Rocaglates as Antivirals: Comparing the Effects on Viral Resistance, Anti-Coronaviral Activity, RNA-Clamping on eIF4A and Immune Cell Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viruses
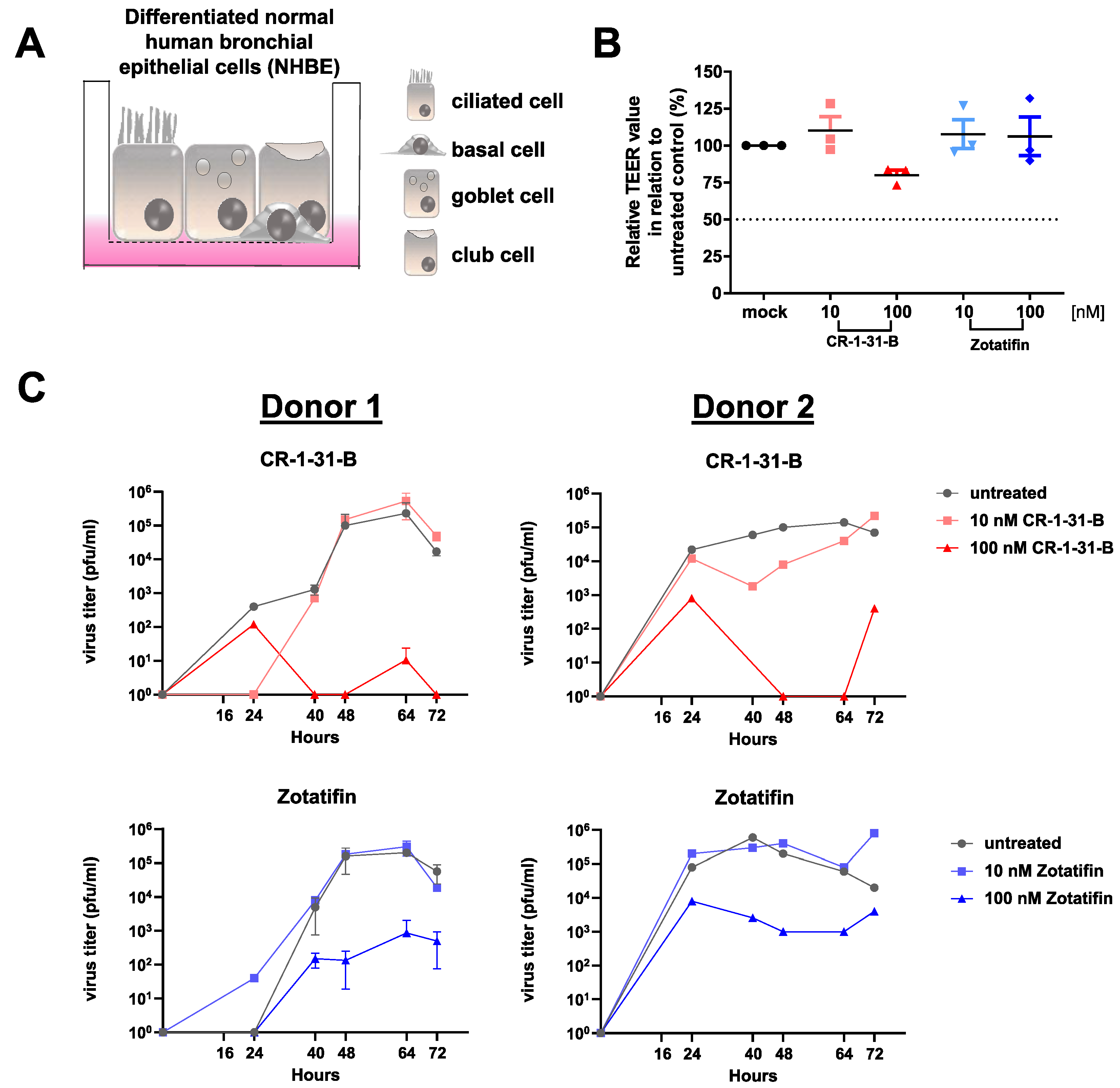
2.2. Human Airway Epithelial Cells
2.3. Reagents
2.4. Cell Toxicity
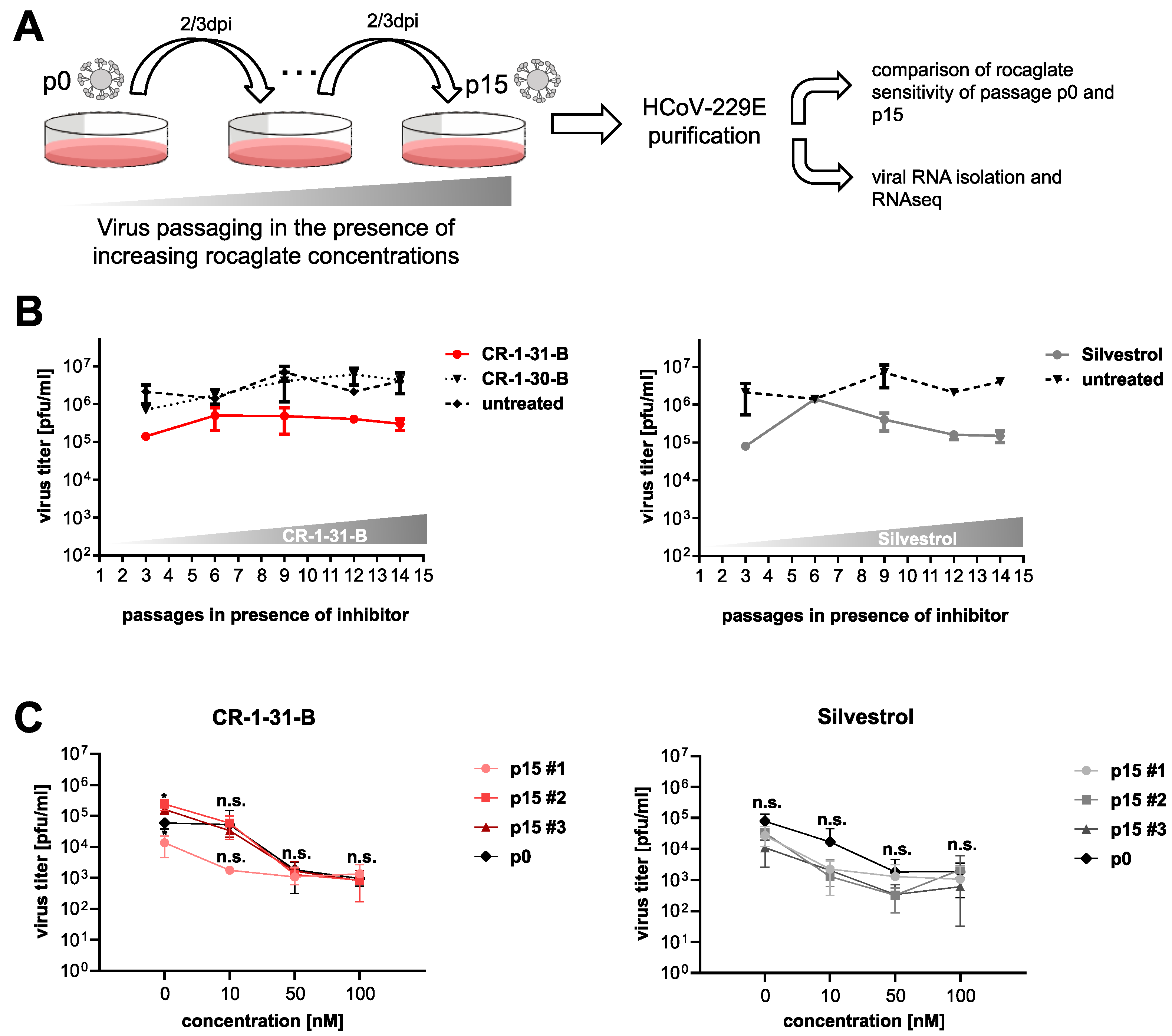
2.5. Serial Passaging of Virus
2.6. Antiviral Activity
2.7. Dual-Luciferase Reporter Assay
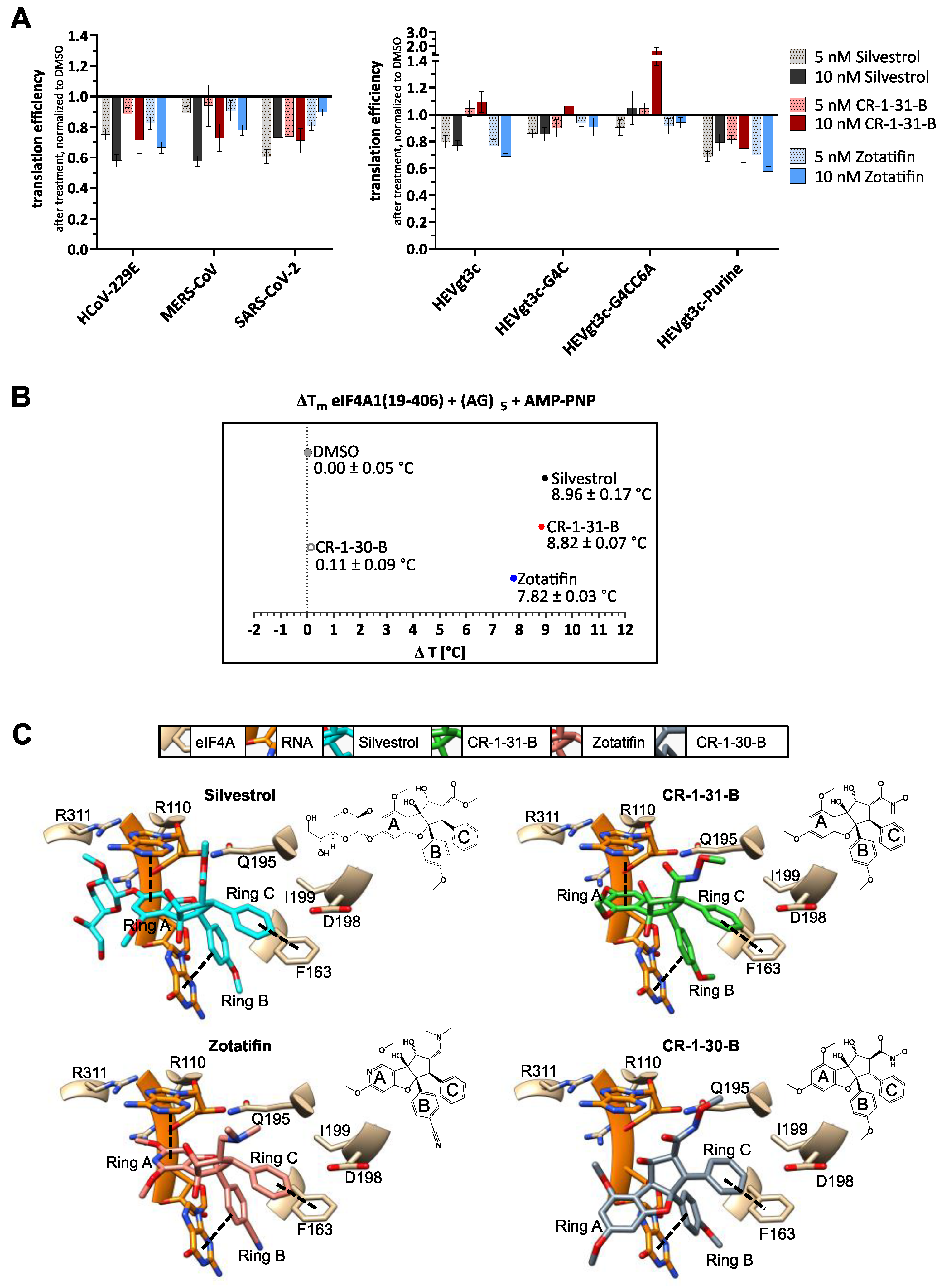
2.8. Thermal Shift Assay
2.9. Docking Analysis
3. Results
3.1. Serial Passaging of HCoV-229E in the Presence of Rocaglates Does Not Decrease Sensitivity to the Corresponding Rocaglates
3.2. Silvestrol, CR-1-31-B and Zotatifin Show Dose-Dependent Inhibition of In Vitro Translation
3.3. Effects on Viability of Silvestrol, CR-1-31-B and Zotatifin in Primary Human Immune Cells
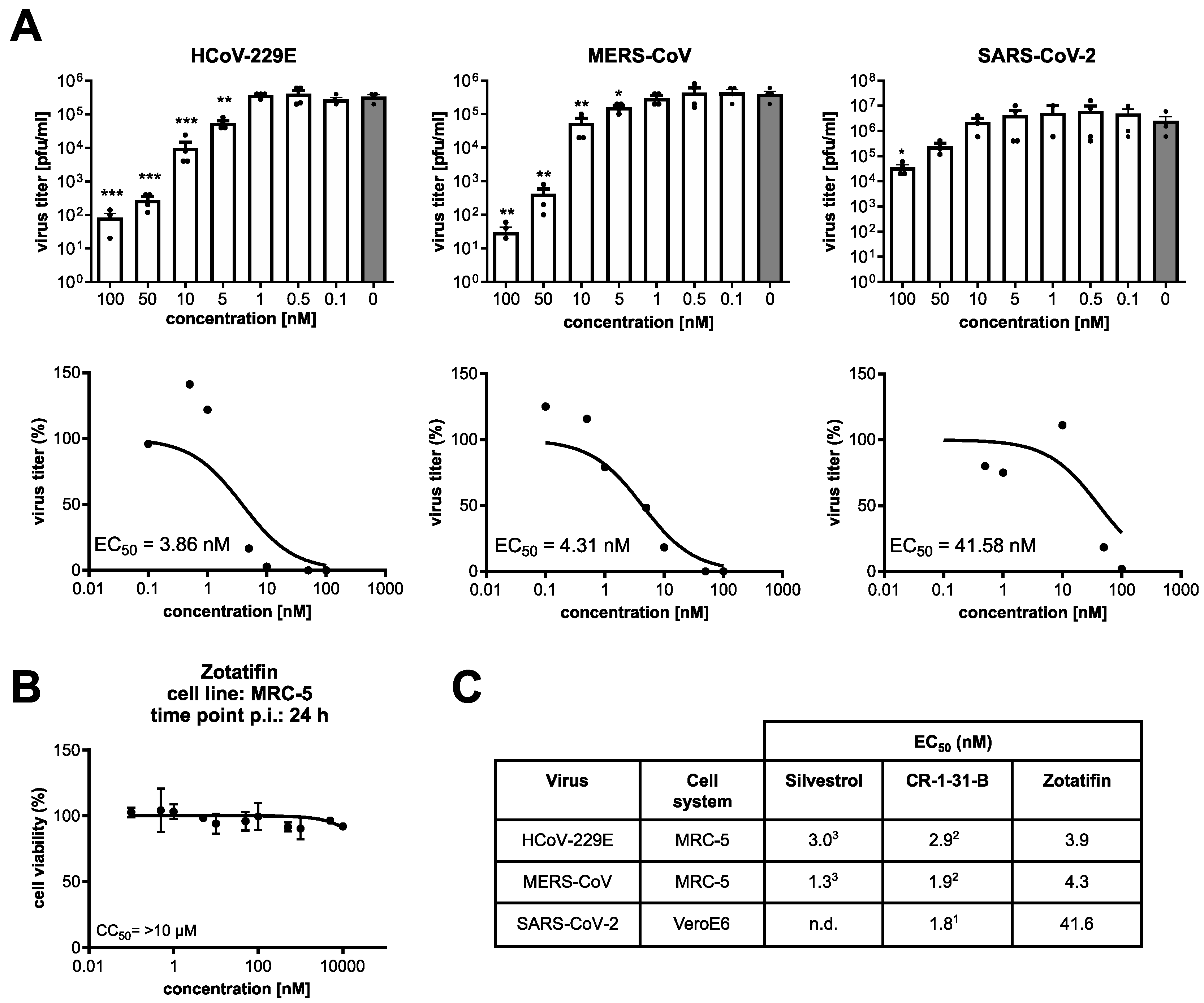
3.4. Zotatifin Reduces Coronavirus Replication In Vitro
3.5. Synthetic Rocaglates Zotatifin and CR-1-31-B Efficiently Reduce Mers-Cov Replication in a Human Airway Epithelial Cell Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Holland, J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997, 51, 151–178. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Sanjuan, R. Adaptive value of high mutation rates of RNA viruses: Separating causes from consequences. J. Virol. 2005, 79, 11555–11558. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.; Galvin, H.D.; Haw, T.Y.; Nutsford, A.N.; Husain, M. Drug resistance in influenza A virus: The epidemiology and management. Infect. Drug Resist. 2017, 10, 121–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar]
- Maher, M.C.; Bartha, I.; Weaver, S.; Di Iulio, J.; Ferri, E.; Soriaga, L.; Lempp, F.A.; Hie, B.L.; Bryson, B.; Berger, B.; et al. Predicting the mutational drivers of future SARS-CoV-2 variants of concern. Sci. Transl. Med. 2022, 14, abk3445. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Patients. Med. Sci. Monit. 2022, 28, e935952. [Google Scholar] [CrossRef]
- Ader, F.; Bouscambert-Duchamp, M.; Hites, M.; Peiffer-Smadja, N.; Poissy, J.; Belhadi, D.; Diallo, A.; Lê, M.-P.; Peytavin, G.; Staub, T.; et al. Remdesivir plus standard of care versus standard of care alone for the treatment of patients admitted to hospital with COVID-19 (DisCoVeRy): A phase 3, randomised, controlled, open-label trial. Lancet. Infect. Dis. 2021, 22, 209–221. [Google Scholar] [CrossRef]
- Bauer, L.; Lyoo, H.; van der Schaar, H.M.; Strating, J.; van Kuppeveld, F.J. Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr. Opin. Virol. 2017, 24, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug. Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Taroncher-Oldenburg, G.; Müller, C.; Obermann, W.; Ziebuhr, J.; Hartmann, R.; Grünweller, A. Targeting the DEAD-Box RNA Helicase eIF4A with Rocaglates-A Pan-Antiviral Strategy for Minimizing the Impact of Future RNA Virus Pandemics. Microorganisms 2021, 9, 540. [Google Scholar] [CrossRef] [PubMed]
- Toribio, R.; Diaz-Lopez, I.; Ventoso, I. New insights into the topology of the scanning ribosome during translation initiation: Lessons from viruses. RNA Biol. 2016, 13, 1223–1227. [Google Scholar] [CrossRef] [Green Version]
- Biedenkopf, N.; Lange-Grünweller, K.; Schulte, F.W.; Weißer, A.; Müller, C.; Becker, D.; Becker, S.; Hartmann, R.K.; Grünweller, A. The natural compound silvestrol is a potent inhibitor of Ebola virus replication. Antiviral Res. 2017, 137, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Elgner, F.; Sabino, C.; Basic, M.; Ploen, D.; Grünweller, A.; Hildt, E. Inhibition of Zika Virus Replication by Silvestrol. Viruses 2018, 10, 149. [Google Scholar] [CrossRef] [Green Version]
- Glitscher, M.; Himmelsbach, K.; Woytinek, K.; Johne, R.; Reuter, A.; Spiric, J.; Schwaben, L.; Grünweller, A.; Hildt, E. Inhibition of Hepatitis E Virus Spread by the Natural Compound Silvestrol. Viruses 2018, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Henss, L.; Scholz, T.; Grünweller, A.; Schnierle, B.S. Silvestrol Inhibits Chikungunya Virus Replication. Viruses 2018, 10, 592. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.; Schulte, F.W.; Lange-Grünweller, K.; Obermann, W.; Madhugiri, R.; Pleschka, S.; Ziebuhr, J.; Hartmann, R.K.; Grünweller, A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018, 150, 123–129. [Google Scholar] [CrossRef]
- Müller, C.; Obermann, W.; Schulte, F.W.; Lange-Grünweller, K.; Oestereich, L.; Elgner, F.; Glitscher, M.; Hildt, E.; Singh, K.; Wendel, H.-G.; et al. Comparison of broad-spectrum antiviral activities of the synthetic rocaglate CR-31-B (-) and the eIF4A-inhibitor Silvestrol. Antivir. Res. 2020, 175, 104706. [Google Scholar] [CrossRef]
- Müller, C.; Obermann, W.; Karl, N.; Wendel, H.G.; Taroncher-Oldenburg, G.; Pleschka, S.; Hartmann, R.K.; Grünweller, A.; Ziebuhr, J. The rocaglate CR-31-B (-) inhibits SARS-CoV-2 replication at non-cytotoxic, low nanomolar concentrations in vitro and ex vivo. Antiviral Res 2021, 186, 105012. [Google Scholar] [CrossRef] [PubMed]
- Todt, D.; Moeller, N.; Praditya, D.; Kinast, V.; Friesland, M.; Engelmann, M.; Verhoye, L.; Sayed, I.M.; Behrendt, P.; Thi, V.L.D.; et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antivir. Res. 2018, 157, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Slaine, P.D.; Kleer, M.; Smith, N.K.; Khaperskyy, D.A.; McCormick, C. Stress Granule-Inducing Eukaryotic Translation Initiation Factor 4A Inhibitors Block Influenza A Virus Replication. Viruses 2017, 9, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, J.; Zhang, W.; Cencic, R.; Devine, W.G.; Beglov, D.; Henkel, T.; Brown, L.E.; Vajda, S.; Porco, J.A.; Pelletier, J. Amidino-Rocaglates: A Potent Class of eIF4A Inhibitors. Cell Chem. Biol. 2019, 26, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.T.; Thompson, P.A.; Nilewski, C.; Sprengeler, P.A.; Sperry, S.; Packard, G.; Michels, T.; Xiang, A.; Tran, C.; Wegerski, C.J.; et al. Design of Development Candidate eFT226, a First in Class Inhibitor of Eukaryotic Initiation Factor 4A RNA Helicase. J. Med. Chem. 2020, 63, 5879–5955. [Google Scholar] [CrossRef]
- Bordeleau, M.-E.; Robert, F.; Gerard, B.; Lindqvist, L.; Chen, S.M.; Wendel, H.-G.; Brem, B.; Greger, H.; Lowe, S.W.; Porco, J.A.; et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Investig. 2008, 118, 2651–2660. [Google Scholar] [CrossRef] [Green Version]
- Kogure, T.; Kinghorn, A.D.; Yan, I.; Bolon, B.; Lucas, D.M.; Grever, M.R.; Patel, T. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS ONE 2013, 8, e76136. [Google Scholar] [CrossRef]
- Patton, J.T.; Lustberg, M.E.; Lozanski, G.; Garman, S.L.; Towns, W.H.; Drohan, C.M.; Lehman, A.; Zhang, X.; Bolon, B.; Pan, L.; et al. The translation inhibitor silvestrol exhibits direct anti-tumor activity while preserving innate and adaptive immunity against EBV-driven lymphoproliferative disease. Oncotarget 2015, 6, 2693–2708. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar]
- Hoffmann, M.; Hannah, K.-W.; Simon, S.; Nadine, K.; Tanja, H.; Sandra, E.; Tobias, S.S.; Georg, H.N.-H.W.; Andreas, N.; Muller, M.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van Der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigo, C.M.; Cencic, R.; Roche, S.P.; Pelletier, J.; Porco, J.J.A. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: Synthetic and biological studies. J. Med. Chem. 2012, 55, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Rajabi, H.; Rodrigo, C.M.; A Porco, J.; Kufe, D. Targeting the eIF4A RNA helicase blocks translation of the MUC1-C oncoprotein. Oncogene 2013, 32, 2179–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, L.; Geisslinger, G.; Parnham, M.J.; Grünweller, A.; Schiffmann, S. Natural antiviral compound silvestrol modulates human monocyte-derived macrophages and dendritic cells. J. Cell Mol. Med. 2020, 24, 6988–6999. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Chu, J.; Zhang, W.; Cencic, R.; O’Connor, P.B.; Robert, F.; Devine, W.G.; Selznick, A.; Henkel, T.; Merrick, W.C.; Brown, L.E.; et al. Rocaglates Induce Gain-of-Function Alterations to eIF4A and eIF4F. Cell. Rep. 2020, 30, 2481–2488.e5. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, S.; Iwasaki, W.; Takahashi, M.; Sakamoto, A.; Watanabe, C.; Shichino, Y.; Floor, S.; Fujiwara, K.; Mito, M.; Dodo, K.; et al. The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA. Mol. Cell. 2019, 73, 738–748.e9. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.V.; Sass, E.J.; Davis, M.E.; Edwards, R.B.; Lozanski, G.; Heerema, N.A.; Lehman, A.; Zhang, X.; Jarjoura, D.; Byrd, J.C.; et al. Resistance to the translation initiation inhibitor silvestrol is mediated by ABCB1/P-glycoprotein overexpression in acute lymphoblastic leukemia cells. AAPS J. 2011, 13, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Szemiel, A.M.; Merits, A.; Orton, R.J.; MacLean, O.A.; Pinto, R.M.; Wickenhagen, A.; Lieber, G.; Matthew, L.T.; Wang, S.; Furnon, W.; et al. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021, 17, e1009929. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Shaban, M.S.; Müller, C.; Mayr-Buro, C.; Weiser, H.; Meier-Soelch, J.; Albert, B.V.; Weber, A.; Linne, U.; Hain, T.; Babayev, I.; et al. Multi-level inhibition of coronavirus replication by chemical ER stress. Nat. Commun. 2021, 12, 5536. [Google Scholar] [CrossRef] [PubMed]

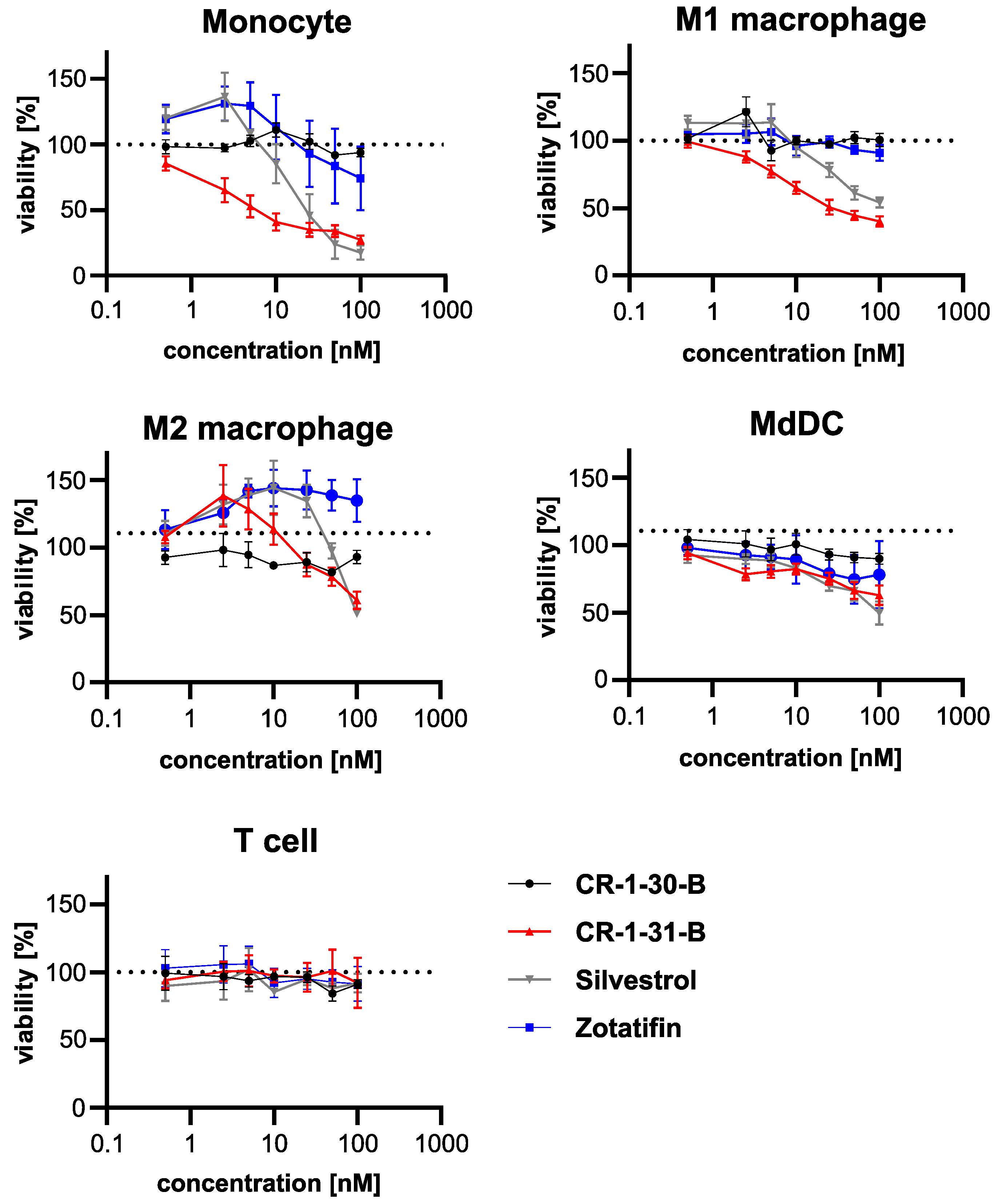
| Cell Type | CC50 (nM) | |||
|---|---|---|---|---|
| Silvestrol | CR-1-31-B | CR-1-30-B | Zotatifin | |
| monocytes | 29.0 | 2.6 | >100 | 78.1 |
| M1 macrophages | 45.6 | 8.8 | >100 | >100 |
| M2 macrophages | >100 | >100 | >100 | >100 |
| dendritic cells | >100 | >100 | >100 | >100 |
| T cells | >100 | >100 | >100 | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obermann, W.; Friedrich, A.; Madhugiri, R.; Klemm, P.; Mengel, J.P.; Hain, T.; Pleschka, S.; Wendel, H.-G.; Hartmann, R.K.; Schiffmann, S.; et al. Rocaglates as Antivirals: Comparing the Effects on Viral Resistance, Anti-Coronaviral Activity, RNA-Clamping on eIF4A and Immune Cell Toxicity. Viruses 2022, 14, 519. https://doi.org/10.3390/v14030519
Obermann W, Friedrich A, Madhugiri R, Klemm P, Mengel JP, Hain T, Pleschka S, Wendel H-G, Hartmann RK, Schiffmann S, et al. Rocaglates as Antivirals: Comparing the Effects on Viral Resistance, Anti-Coronaviral Activity, RNA-Clamping on eIF4A and Immune Cell Toxicity. Viruses. 2022; 14(3):519. https://doi.org/10.3390/v14030519
Chicago/Turabian StyleObermann, Wiebke, Alexandra Friedrich, Ramakanth Madhugiri, Paul Klemm, Jan Philipp Mengel, Torsten Hain, Stephan Pleschka, Hans-Guido Wendel, Roland K. Hartmann, Susanne Schiffmann, and et al. 2022. "Rocaglates as Antivirals: Comparing the Effects on Viral Resistance, Anti-Coronaviral Activity, RNA-Clamping on eIF4A and Immune Cell Toxicity" Viruses 14, no. 3: 519. https://doi.org/10.3390/v14030519
APA StyleObermann, W., Friedrich, A., Madhugiri, R., Klemm, P., Mengel, J. P., Hain, T., Pleschka, S., Wendel, H.-G., Hartmann, R. K., Schiffmann, S., Ziebuhr, J., Müller, C., & Grünweller, A. (2022). Rocaglates as Antivirals: Comparing the Effects on Viral Resistance, Anti-Coronaviral Activity, RNA-Clamping on eIF4A and Immune Cell Toxicity. Viruses, 14(3), 519. https://doi.org/10.3390/v14030519








