Multiorgan and Vascular Tropism of SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Data and Autopsy
2.2. Histopathology
2.3. Viral Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Gauchotte, G.; Venard, V.; Segondy, M.; Cadoz, C.; Esposito-Fava, A.; Barraud, D.; Louis, G. SARS-Cov-2 fulminant myocarditis: An autopsy and histopathological case study. Int. J. Legal Med. 2021, 135, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Basso, C.; Leone, O.; Rizzo, S.; De Gaspari, M.; Van Der Wal, A.C.; Aubry, M.C.; Bois, M.C.; Lin, P.T.; Maleszewski, J.J.; Stone, J.R. Pathological features of COVID-19-associated myocardial injury: A multicentre cardiovascular pathology study. Eur. Heart J. 2020, 41, 3827–3835. [Google Scholar] [CrossRef] [PubMed]
- Guéant, J.; Guéant-Rodriguez, R.; Fromonot, J.; Oussalah, A.; Louis, H.; Chery, C.; Gette, M.; Gleye, S.; Callet, J.; Raso, J.; et al. Elastase and exacerbation of neutrophil innate immunity are involved in multi-visceral manifestations of COVID-19. Allergy 2021, 76, 1846–1858. [Google Scholar] [CrossRef]
- Delgado-Gonzalez, P.; Gonzalez-Villarreal, C.A.; Roacho-Perez, J.A.; Quiroz-Reyes, A.G.; Islas, J.F.; Delgado-Gallegos, J.L.; Arellanos-Soto, D.; Galan-Huerta, K.A.; Garza-Treviño, E.N. Inflammatory effect on the gastrointestinal system associated with COVID-19. World J. Gastroenterol. 2021, 27, 4160–4171. [Google Scholar] [CrossRef]
- McGrowder, D.A.; Miller, F.; Anderson Cross, M.; Anderson-Jackson, L.; Bryan, S.; Dilworth, L. Abnormal Liver Biochemistry Tests and Acute Liver Injury in COVID-19 Patients: Current Evidence and Potential Pathogenesis. Diseases 2021, 9, 50. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 2–5. [Google Scholar] [CrossRef]
- Bhatnagar, J.; Gary, J.; Reagan-Steiner, S.; Estetter, L.B.; Tong, S.; Tao, Y.; Denison, A.M.; Lee, E.; Deleon-Carnes, M.; Li, Y.; et al. Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 Replication and Tropism in the Lungs, Airways, and Vascular Endothelium of Patients with Fatal Coronavirus Disease 2019: An Autopsy Case Series. J. Infect. Dis. 2021, 223, 752–764. [Google Scholar] [CrossRef]
- Casagrande, M.; Fitzek, A.; Spitzer, M.; Püschel, K.; Glatzel, M.; Krasemann, S.; Aepfelbacher, M.; Nörz, D.; Lütgehetmann, M.; Pfefferle, S.; et al. Detection of SARS-CoV-2 genomic and subgenomic RNA in retina and optic nerve of patients with COVID-19. Br. J. Ophthalmol. 2021, 1–5. [Google Scholar] [CrossRef]
- Campos, R.K.; Camargos, V.N.; Azar, S.R.; Haines, C.A.; Eyzaguirre, E.J.; Rossi, S.L. Sars-cov-2 infects hamster testes. Microorganisms 2021, 9, 1318. [Google Scholar] [CrossRef]
- Ardestani Zadeh, A.; Arab, D. COVID-19 and male reproductive system: Pathogenic features and possible mechanisms. J. Mol. Histol. 2021, 2, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Etievant, S.; Bal, A.; Escuret, V.; Brengel-Pesce, K.; Bouscambert, M.; Cheynet, V.; Generenaz, L.; Oriol, G.; Destras, G.; Billaud, G.; et al. Performance Assessment of SARS-CoV-2 PCR Assays Developed by WHO Referral Laboratories. J. Clin. Med. 2020, 9, 1871. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Kamar, N.; Izopet, J.; Cintas, P.; Garrouste, C.; Uro-Coste, E.; Cointault, O.; Rostaing, L. Hepatitis E Virus-Induced Neurological Symptoms in a Kidney-Transplant Patient with Chronic Hepatitis. Am. J. Transplant. 2010, 10, 1321–1324. [Google Scholar] [CrossRef] [PubMed]
- Dunfee, R.L.; Thomas, E.R.; Gorry, P.R.; Wang, J.; Taylor, J.; Kunstman, K.; Wolinsky, S.M.; Gabuzda, D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. USA 2006, 103, 15160–15165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, S.; Saksena, S.; Sadri-Ardekani, H. ACE2 receptor expression in testes: Implications in coronavirus disease 2019 pathogenesis. Biol. Reprod. 2020, 103, 449–451. [Google Scholar] [CrossRef]
- Poma, A.M.; Bonuccelli, D.; Giannini, R.; Macerola, E.; Vignali, P.; Ugolini, C.; Torregrossa, L.; Proietti, A.; Pistello, M.; Basolo, A.; et al. COVID-19 autopsy cases: Detection of virus in endocrine tissues. J. Endocrinol. Investig. 2021, 45, 209–214. [Google Scholar] [CrossRef]
- Verma, R.; Kim, E.; Martínez-Colón, G.J.; Jagannathan, P.; Rustagi, A.; Parsonnet, J.; Bonilla, H.; Khosla, C.; Holubar, M.; Subramanian, A.; et al. SARS-CoV-2 Subgenomic RNA Kinetics in Longitudinal Clinical Samples. Open Forum Infect. Dis. 2021, 8, ofab310. [Google Scholar] [CrossRef]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef]
- Veronese, G.; Ammirati, E.; Chen, C.; Klingel, K.; Suzuki, M.; Okumura, T.; Maisch, B.; Zuo, H.; Ni, L.; Jiang, J.; et al. Management perspectives from the 2019 Wuhan international workshop on fulminant myocarditis. Int. J. Cardiol. 2021, 324, 131–138. [Google Scholar] [CrossRef]
- Wagner, J.U.G.; Bojkova, D.; Shumliakivska, M.; Luxán, G.; Nicin, L.; Aslan, G.S.; Milting, H.; Kandler, J.D.; Dendorfer, A.; Heumueller, A.W.; et al. Increased susceptibility of human endothelial cells to infections by SARS-CoV-2 variants. Basic Res. Cardiol. 2021, 116, 42. [Google Scholar] [CrossRef] [PubMed]
- Rueca, M.; Bartolini, B.; Gruber, C.E.M.; Piralla, A.; Baldanti, F.; Giombini, E.; Messina, F.; Marchioni, L.; Ippolito, G.; Di Caro, A.; et al. Compartmentalized Replication of SARS-Cov-2 in Upper vs. Lower Respiratory Tract Assessed by Whole Genome Quasispecies Analysis. Microorganisms 2020, 8, 1302. [Google Scholar] [CrossRef]
- Jary, A.; Leducq, V.; Malet, I.; Marot, S.; Klement-Frutos, E.; Teyssou, E.; Soulié, C.; Abdi, B.; Wirden, M.; Pourcher, V.; et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020, 26, 1560.e1–1560.e4. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Liu, H.; Wu, N.C.; Wilson, I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021, 538, 192–203. [Google Scholar] [CrossRef]
- Mahmoudi Gomari, M.; Rostami, N.; Omidi-Ardali, H.; Arab, S.S. Insight into molecular characteristics of SARS-CoV-2 spike protein following D614G point mutation, a molecular dynamics study. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Tiwari, R.; Dhama, K. SARS-CoV-2 in semen: Potential for sexual transmission in COVID-19. Int. J. Surg. 2020, 84, 156–158. [Google Scholar] [CrossRef]
- Omolaoye, T.S.; Adeniji, A.A.; Cardona Maya, W.D.; du Plessis, S. SARS-COV-2 (Covid-19) and male fertility: Where are we? Reprod. Toxicol. 2020, 99, 65–70. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Ren, J.; Zhao, Y.; Chen, J.; Chen, X. Effect of COVID-19 on Male Reproductive System—A Systematic Review. Front. Endocrinol. (Lausanne) 2021, 12, 677701. [Google Scholar] [CrossRef]
- Holtmann, N.; Edimiris, P.; Andree, M.; Doehmen, C. Assessment of SARS-CoV-2 in human semen—A cohort study. Fertil. Steril. 2020, 114, 233–238. [Google Scholar] [CrossRef]
- Vahedian-Azimi, A.; Karimi, L.; Makvandi, S.; Jamialahmadi, T.; Sahebkar, A. Does SARS-CoV-2 Threaten Male Fertility? In Clinical, Biological and Molecular Aspects of COVID-19; Guest, P.C., Ed.; Advances in Experimental Medicine and Biology Series; Springer: Cham, Switzerland, 2021; pp. 139–146. [Google Scholar]

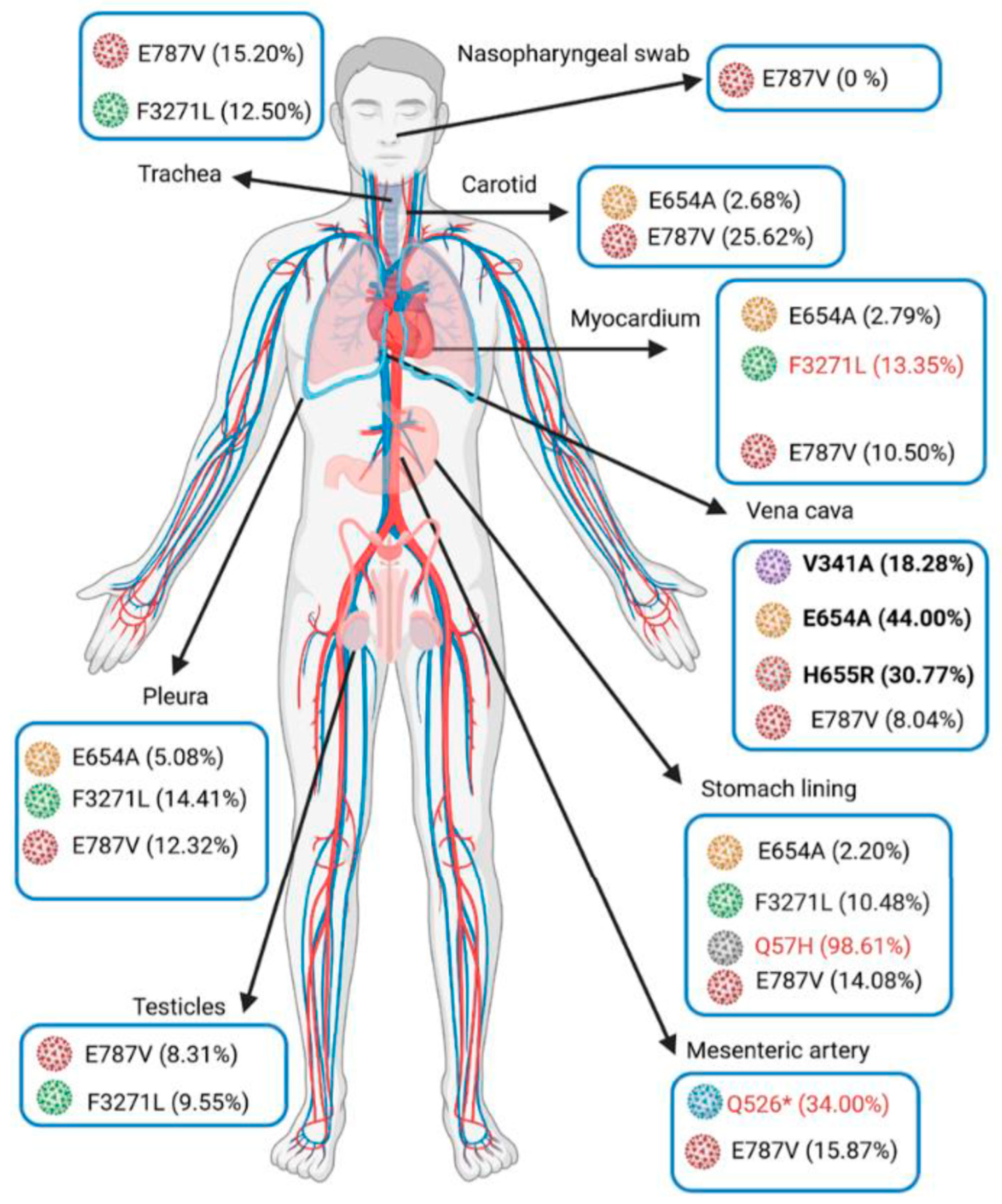
| Specimen | Nasopharynx (9 Days before Death) | Trachea | Lung | Pleura | Mediastenal Lymph Nodes | Stomach | Pancreas | Adrenal Glands | Testicle | Myocardium | Inferior Vena Cava | Mesenteric Artery | Carotid Artery | Aorta | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-Co-2 genome detection (IP4-Cq) | 27.7 | 24.6 | 23.5 | 24.2 | 32.4 | 23.6 | 32 | 27.3 | 30 | 25.5 | 28.8 | 22.8 | 20.4 | 31.9 | |
| Subgneomic RNA detection (Cq) | 31.3 | 33.9 | 32.1 | 33.6 | 37.1 | 31.7 | ND | 34.7 | 32.1 | 34.7 | ND | 32.3 | 29.1 | ND | |
| Mutations | |||||||||||||||
| ORF1a | R43H | 0.00% | 0.00% | ND | 0.00% | failed | 14.89% | failed | failed | 0.00% | 0.00% | ND | 0.00% | 0.00% | failed |
| Q526* | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 34.00% | 0.00% | |||||
| A537V | 0.00% | 0.00% | 0.00% | 18.75% | 0.00% | 0.00% | 9.15% | 3.03% | 0.00% | 22.82% | |||||
| V1968M | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | ND | 0.00% | 21.66% | |||||
| M3087I | 72.73% | 96.77% | 100.00% | 97.12% | 98.39% | 98.58% | 97.81% | ND | ND | ND | |||||
| F3271L | 0.00% | 12.50% | 0.00% | 14.41% | 10.48% | 9.55% | 13.35% | 0.00% | 0.00% | 0.00% | |||||
| Y3300H | 97.81% | 96.56% | 98.89% | 97.58% | 98.65% | 98.27% | 97.68% | 97.54% | 99.48% | 99.24% | |||||
| D3703H | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | ND | ND | 24.44% | |||||
| G4374D | 26.62% | 8.41% | 0.00% | 0.00% | 27.49% | 10.25% | 0.00% | ND | 0.00% | 0.00% | |||||
| ORF1b | A176S | 100.00% | 0.00% | ND | 100.00% | 0.00% | 100.00% | 92.86% | ND | ND | ND | ||||
| P314L | 100.00% | 92.86% | ND | 100.00% | 100.00% | 76.47% | 100.00% | ND | ND | ND | |||||
| V767L | 98.25% | 97.01% | 98.40% | 98.68% | 98.20% | 98.93% | 98.71% | 99.07% | 99.30% | 99.22% | |||||
| E787V | 0.00% | 15.20% | 15.13% | 12.32% | 14.08% | 8.31% | 10.50% | 8.04% | 15.87% | 25.62% | |||||
| K1141R | 100.00% | 92.06% | 100.00% | 94.44% | 100.00% | 89.47% | 96.00% | ND | 100.00% | 98.80% | |||||
| E1184D | 90.91% | 100.00% | 100.00% | 84.00% | 97.01% | 93.75% | 92.93% | ND | 100.00% | 100.00% | |||||
| Spike | V341A | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 18.28% | 0.00% | 0.00% | ||||
| S477N | 100.00% | 98.76% | 100.00% | 97.98% | 98.94% | 100.00% | 98.98% | 100.00% | 97.30% | 100.00% | |||||
| D614G | 98.82% | 98.29% | 98.77% | 98.98% | 99.29% | 98.50% | 99.02% | 98.98% | 99.60% | 99.51% | |||||
| E654A | 0.00% | 0.00% | 0.00% | 5.08% | 2.20% | 0.00% | 2.79% | 44.00% | 0.00% | 2.68% | |||||
| E654G | 9.14% | 16.30% | 0.00% | 0.00% | 18.18% | 7.05% | 15.08% | 0.00% | 14.35% | 18.21% | |||||
| H655R | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 30.77% | 0.00% | 0.00% | |||||
| I1172T | 0.00% | 0.00% | 0.00% | 10.91% | 10.82% | 10.46% | 8.55% | 0.00% | 0.00% | 0.00% | |||||
| ORF3a | Q57H | 0.00% | 0.00% | 0.00% | 0.00% | 98.61% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | ||||
| P207S | ND | ND | ND | ND | 0.00% | ND | ND | ND | 25.00% | ND | |||||
| N | K169N | 0.00% | 0.00% | 0.00% | 13.10% | 3.32% | 5.27% | 2.80% | 0.00% | 0.00% | 0.00% | ||||
| M234I | 94.12% | 97.33% | 81.25% | 96.64% | 99.17% | 98.71% | 98.95% | 100.00% | 98.61% | 99.61% | |||||
| K256* | ND | 0.00% | 10.27% | 10.53% | 10.18% | 11.81% | 7.76% | 13.51% | 0.00% | 8.76% | |||||
| A376T | 100.00% | 100.00% | 100.00% | 98.00% | 98.96% | 100.00% | 98.98% | 100.00% | 98.18% | 99.25% | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartard, C.; Chaqroun, A.; Settembre, N.; Gauchotte, G.; Lefevre, B.; Marchand, E.; Mazeaud, C.; Nguyen, D.T.; Martrille, L.; Koscinski, I.; et al. Multiorgan and Vascular Tropism of SARS-CoV-2. Viruses 2022, 14, 515. https://doi.org/10.3390/v14030515
Hartard C, Chaqroun A, Settembre N, Gauchotte G, Lefevre B, Marchand E, Mazeaud C, Nguyen DT, Martrille L, Koscinski I, et al. Multiorgan and Vascular Tropism of SARS-CoV-2. Viruses. 2022; 14(3):515. https://doi.org/10.3390/v14030515
Chicago/Turabian StyleHartard, Cédric, Ahlam Chaqroun, Nicla Settembre, Guillaume Gauchotte, Benjamin Lefevre, Elodie Marchand, Charles Mazeaud, Duc Trung Nguyen, Laurent Martrille, Isabelle Koscinski, and et al. 2022. "Multiorgan and Vascular Tropism of SARS-CoV-2" Viruses 14, no. 3: 515. https://doi.org/10.3390/v14030515
APA StyleHartard, C., Chaqroun, A., Settembre, N., Gauchotte, G., Lefevre, B., Marchand, E., Mazeaud, C., Nguyen, D. T., Martrille, L., Koscinski, I., Malikov, S., & Schvoerer, E. (2022). Multiorgan and Vascular Tropism of SARS-CoV-2. Viruses, 14(3), 515. https://doi.org/10.3390/v14030515









