Characterization of Bovine Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Recruiting ESCRT for Budding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Plasmid Constructs
2.3. siRNA Construction
2.4. Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
2.5. Purification of BFV Virus-like Particles (VLPs)
2.6. Western Blotting Analysis
2.7. Immunofluorescent Assay
2.8. Co-Immunoprecipitation
2.9. Separation of Cell Nucleus and Cell Cytoplasm
2.10. Antibodies
2.11. Statistical Analysis
3. Results
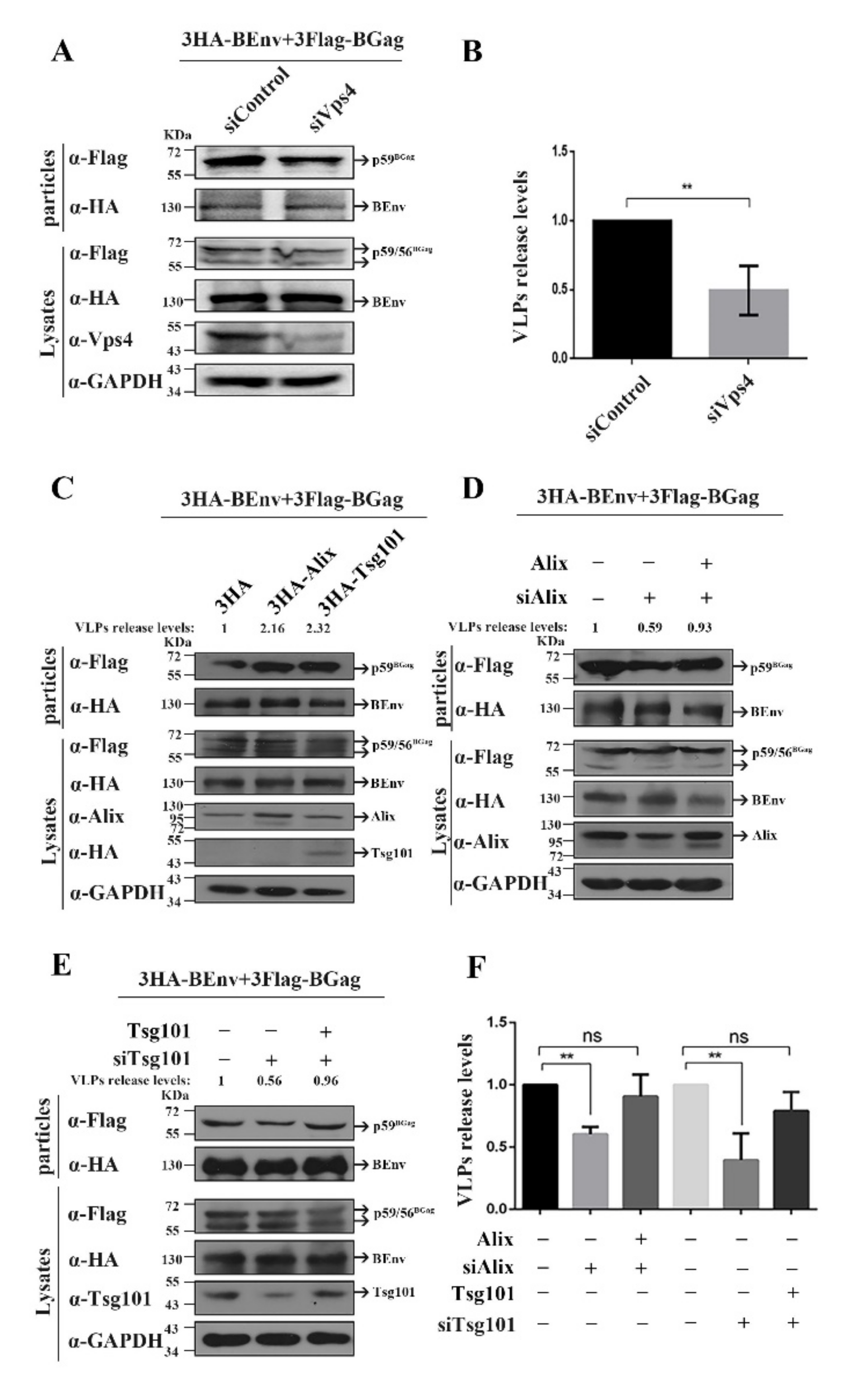
3.1. Alix and Tsg101 Are Necessary for the Budding of BFV VLPs
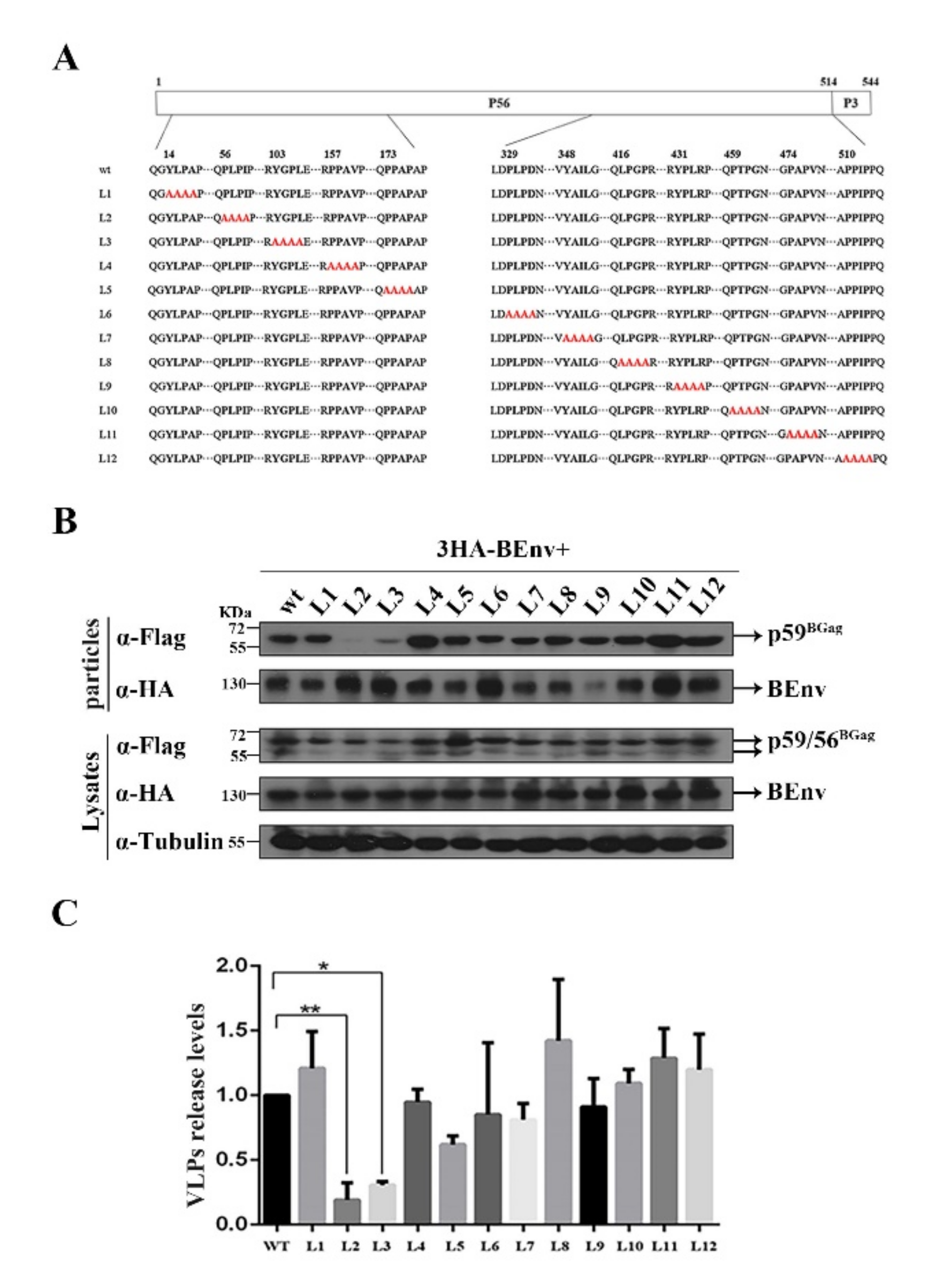
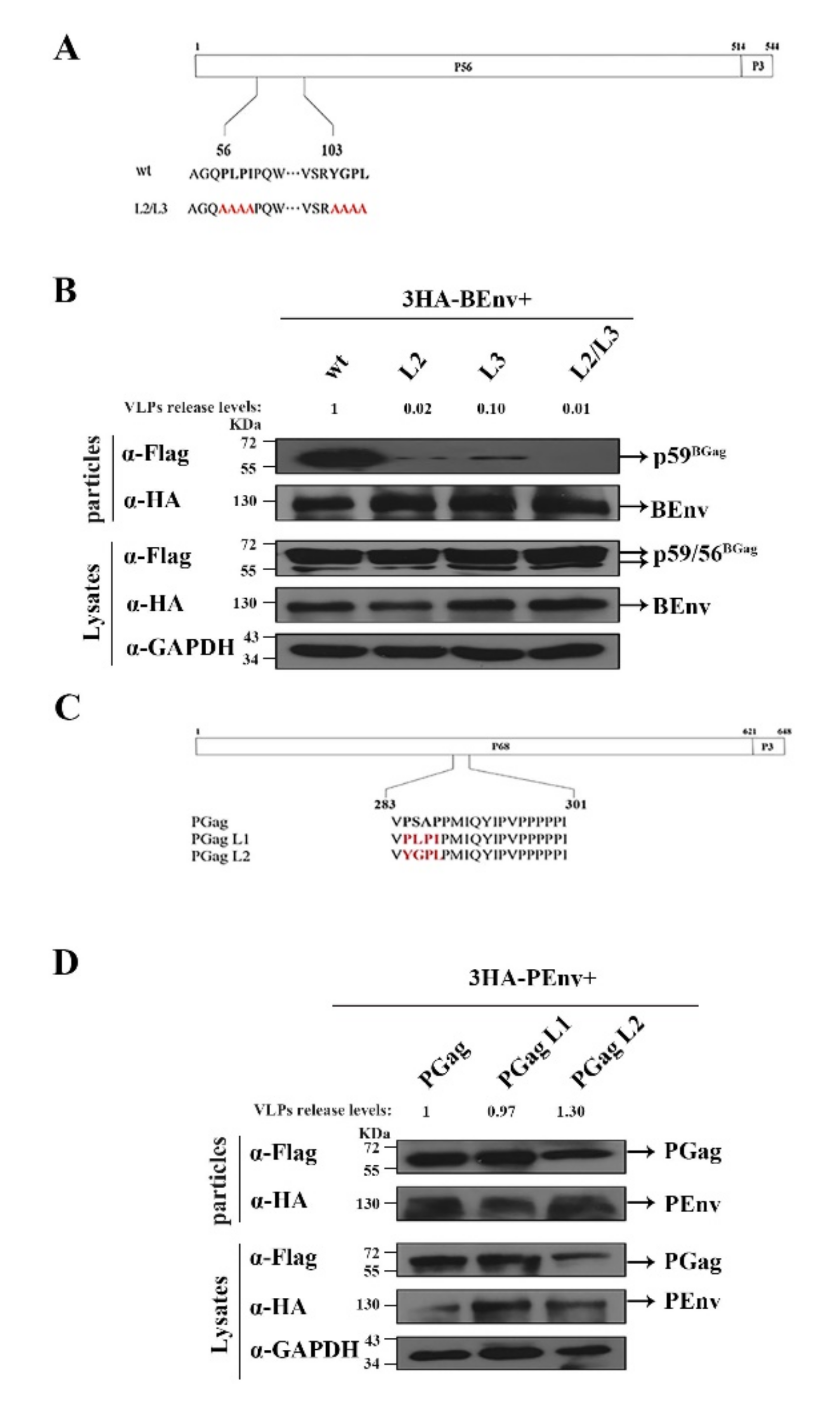
3.2. Identification of L Domain Sequences on the BGag Protein
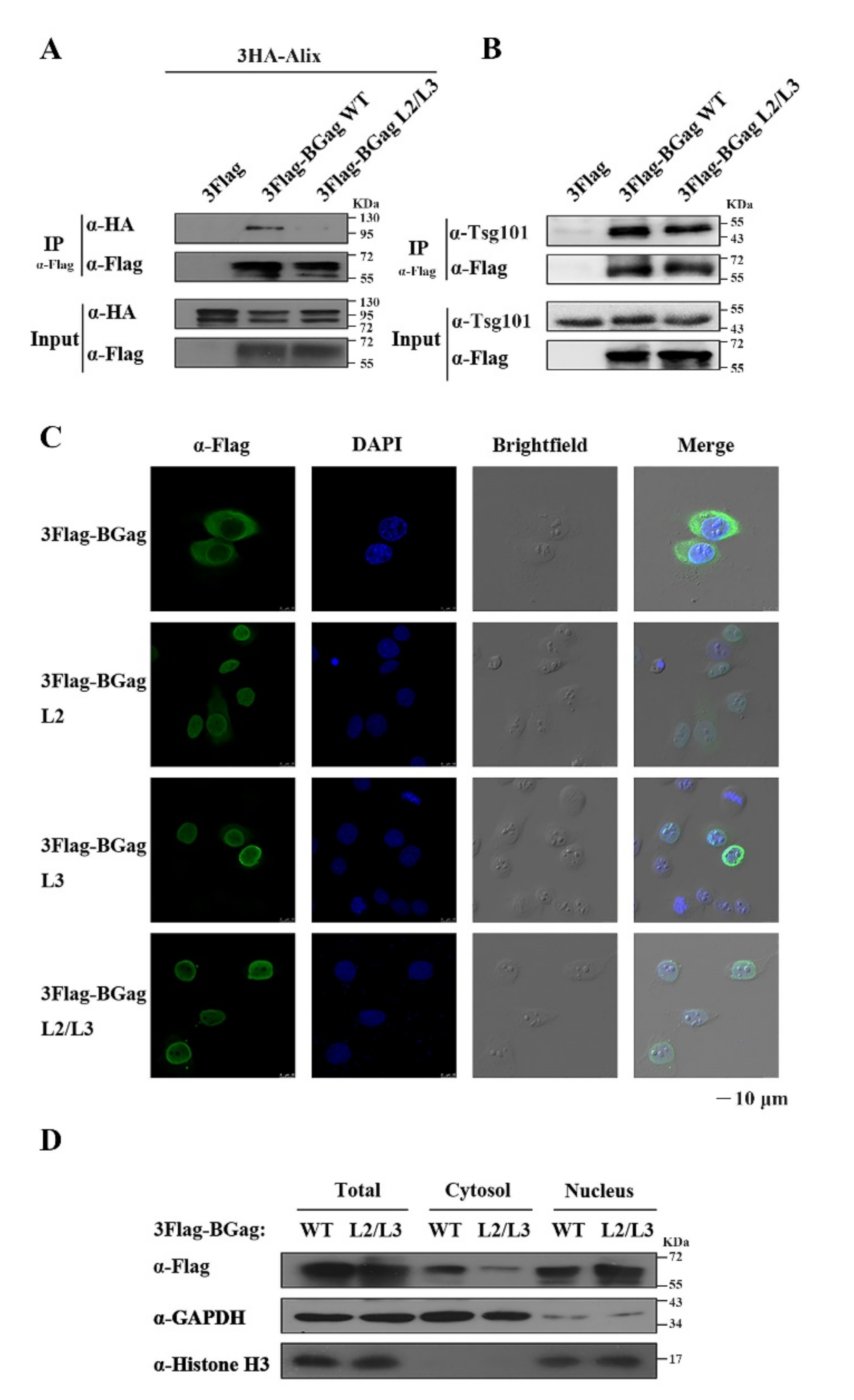
3.3. The Two L Domains of BGag Are Required for Its Cytoplasmic Localization
3.4. V498 in the V Domain of Alix Is Crucial for BFV VLP Budding
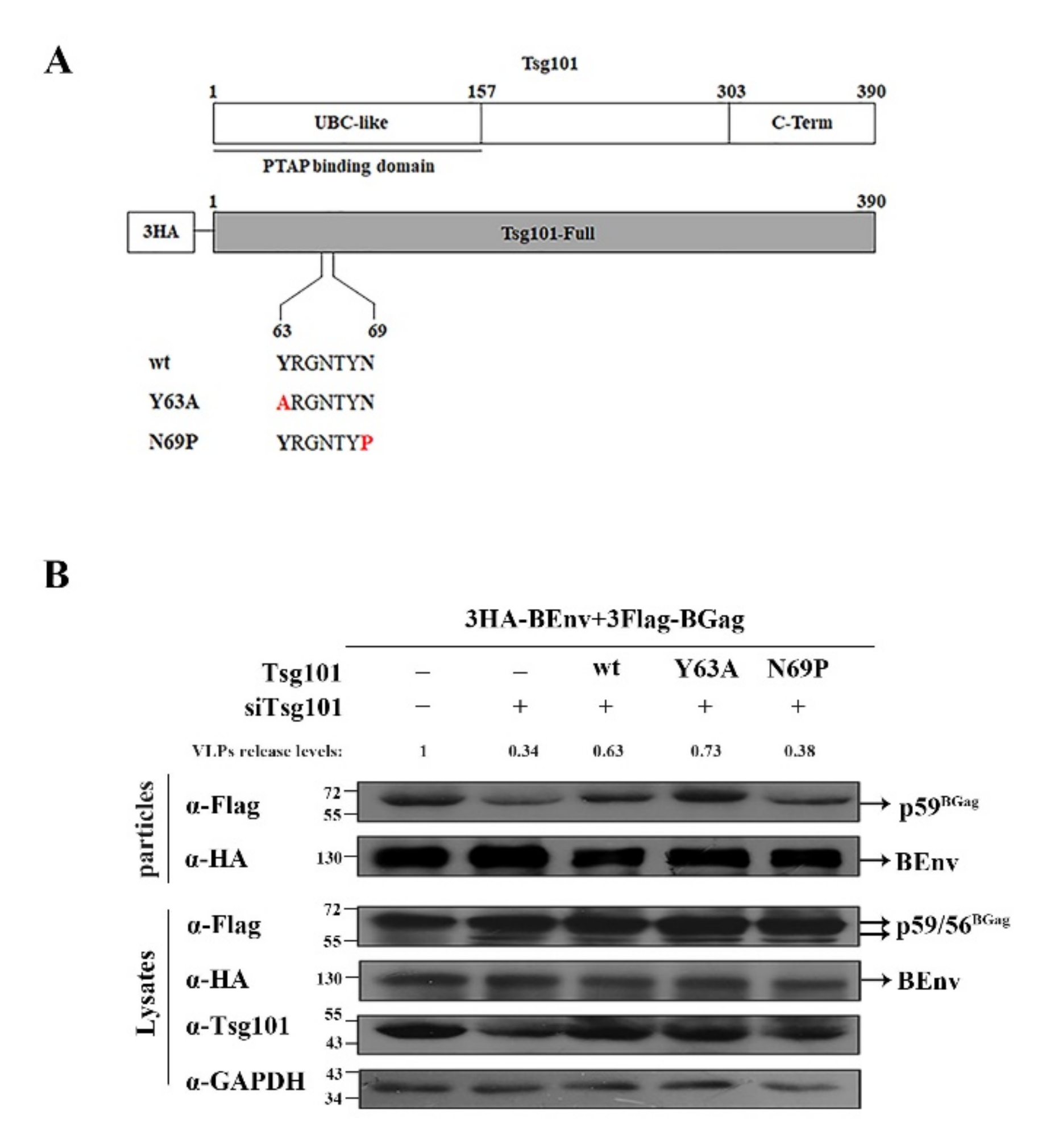
3.5. N69 in the UBC-like Domain of Tsg101 Is Crucial for BFV VLP Budding
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Linial, M.L. Foamy viruses are unconventional retroviruses. J. Virol. 1999, 73, 1747–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rethwilm, A. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.F.; Baldwin, D.N.; Gwynn, S.R.; Yendapalli, S.; Linial, M.L. Human foamy virus replication: A pathway distinct from that of retroviruses and hepadnaviruses. Science 1996, 271, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Lochelt, M. Foamy virus transactivation and gene expression. Curr. Top. Microbiol. Immunol. 2003, 277, 27–61. [Google Scholar] [CrossRef]
- Hutter, S.; Zurnic, I.; Lindemann, D. Foamy virus budding and release. Viruses 2013, 5, 1075–1098. [Google Scholar] [CrossRef]
- Eastman, S.W.; Linial, M.L. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 2001, 75, 6857–6864. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.H.; Oginnusi, A.A.; Ladds, P.W. Isolations and serology of bovine spumavirus. Aust. Vet. J. 1983, 60, 147. [Google Scholar] [CrossRef]
- Broussard, S.R.; Comuzzie, A.G.; Leighton, K.L.; Leland, M.M.; Whitehead, E.M.; Allan, J.S. Characterization of new simian foamy viruses from African nonhuman primates. Virology 1997, 237, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, M.; Akuzawa, M.; Nagatomo, H. Serological survey of the Iriomote cat (Felis iriomotensis) in Japan. J. Wildl. Dis. 1990, 26, 236–245. [Google Scholar] [CrossRef]
- Tobaly-Tapiero, J.; Bittoun, P.; Neves, M.; Guillemin, M.C.; Lecellier, C.H.; Puvion-Dutilleul, F.; Gicquel, B.; Zientara, S.; Giron, M.L.; de The, H.; et al. Isolation and characterization of an equine foamy virus. J. Virol. 2000, 74, 4064–4073. [Google Scholar] [CrossRef] [Green Version]
- Martin-Serrano, J.; Neil, S.J. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011, 9, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Morita, E. Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. FEBS J. 2012, 279, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Bieniasz, P.D. Late budding domains and host proteins in enveloped virus release. Virology 2006, 344, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.J.; Lamb, R.A. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology 2008, 372, 221–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Orenstein, J.M.; Martin, M.A.; Freed, E.O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995, 69, 6810–6818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puffer, B.A.; Parent, L.J.; Wills, J.W.; Montelaro, R.C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 1997, 71, 6541–6546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wills, J.W.; Cameron, C.E.; Wilson, C.B.; Xiang, Y.; Bennett, R.P.; Leis, J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 1994, 68, 6605–6618. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Cameron, C.E.; Wills, J.W.; Leis, J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 1996, 70, 5695–5700. [Google Scholar] [CrossRef] [Green Version]
- Strack, B.; Calistri, A.; Craig, S.; Popova, E.; Gottlinger, H.G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003, 114, 689–699. [Google Scholar] [CrossRef]
- Seo, E.J.; Leis, J. Budding of enveloped viruses: Interferon-induced ISG15-antivirus mechanisms targeting the release process. Adv. Virol. 2012, 2012, 532723. [Google Scholar] [CrossRef] [Green Version]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Cote, M.; Rich, R.L.; et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [Green Version]
- VerPlank, L.; Bouamr, F.; la Grassa, T.J.; Agresta, B.; Kikonyogo, A.; Leis, J.; Carter, C.A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 2001, 98, 7724–7729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, O.; Rainbow, L.; Tilburn, J.; Arst, H.N., Jr.; Penalva, M.A. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell Biol. 2003, 23, 1647–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harty, R.N.; Paragas, J.; Sudol, M.; Palese, P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: Implications for viral budding. J. Virol. 1999, 73, 2921–2929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blot, V.; Perugi, F.; Gay, B.; Prevost, M.C.; Briant, L.; Tangy, F.; Abriel, H.; Staub, O.; Dokhelar, M.C.; Pique, C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 2004, 117, 2357–2367. [Google Scholar] [CrossRef] [Green Version]
- Stange, A.; Mannigel, I.; Peters, K.; Heinkelein, M.; Stanke, N.; Cartellieri, M.; Gottlinger, H.; Rethwilm, A.; Zentgraf, H.; Lindemann, D. Characterization of prototype foamy virus gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 2005, 79, 5466–5476. [Google Scholar] [CrossRef] [Green Version]
- Gc, J.B.; Johnson, K.A.; Husby, M.L.; Frick, C.T.; Gerstman, B.S.; Stahelin, R.V.; Chapagain, P.P. Interdomain salt-bridges in the Ebola virus protein VP40 and their role in domain association and plasma membrane localization. Protein Sci. 2016, 25, 1648–1658. [Google Scholar] [CrossRef] [Green Version]
- Stange, A.; Luftenegger, D.; Reh, J.; Weissenhorn, W.; Lindemann, D. Subviral particle release determinants of prototype foamy virus. J. Virol. 2008, 82, 9858–9869. [Google Scholar] [CrossRef] [Green Version]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Meng, B.; Lever, A.M.L. The interplay between ESCRT and viral factors in the enveloped virus life cycle. Viruses 2021, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Materniak-Kornas, M.; Tan, J.; Heit-Mondrzyk, A.; Hotz-Wagenblatt, A.; Lochelt, M. Bovine foamy virus: Shared and unique molecular features in vitro and in vivo. Viruses 2019, 11, 84. [Google Scholar] [CrossRef] [Green Version]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurley, J.H.; Hanson, P.I. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 2010, 11, 556–566. [Google Scholar] [CrossRef]
- Garcia, M.L.; Reynolds, T.D.; Mothes, W.; Robek, M.D. Functional characterization of the putative hepatitis B virus core protein late domain using retrovirus chimeras. PLoS ONE 2013, 8, e72845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, L.J.; Bennett, R.P.; Craven, R.C.; Nelle, T.D.; Krishna, N.K.; Bowzard, J.B.; Wilson, C.B.; Puffer, B.A.; Montelaro, R.C.; Wills, J.W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 1995, 69, 5455–5460. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Campbell, S.; Bacharach, E.; Rein, A.; Goff, S.P. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 2000, 74, 7250–7260. [Google Scholar] [CrossRef] [Green Version]
- Mullers, E. The foamy virus Gag proteins: What makes them different? Viruses 2013, 5, 1023–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Z.; Madara, J.J.; Liu, Y.; Liu, W.; Ruthel, G.; Freedman, B.D.; Harty, R.N. ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of ebola VP40: A role for ALIX in ebola virus egress. J. Infect. Dis. 2015, 212 (Suppl. 2), S138–S145. [Google Scholar] [CrossRef]
- Fisher, R.D.; Chung, H.Y.; Zhai, Q.; Robinson, H.; Sundquist, W.I.; Hill, C.P. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 2007, 128, 841–852. [Google Scholar] [CrossRef] [Green Version]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. Role of ESCRT-I in retroviral budding. J. Virol. 2003, 77, 4794–4804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, Y.J.; Kuo, L.; Ren, X.; Burgos, P.V.; Zhao, X.Z.; Liu, F.; Burke, T.R., Jr.; Bonifacino, J.S.; Freed, E.O.; Hurley, J.H. Crystallographic and functional analysis of the ESCRT-I /HIV-1 Gag PTAP interaction. Structure 2010, 18, 1536–1547. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Popov, S.; Popova, E.; Inoue, M.; Weissenhorn, W.H.G.G. The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 2009, 37, 181–184. [Google Scholar] [CrossRef]
- Mannigel, I.; Stange, A.; Zentgraf, H.; Lindemann, D. Correct capsid assembly mediated by a conserved YXXLGL motif in prototype foamy virus Gag is essential for infectivity and reverse transcription of the viral genome. J. Virol. 2007, 81, 3317–3326. [Google Scholar] [CrossRef] [Green Version]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar] [CrossRef] [Green Version]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [Green Version]
- Dupont, S.; Sharova, N.; de Horatius, C.; Virbasius, C.M.; Zhu, X.; Bukrinskaya, A.G.; Stevenson, M.; Green, M.R. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 1999, 402, 681–685. [Google Scholar] [CrossRef]
- Beyer, A.R.; Bann, D.V.; Rice, B.; Pultz, I.S.; Kane, M.; Goff, S.P.; Golovkina, T.V.; Parent, L.J. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 2013, 87, 1069–1082. [Google Scholar] [CrossRef] [Green Version]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, R.; Liu, C.; Qiao, W.; Tan, J. Characterization of Bovine Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Recruiting ESCRT for Budding. Viruses 2022, 14, 522. https://doi.org/10.3390/v14030522
Wang Z, Li R, Liu C, Qiao W, Tan J. Characterization of Bovine Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Recruiting ESCRT for Budding. Viruses. 2022; 14(3):522. https://doi.org/10.3390/v14030522
Chicago/Turabian StyleWang, Zhaohuan, Rui Li, Chenxi Liu, Wentao Qiao, and Juan Tan. 2022. "Characterization of Bovine Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Recruiting ESCRT for Budding" Viruses 14, no. 3: 522. https://doi.org/10.3390/v14030522
APA StyleWang, Z., Li, R., Liu, C., Qiao, W., & Tan, J. (2022). Characterization of Bovine Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Recruiting ESCRT for Budding. Viruses, 14(3), 522. https://doi.org/10.3390/v14030522




