna-Seq Analysis of Resistant and Susceptible Potato Varieties During the Early Stages of Potato virus Y Infection. BMC Genom. 2015, 16, 472. [Google Scholar] [CrossRef] [PubMed]
- Baebler, Š.; Coll, A.; Gruden, K. Plant Molecular Responses to Potato virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses 2020, 12, 217. [Google Scholar] [CrossRef] [PubMed]
- Nolte, P.; Whitworth, J.L.; Thornton, M.K.; McIntosh, C.S. Effect of Seedborne Potato virus Y on Performance of Russet Burbank, Russet Norkotah, and Shepody Potato. Plant Dis. 2004, 88, 248–252. [Google Scholar] [CrossRef]
- Lauring, A.S.; Frydman, J.; Andino, R. The Role of Mutational Robustness in RNA Virus Evolution. Nat. Rev. Microbiol. 2013, 11, 327–336. [Google Scholar] [CrossRef]
- Karasev, A.V.; Gray, S.M. Genetic Diversity of Potato virus Y Complex. Am. J. Potato Res. 2013, 90, 7–13. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant Immunity against Viruses: Antiviral Immune Receptors in Focus. Ann. Bot. 2016, 119, 711–723. [Google Scholar] [CrossRef]
- Kourelis, J.; Van Der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Ross, B.T.; Zidack, N.K.; Flenniken, M.L. Extreme Resistance to Viruses in Potato and Soybean. Front. Plant Sci. 2021, 12, 658981. [Google Scholar] [CrossRef]
- Kumar, D. Salicylic Acid Signaling in Disease Resistance. Plant Sci. 2014, 228, 127–134. [Google Scholar] [CrossRef]
- Tian, Y.-P.; Valkonen, J.P.T. Genetic Determinants of Potato virus Y Required to Overcome or Trigger Hypersensitive Resistance to PVY Strain Group O Controlled by the Gene Ny in Potato. Mol. Plant Microbe Interact. 2013, 26, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solińska, J.; Urbanowicz, A.; Figlerowicz, M.; Bujarski, J.J. RNA-RNA Recombination in Plant Virus Replication and Evolution. Annu. Rev. Phytopathol. 2011, 49, 415–443. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Gratias, A.; Meyers, B.C.; Geffroy, V. Molecular Mechanisms That Limit the Costs of NLR-mediated Resistance in Plants. Mol. Plant Pathol. 2018, 19, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Eulgem, T. Transcript-Level Expression Control of Plant NLR Genes. Mol. Plant Pathol. 2018, 19, 1267–1281. [Google Scholar] [CrossRef]
- Chang, C.; Zhang, L.; Shen, Q.H. Partitioning, Repressing and Derepressing: Dynamic Regulations in Mla Immune Receptor Triggered Defense Signaling. Front. Plant Sci. 2013, 4, 396. [Google Scholar] [CrossRef] [PubMed]
- Stokes, T.L. Epigenetic Variation in Arabidopsis Disease Resistance. Genes Dev. 2002, 16, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Soosaar, J.L.M.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of Plant Resistance to Viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Galun, E. RNA Silencing in Plants. In Vitro Cell. Dev. Biol. Plant 2005, 41, 113–123. [Google Scholar] [CrossRef]
- Vaucheret, H. Plant Argonautes. Trends Plant Sci. 2008, 13, 350–358. [Google Scholar] [CrossRef]
- Ding, S.-W.; Voinnet, O. Antiviral Immunity Directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Fang, X.; Qi, Y. RNAi in Plants: An Argonaute-Centered View. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef]
- Szittya, G.; Burgyán, J. RNA Interference-Mediated Intrinsic Antiviral Immunity in Plants; Springer: Berlin/Heidelberg, Germany, 2013; pp. 153–181. [Google Scholar]
- Axtell, M.J.; Bartel, D.P. Antiquity of MicroRNAs and Their Targets in Land Plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; Gonzalez, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. Micrornas as Master Regulators of the Plant NB-LRR Defense Gene Family Via the Production of Phased, Trans-Acting Sirnas. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A MicroRNA Superfamily Regulates Nucleotide Binding Site-Leucine-Rich Repeats and Other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef]
- Tameling, W.I.L.; Elzinga, S.D.J.; Darmin, P.S.; Vossen, J.H.; Takken, F.L.W.; Haring, M.A.; Cornelissen, B.J.C. The Tomato R Gene Products I-2 and Mi-1 are Functional ATP Binding Proteins with ATPase Activity. Plant Cell 2002, 14, 2929–2939. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.-W.; et al. Ligand-Triggered Allosteric ADP Release Primes a Plant NLR Complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Bethke, P.C.; Nassar, A.M.K.; Kubow, S.; Leclerc, Y.N.; Li, X.-Q.; Haroon, M.; Molen, T.; Bamberg, J.; Martin, M.; Donnelly, D.J. History and Origin of Russet Burbank (Netted Gem) a Sport of Burbank. Am. J. Potato Res. 2014, 91, 594–609. [Google Scholar] [CrossRef]
- Boyd, E.; Carpenter, E.; Ross, B.T.; Zidack, N.; Flenniken, M.L. Potato Cultivar and Seed Type Affect the Development of Systemic Potato virus Y (PVYN-Wi) Infection. Am. J. Potato Res. 2018, 95, 183–190. [Google Scholar] [CrossRef]
- Baebler, S.; Krecic-Stres, H.; Rotter, A.; Kogovsek, P.; Cankar, K.; Kok, E.J.; Gruden, K.; Kovac, M.; Zel, J.; Pompe-Novak, M.; et al. PVY(NTN) Elicits a Diverse Gene Expression Response in Different Potato Genotypes in the First 12 H after Inoculation. Mol. Plant Pathol. 2009, 10, 263–275. [Google Scholar] [CrossRef]
- Baebler, Š.; Witek, K.; Petek, M.; Stare, K.; Tušek-Žnidarič, M.; Pompe-Novak, M.; Renaut, J.; Szajko, K.; Strzelczyk-Żyta, D.; Marczewski, W.; et al. Salicylic Acid Is an Indispensable Component of the Ny-1 Resistance-Gene-Mediated Response against Potato virus Y Infection in Potato. J. Exp. Bot. 2014, 65, 1095–1109. [Google Scholar] [CrossRef]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.G.; Hennig, J. Extreme Resistance to Potato virus Y in Potato Carrying the Ry Sto Gene Is Mediated by a TIR-NLR Immune Receptor. Plant Biotechnol. J. 2020, 18, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Brigneti, G.; Garcia-Mas, J.; Baulcombe, D.C. Molecular Mapping of the Potato virus Y Resistance Gene Rysto in Potato. Theor. Appl. Genet. 1997, 94, 198–203. [Google Scholar] [CrossRef]
- Kondrák, M.; Kopp, A.; Uri, C.; Sós-Hegedűs, A.; Csákvári, E.; Schiller, M.; Barta, E.; Cernák, I.; Polgár, Z.; Taller, J.; et al. Mapping and DNA Sequence Characterisation of the Rysto Locus Conferring Extreme Virus Resistance to Potato Cultivar ‘White Lady’. PLoS ONE 2020, 15, e0224534. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene Quantification Using Real-Time Quantitative PCR. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with Deseq2. Gen. Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lunardon, A.; Johnson, N.R.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated Annotations and Analyses of Small RNA–Producing Loci from 47 Diverse Plants. Gen. Res. 2020, 30, 497–513. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISTAT, Stringtie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Pham, G.M.; Hamilton, J.P.; Wood, J.C.; Burke, J.T.; Zhao, H.; Vaillancourt, B.; Ou, S.; Jiang, J.; Buell, C.R. Construction of a Chromosome-Scale Long-Read Reference Genome Assembly for Potato. GigaScience 2020, 9, giaa100. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx Gene from Potato Controls Separate Virus Resistance and Cell Death Responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Teng, C.; Xia, R.; Meyers, B.C. Phasirnas in Plants: Their Biogenesis, Genic Sources, and Roles in Stress Responses, Development, and Reproduction. Plant Cell 2020, 32, 3059–3080. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Takeuchi, S.; Kiba, A.; Tsuda, S.; Suzuki, K.; Hikichi, Y.; Okuno, T. Timing and Extent of Hypersensitive Response Are Critical to Restrict Local and Systemic Spread of Pepper Mild Mottle Virus in Pepper Containing the L3 Gene. J. Gen. Plant Pathol. 2005, 71, 90–94. [Google Scholar] [CrossRef]
- Sasaki, N.; Murakami, T.; Yoshimoto, N.; Komatsu, K.; Matsushita, Y.; Nyunoya, H. Cell-Death-Independent Antiviral Response Mediated by N Resistance Factor in Nicotiana Benthamiana Involves Inhibited Localization of Tobamovirus Movement Protein to Plasmodesmata. J. Gen. Plant Pathol. 2021, 87, 170–177. [Google Scholar] [CrossRef]
- Fan, W.; Dong, X. In Vivo Interaction between NPR1 and Transcription Factor TGA2 Leads to Salicylic Acid–Mediated Gene Activation in Arabidopsis. Plant Cell 2002, 14, 1377–1389. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Chern, M.; Fitzgerald, H.A.; Canlas, P.E.; Navarre, D.A.; Ronald, P.C. Overexpression of a Rice NPR1 Homolog Leads to Constitutive Activation of Defense Response and Hypersensitivity to Light. Mol. Plant Microbe Interact. 2005, 18, 511–520. [Google Scholar] [CrossRef]
- Fitzgerald, H.A.; Chern, M.-S.; Navarre, R.; Ronald, P.C. Overexpression of (At)NPR1 in Rice Leads to a BTH- and Environment-Induced Lesion-Mimic/Cell Death Phenotype. Mol. Plant Microbe Interact. 2004, 17, 140–151. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Lu, S.; Faris, J.D.; Sherwood, R.; Edwards, M.C. Dimerization and Protease Resistance: New Insight into the Function of PR-1. J. Plant Physiol. 2013, 170, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Kogovšek, P.; Pompe-Novak, M.; Baebler, Š.; Rotter, A.; Gow, L.; Gruden, K.; Foster, G.D.; Boonham, N.; Ravnikar, M. Aggressive and Mild Potato virus Y Isolates Trigger Different Specific Responses in Susceptible Potato Plants. Plant Pathol. 2010, 59, 1121–1132. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of Alternative Splicing Regulation: Insights from Molecular and Genomics Approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, R.A. Plant Terpenes: Defense Responses, Phylogenetic Analysis, Regulation and Clinical Applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene Synthases and the Regulation, Diversity and Biological Roles of Terpene Metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef]
- Memelink, J.; Verpoorte, R.; Kijne, J.W. Orcanization of Jasmonate-Responsive Gene Expression in Alkaloid Metabolism. Trends Plant Sci. 2001, 6, 212–219. [Google Scholar] [CrossRef]
- Niki, T.; Mitsuhara, I.; Seo, S.; Ohtsubo, N.; Ohashi, Y. Antagonistic Effect of Salicylic Acid and Jasmonic Acid on the Expression of Pathogenesis-Related (PR) Protein Genes in Wounded Mature Tobacco Leaves. Plant Cell Physiol. 1998, 39, 500–507. [Google Scholar] [CrossRef]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.-W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.-K.; Choi, Y.D. AtMYB44 regulates WRKY70 and Modulates Antagonistic Interaction between Salicylic Acid and Jasmonic Acid Signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Chakraborty, S. Chloroplast: The Trojan Horse in Plant–Virus Interaction. Mol. Plant Pathol. 2018, 19, 504–518. [Google Scholar] [CrossRef]
- Stare, T.; Ramšak, Ž.; Blejec, A.; Stare, K.; Turnšek, N.; Weckwerth, W.; Wienkoop, S.; Vodnik, D.; Gruden, K. Bimodal Dynamics of Primary Metabolism-Related Responses in Tolerant Potato-Potato virus Y Interaction. BMC Genom. 2015, 16, 716. [Google Scholar] [CrossRef]
- Križnik, M.; Baebler, Š.; Gruden, K. Roles of Small Rnas in the Establishment of Tolerant Interaction between Plants and Viruses. Curr. Opin. Virol. 2020, 42, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Križnik, M.; Petek, M.; Dobnik, D.; Ramšak, Ž.; Baebler, Š.; Pollmann, S.; Kreuze, J.F.; Žel, J.; Gruden, K. Salicylic Acid Perturbs sRNA-Gibberellin Regulatory Network in Immune Response of Potato to Potato virus Y Infection. Front. Plant Sci. 2017, 8, 2192. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Križnik, M.; De Paola, D.; Catalano, D.; Gruden, K.; Finetti-Sialer, M.M.; Cillo, F. Potato virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato. Viruses 2019, 11, 1100. [Google Scholar] [CrossRef]
- Li, Y.; Cui, H.; Cui, X.; Wang, A. The Altered Photosynthetic Machinery During Compatible Virus Infection. Curr. Opin. Virol. 2016, 17, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Huang, T.-S.; McNeil, J.; Laliberté, J.-F.O.; Hong, J.; Nelson, R.S.; Wang, A. Sequential Recruitment of the Endoplasmic Reticulum and Chloroplasts for Plant Potyvirus Replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef]
- Aguilar, E.; Garnelo Gomez, B.; Lozano-Duran, R. Recent Advances on the Plant Manipulation by Geminiviruses. Curr. Opin. Plant Biol. 2020, 56, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Laliberté, J.-F.; Sanfaçon, H. Cellular Remodeling During Plant Virus Infection. Annu. Rev. Phytopathol. 2010, 48, 69–91. [Google Scholar] [CrossRef]
- Laliberté, J.-F.; Zheng, H. Viral Manipulation of Plant Host Membranes. Annu. Rev. Virol. 2014, 1, 237–259. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Lozano-Duran, R. Tailoring the Cell: A Glimpse of How Plant Viruses Manipulate Their Hosts. Curr. Opin. Plant Biol. 2019, 52, 164–173. [Google Scholar] [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of Susceptibility as a Novel Breeding Strategy for Durable and Broad-Spectrum Resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef]
- Kanehisa, M. Kegg: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Gorovits, R.; Moshe, A.; Ghanim, M.; Czosnek, H. Recruitment of the Host Plant Heat Shock Protein 70 by Tomato Yellow Leaf Curl Virus Coat Protein Is Required for Virus Infection. PLoS ONE 2013, 8, e70280. [Google Scholar] [CrossRef]
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential Roles of Hsp70 and Hsp90 in the Assembly of the Replicase Complex of a Positive-Strand RNA Plant Virus. J. Virol. 2012, 86, 12091–12104. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Saitoh, H.; Ito, A.; Fujisawa, S.; Kamoun, S.; Katou, S.; Yoshioka, H.; Terauchi, R. Cytosolic HSP90 and HSP70 Are Essential Components of INF1-Mediated Hypersensitive Response and Non-Host Resistance to Pseudomonas Cichorii in Nicotiana Benthamiana. Mol. Plant Pathol. 2003, 4, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. Kunitz Trypsin Inhibitor: An Antagonist of Cell Death Triggered by Phytopathogens and Fumonisin B1 in Arabidopsis. Mol. Plant 2008, 1, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The Expanding World of Small RNAs in Plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Millar, A.A. The Function of miRNAs in Plants. Plants 2020, 9, 198. [Google Scholar] [CrossRef]
- Daxinger, L.; Kanno, T.; Bucher, E.; Van Der Winden, J.; Naumann, U.; Matzke, A.J.M.; Matzke, M. A Stepwise Pathway for Biogenesis of 24-Nt Secondary Sirnas and Spreading of DNA Methylation. EMBO J. 2009, 28, 48–57. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, H.-L.; Daxinger, L.; Pontes, O.; He, X.; Qian, W.; Lin, H.; Xie, M.; Lorkovic, Z.J.; Zhang, S.; et al. An RNA Polymerase-II- and AGO4-Associated Protein Acts in RNA-Directed DNA Methylation. Nature 2010, 465, 106–109. [Google Scholar] [CrossRef]
- Moyo, L.; Ramesh, S.V.; Kappagantu, M.; Mitter, N.; Sathuvalli, V.; Pappu, H.R. The Effects of Potato Virus Y-Derived Virus Small Interfering RNAs of Three Biologically Distinct Strains on Potato (Solanum Tuberosum) Transcriptome. Virol. J. 2017, 14, 129. [Google Scholar] [CrossRef]
- Deng, P.; Muhammad, S.; Cao, M.; Wu, L. Biogenesis and Regulatory Hierarchy of Phased Small Interfering RNAs in Plants. Plant Biotechnol. J. 2018, 16, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A Role for Small RNA in Regulating Innate Immunity During Plant Growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef] [PubMed]
- Jupe, F.; Witek, K.; Verweij, W.; Śliwka, J.; Pritchard, L.; Etherington, G.J.; Maclean, D.; Cock, P.J.; Leggett, R.M.; Bryan, G.J.; et al. Resistance Gene Enrichment Sequencing (RenSeq) Enables Reannotation of the NB-LRR Gene Family from Sequenced Plant Genomes and Rapid Mapping of Resistance Loci in Segregating Populations. Plant J. 2013, 76, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Diezma-Navas, L.; Pérez-González, A.; Artaza, H.; Alonso, L.; Caro, E.; Llave, C.; Ruiz-Ferrer, V. Crosstalk between Epigenetic Silencing and Infection By tobacco Rattle Virus in Arabidopsis. Mol. Plant Pathol. 2019, 20, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Axtell, M.J. Identification and Annotation of Small RNA Genes Using Shortstack. Methods 2014, 67, 20–27. [Google Scholar] [CrossRef]
- Iqbal, M.S.; Hafeez, M.N.; Wattoo, J.I.; Ali, A.; Sharif, M.N.; Rashid, B.; Tabassum, B.; Nasir, I.A. Prediction of Host-Derived miRNAs with the Potential to Target PVY in Potato Plants. Front. Genet. 2016, 7, 159. [Google Scholar] [CrossRef][Green Version]
- Kidner, C.A.; Martienssen, R.A. The Developmental Role of microRNA in Plants. Curr. Opin. Plant Biol. 2005, 8, 38–44. [Google Scholar] [CrossRef]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; Macpherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an Evolutionarily Conserved Target of miR167, Mediates Arabidopsis Root Architecture Changes during High Osmotic Stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef]
- Kuo, Y.-W.; Lin, J.-S.; Li, Y.-C.; Jhu, M.-Y.; King, Y.-C.; Jeng, S.-T. Micror408 Regulates Defense Response Upon Wounding in Sweet Potato. J. Exp. Bot. 2019, 70, 469–483. [Google Scholar] [CrossRef]
- Pagán, I.; García-Arenal, F. Tolerance of Plants to Pathogens: A Unifying View. Annu. Rev. Phytopathol. 2020, 58, 77–96. [Google Scholar] [CrossRef]
- Coruh, C.; Shahid, S.; Axtell, M.J. Seeing the Forest for the Trees: Annotating Small RNA Producing Genes in Plants. Curr. Opin. Plant Biol. 2014, 18, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical Action and Inhibition of Plant Dicer-Like Proteins in Antiviral Defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ullah, Z.; Grumet, R. Interaction between Zucchini Yellow Mosaic Potyvirus RNA-Dependent RNA Polymerase and Host Poly-(A) Binding Protein. Virology 2000, 275, 433–443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Graveley, B.R. Alternative Splicing: Increasing Diversity in the Proteomic World. Trends Genet. 2001, 17, 100–107. [Google Scholar] [CrossRef]
- Wang, B.B.; Brendel, V. Genomewide Comparative Analysis of Alternative Splicing in Plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Vitting-Seerup, K.; Sandelin, A. Isoformswitchanalyzer: Analysis of Changes in Genome-Wide Patterns of Alternative Splicing and Its Functional Consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, S.; Dou, Y.; Zhang, C.; Chen, X.; Yu, B.; Ren, G. Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3′ Tailing of Small RNAs in Arabidopsis. PLoS Genet. 2015, 11, e1005091. [Google Scholar] [CrossRef]
- Lipka, V.; Kwon, C.; Panstruga, R. Snare-Ware: The Role of Snare-Domain Proteins in Plant Biology. Annu. Rev. Cell Dev. Biol. 2007, 23, 147–174. [Google Scholar] [CrossRef]
- Zhang, Z.; Lenk, A.; Andersson, M.X.; Gjetting, T.; Pedersen, C.; Nielsen, M.E.; Newman, M.-A.; Hou, B.-H.; Somerville, S.C.; Thordal-Christensen, H. A Lesion-Mimic Syntaxin Double Mutant in Arabidopsis Reveals Novel Complexity of Pathogen Defense Signaling. Mol. Plant 2008, 1, 510–527. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. Salt Stress Responses in Arabidopsis Utilize a Signal Transduction Pathway Related to Endoplasmic Reticulum Stress Signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Crick, S.L.; Pappu, R.V. Intrinsic Disorder in the Basic Regions of bZIP Transcription Factors: What It Means to Be Disordered and Why It Might Matter! Biophys. J. 2011, 100, 519a. [Google Scholar] [CrossRef][Green Version]
- Das, R.K.; Crick, S.L.; Pappu, R.V. N-Terminal Segments Modulate the A-Helical Propensities of the Intrinsically Disordered Basic Regions of bZIP Proteins. J. Mol. Biol. 2012, 416, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human Transcription Factors Contain a High Fraction of Intrinsically Disordered Regions Essential for Transcriptional Regulation. J. Mol. Biol. 2006, 359, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-S.; Gupta, J.D.; Hunt, A.G. A Plant Poly(a) Polymerase Requires a Novel RNA-Binding Protein for Activity. J. Biol. Chem. 1996, 271, 19831–19835. [Google Scholar] [CrossRef]
- Chang, Y.-F.; Imam, J.S.; Wilkinson, M.F. The Nonsense-Mediated Decay RNA Surveillance Pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef]
- Trivedi, P.; Schenk, P.M.; Wallenstein, M.D.; Singh, B.K. Tiny Microbes, Big Yields: Enhancing Food Crop Production with Biological Solutions. Microb. Biotechnol. 2017, 10, 999–1003. [Google Scholar] [CrossRef]
- Arif, I.; Batool, M.; Schenk, P.M. Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ross, B.T.; Zidack, N.; McDonald, R.; Flenniken, M.L. Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank. Viruses 2022, 14, 523. https://doi.org/10.3390/v14030523
Ross BT, Zidack N, McDonald R, Flenniken ML. Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank. Viruses. 2022; 14(3):523. https://doi.org/10.3390/v14030523
Chicago/Turabian StyleRoss, Brian T., Nina Zidack, Rose McDonald, and Michelle L. Flenniken. 2022. "Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank" Viruses 14, no. 3: 523. https://doi.org/10.3390/v14030523
APA StyleRoss, B. T., Zidack, N., McDonald, R., & Flenniken, M. L. (2022). Transcriptome and Small RNA Profiling of Potato Virus Y Infected Potato Cultivars, Including Systemically Infected Russet Burbank. Viruses, 14(3), 523. https://doi.org/10.3390/v14030523





