Abstract
Bacillus subtilis is an important bacterial species due to its various industrial, medicinal, and agricultural applications. Prophages are known to play vital roles in host properties. Nevertheless, studies on the prophages and temperate phages of B. subtilis are relatively limited. In the present study, an in silico analysis was carried out in sequenced B. subtilis strains to investigate their prevalence, diversity, insertion sites, and potential roles. In addition, the potential for UV induction and prevalence was investigated. The in silico prophage analysis of 164 genomes of B. subtilis strains revealed that 75.00% of them contained intact prophages that exist as integrated and/or plasmid forms. Comparative genomics revealed the rich diversity of the prophages distributed in 13 main clusters and four distinct singletons. The analysis of the putative prophage proteins indicated the involvement of prophages in encoding the proteins linked to the immunity, bacteriocin production, sporulation, arsenate, and arsenite resistance of the host, enhancing its adaptability to diverse environments. An induction study in 91 B. subtilis collections demonstrated that UV-light treatment was instrumental in producing infective phages in 18.68% of them, showing a wide range of host specificity. The high prevalence and inducibility potential of the prophages observed in this study implies that prophages may play vital roles in the B. subtilis host.
1. Introduction
All bacteria are prone to infection by the most abundant viruses on earth, which are called bacteriophages (also known as phages for short) [1]. Based on their lifestyles, phages are grouped into either virulent (phages that cause cell lysis after infection) or temperate (phages that cause lysogeny or cell lysis after infection) [2]. In the lysogenic lifestyle, the DNA of temperate phages is maintained as prophages within the host and is replicated as its chromosomal integrated form (integrated prophages) or in its circular or linear plasmid forms (plasmid prophages) to render the host as a lysogen [3,4]. The lytic genes of prophages are repressed, while some regulatory and lysogenic conversion genes continue to be expressed [5] to maintain the lysogenic state and to induce prophage-mediated host-phenotypic changes, such as super-infection exclusion, increasing pathogenicity, and expanding the metabolic capacities of the host [6,7].
About half of the sequenced bacteria harbor prophages [7] whose genome accounts from less than 1.00% up to 20.00%, depending on the type of the species [8]. The proportion of prophages in the genomes of Bacillus thuringiensis is 10.18% [9], while that of Group A Streptococcus (GAS) ranges from 7.10 to 12.40% [10]. The prophage of Escherichia coli O157:H7 strain Sakai accounts 16.00% [11], while that of Streptococcus thermophilus and Streptococcus pyogenes are 0.40 and 9.50%, respectively [12]. Moreover, the prophages of Pectobacterium spp. and Dickeya spp. represent less than 2.00% [13]. Not all prophages are able to induce infective phages because some may not be fully functional [14]. The literature shows that the prevalence of inducible functional prophages ranges from 0 (i.e., defective) to 42.00% [15,16].
Bacillus subtilis is a fast-growing, Gram-positive, spore-forming bacterium that is regarded as a model organism due to its importance in the study of the fundamentals of bacterial physiology and metabolism [17]. It is also considered as a key industrial organism due to its natural and engineered use in industrial production application, such as in food production (through fermentation), hydrolytic enzymes (amylases, protases), and fine chemicals (e.g., riboflavin) [18]. Despite the importance of B. subtilis strains in the food industry and biotechnology, the temperate phages SPbeta and phi3T are the two main phages that have been involved in research the most in past decades, and the B. subtilis 168 strain is the most frequently preferred host [19,20]. However, no data are available regarding the prevalence, diversity, and inducibility potential of the prophages in large collections of B. subtilis strains.
Comparative genomics studies can show the diversity of prophages. Studies into the prophage-encoding putative proteins and inducibility potential in B. subtilis can help us to understand their desirable and undesirable effect on their hosts as well as on their applications in the fermentation industry. The present study explored (a) the prevalence, diversity, characterization, and plausible roles of prophages in 164 sequenced B. subtilis strains via in silico analysis methods and (b) the UV-light inducibility potential of prophages in 91 B. subtilis collections and their prevalence. This work represents the first comprehensive study on the prevalence, diversity, and UV-light inducibility potential of prophages in a large collection of B. subtilis strains.
2. Materials and Methods
2.1. Collection of B. subtilis Genome Dataset and In Silico Prediction of Prophages
Complete genome sequences of 194 B. subtilis strains deposited in NCBI (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/665/) (accessed on 5 February 2021) were retrieved for putative prophage prediction. Following manual analysis, the strains of 30 genomes were dropped from further analyses for multiple reasons, such as prophage predictions that comprised >99.80% of the host’s genome (BS38 (NZ_CP017314.1), HJ0-6 (NZ_CP016894.1), and SG6 (NZ_CP009796.1)), the prediction of prophages with ‘N’ nucleotides (FB6-3 (CP032089.1)) and when strains shared identical strain names (with >98.00% symmetric identity) coupled with the prediction of identical/nearly identical prophages. Thus, the genomes of 164 strains (Table S1) were considered for prophage prediction using two programs: PHASTER [21] and PhiSpy [22]. PHASTER is a web-based program that uses gene/protein hits in a given region to predict prophages as intact prophages (IPs), questionable prophages (QP), and incomplete prophages (InP) based on their completeness. It also annotates the prophages using Glimmer software for structural annotation and BLASTP for functional annotation [21], which were used for the analysis of the putative prophage-encoded proteins.
The integrated intact prophage regions predicted by PHASTER were then checked for whether they could be identified by the second prophage predicting program, PhiSpy, which operates on a Unix-based OSs platform. The PhiSpy program can identify prophages with and without sequence similarities to known phage sequences. The program was run with the default setting and using the B. subtilis_subsp._subtilis_str._168 training set distributed with it. The exact position of the end coordinates of the PhiSpy-predicted regions showed some degree of deviation compared to the PHASTER prediction due to variations in the prediction methods [23]; thus, the significance of their similarity was further checked using BLASTN. Only integrated IPs that were identified by both programs were included in the downstream analysis. Because PHASTER has the ability to evaluate and report the completeness of the prophages, the prophage genome sequences extracted using the end coordinates specified by PHASTER were considered as IP genome sequences.
2.2. Occurrence, Distribution and Prevalence of Prophages in B. subtilis
The occurrence of IPs in the 164 B. subtilis genomes and their distribution as either their integrated or plasmid forms was evaluated. All of the putative prophages were used to study their occurrence and distribution along with the genome sizes of the hosts. Only IPs were considered to estimate the prevalence, abundance (number of IPs per host genome), and proportion (genomic proportion of IPs) and to study the features of their genomes. The genomic proportion of IPs was calculated as “% = (Total IP Length ÷ Host Genome Length) × 100”.
2.3. Bioinformatic Analysis of B. subtilis Prophages
A comparative analysis of the IP genomes was carried out by generating average similarity matrices using Gegenees v3.1 with the 200/100 setting-fragmented BLASTN and TBLASTX comparison methods [24]. The phylogenomic relationships of the IPs were produced using the neighbor-joining method in SplitsTree4 [25]. The phylogenomic tree was then visualized using the iTOL (v5) tool (https://itol.embl.de/ (accessed on 10 November 2021)). A genome-scale dotplot was created using the Genome Pair Rapid Dotter (Gepard) [26].
2.4. Growth Conditions of Bacteria and Bacteriophages
The B. subtilis used for the prophage induction study included strains from the Korean Culture Center of Microorganisms (KCCM), Korean Agricultural Culture Collection (KACC), Korean Collection for Type Cultures (KCTC), American Type Culture Collection (ATCC) as well as lab isolates (Table S2). Of the strains, the genomes of five strains were found in the NCBI database, namely JCM 1465 (NBRC 13719 (NZ_AP019714.1)), KCCM 32835 (NCIB 3610 (NZ_CP020102.1)), SRCM 102751 (NZ_CP028217.1), KCCM 11316 (ATCC 6633 (NZ_CP0349431.1)), and KCTC 2217 (168 (NZ_CP0110052.1)). B. subtilis were cultured aerobically in tryptic soy broth (TSB: BD, Sparks, MD, USA) or TSB agar (TSA: TSB enriched with 1.5% agar) at 37 °C. Lactobacillus strains were cultured in BD Difco™ Lactobacilli MRS media. During the course of the phage isolation and propagation procedures, TA soft agar containing nutrient broth (8 g/L), NaCl (86 mM), MgSO4.7H2O (0.8 mM), MnSO4 (0.3 mM), and CaCl2 (1.0 mM) (pH 6.0), SM buffer containing 50 mM Tris-HCl (pH 7.5), and 100 mM NaCl plus 10 mM MgSO4 were used as described in the works of Bandara et al. (2012) [27].
2.5. UV-Light Survival Rate of B. subtilis
To optimize a protocol for the prophage induction experiment, B. subtilis strains were exposed to a UVC light source (T-8C, 8W with 254 nm range, VILBER) at varying distances (i.e., 8, 9, and 10 cm) and exposure times (i.e., 1, 2, 3, 5, 10, 15, and 20 min) in the dark. After the addition of an equal volume of fresh TSB medium, cultures were kept in the dark for 2 h at 37 °C followed by dilution, plating, and incubation overnight at 37 °C for CFU counting. The survival rate was calculated using the plate count method presented in the reports of Djurdjevic-Milosevic et al. (2011) [28].
2.6. Prophage Induction up on Ultraviolet-Light Treatment of B. subtilis
B. subtilis strains were treated with UV light for the prophage induction experiments and for the supernatant preparations, and the strains served as indicator strains to test the infectivity of each supernatant by means of the dotting assay. An amount of 500 µL of exponentially growing B. subtilis culture was irradiated with UV-light, mixed with an equal volume of TSB, and incubated at 37 °C in dark for 2 h. Finally, the culture was centrifuged at 14,000× g for 5 min, followed by the collection and filter-sterilization (0.2 µm pore size) of the supernatants as outlined by Kim et al. (2012) [29]. All experiments were carried out in triplicate.
2.7. Infectivity, Prevalence of UV-Light Inducible Prophages and Phage Isolation
The infectivity and host spectrum of the induced prophages were determined by dotting the supernatants onto 91 indicator B. subtilis strains using the dotting assay method described by Kropinski et al. (2009) [30] or as indicated otherwise (Table S2). An amount of 10 µL of supernatant was dotted onto pre-solidified TSB agar that had been overlayed with 300 µL of the indicator strain that had been grown overnight after being mixed with 4 mL TA soft agar. All supernatants a showing clear lysis zone (C) and some (n = 12) showing a turbid lysis zone (T) were considered for phage purification and evaluation. Moreover, the UV-light-treated supernatants that eventually produced infective phages were compared against their corresponding non-UV-light treated supernatants to check for any spontaneous prophage inductions. The non-UV-light-treated supernatants were prepared using the same method used for the preparation of UV-treated supernatants described in Section 2.5 above but without UV-light treatment.
2.8. B. subtilis Phage Host Range Analysis
Host range analysis was carried out against the B. subtilis strains and other purposely selected bacteria, including Gram-positive and Gram-negative as well as pathogenic and non-pathogenic species using the dotting assay method outlined by Ghosh et al. (2018) [31], in which 5 µL of phage samples were dotted on pre-solidified TA soft agar containing indicator strains. In addition to B. subtilis, five B. licheniformis (JCM 2505, SCC 125037, SDC 125016, SCC 123050, and SDC 125015), three Lactobacillus spp. (SRCM 100888, KACC 11451, and KACC 13877), five B. cereus groups (ATCC 1611, ATCC 14579, ATCC 13061, ATCC 21768, and ATCC 27348), one B. thuringiensis (ATCC 10792), one B. mycoides (ATCC 21929), one Staphylococcus aureus (ATCC 144458), one Listeria monocytogenes (Scott A), one E. coli (BW 25113), and one Enterobacter sakazakii (KCTC 2949) were included. Phages that showed identical host range lysis patterns were compared using the efficiency of plating (EOP) assay [32] and calculated as (PFU on Target Host Strain ÷ PFU on Propagation Host Strain) × 100.
2.9. B. subtilis Phage Large Scale Propagation Condition Obtimizations
Phage growth conditions such as media types (solid vs. liquid), temperature (30 °C vs. 37 °C), and multiplicity of infection (1 vs. 0.1) were optimized. Large-scale propagated phages were centrifuged at 10,000× g for 30 min and incubated overnight at 4 °C after the addition of 10% (w/v) polyethylene glycol (PEG 8000, Sigma, Saint Louis, MO, USA) and 0.5 M NaCl. They were then precipitated at 10,000× g for 30 min, the pellet re-suspended in SM buffer, filter-sterilized (0.2 µm pore size), and subjected to ultracentrifugation in six-density gradients of CsCl, vis-à-vis 1.7, 1.5, 1.45, 1.4, 1.3, and 1.2 g/mL of SM buffer at 166,900× g for 4 h using an ultracentrifuge (Soravel WX + ULTRA SERIES Centrifuge, Thermo Scientific, Waltham, MA, USA) at the Korea Food Research Institute (KFRI, Korea). Blue bands were withdrawn using a syringe from the centrifuge tubes (06750-AV tubes, Thermo Scientific, Asheville, NC, USA) and dialyzed against SM buffer by means of gentle shaking at 4 °C overnight followed by PFU checking and were stored at 4 °C [33].
2.10. Data Analysis
Data were analyzed using the ggplot2 package [32] provided in R programming [34] with its integrated development environment RStudio [35] (available at https://www.r-project.org/ and https://rstudio.com/ (accessed on 2 February 2022)).
3. Results
3.1. Prevalence and Distribution of Prophages in Sequenced B. subtilis Genomes
Prophages were predicted in 164 sequenced B. subtilis genomes and were used for prevalence and distribution analysis. The analyses showed that all of the strains harbored one or more types of intact prophages (IP), questionable prophages (QP), and incomplete prophages (InP). Whereas 92.07% (n = 151) of the strains contained IP and/or QP, 8% (n = 13) contained InP only. The analyses showed that 75.00% of the (n = 123) B. subtilis strains have IP, while 25.00% (n = 41) of them have no IP (Table S1).
The sizes of the host genomes for most B. subtilis strains are confined from around 3.4 to 4.5 Mb and can contain all types of prophages. A relationship analysis between the type and abundance of putative prophages and host genome size showed no strong association. It is worth mentioning that the IPs were not found for strains with small-sized genomes (Figure 1).
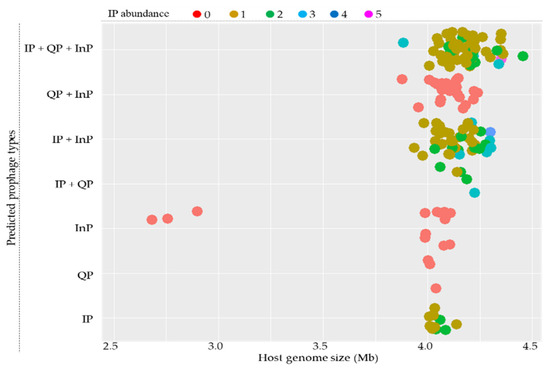
Figure 1.
Distribution of intact prophages (IP), questionable prophages (QP), and incomplete prophages (InP) in 164 sequenced B. subtilis genomes. Host genome size is indicated on the X-axis. The distributions of the types of IPs according to host genome sizes are indicated on the Y-axis. Closed circles represent predicted prophages, and their colors indicate IP abundance, as indicated at the top.
When we look into the locations of the IPs on the replicons of the hosts, 93.50% (n = 115) of the strains have integrated IPs, 2.44% (n = 3) have plasmid IPs, and 4.07% (n = 5) have both integrated and plasmid IPs (Figure 2).
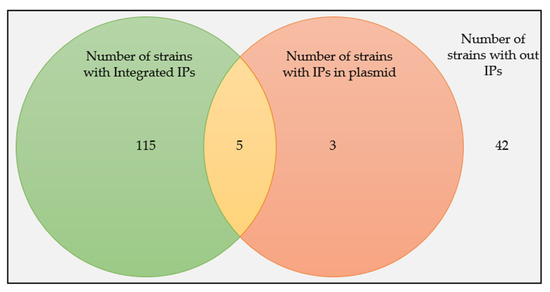
Figure 2.
The prevalence of Ips in B. subtilis strains and their distributions as integrated or plasmid forms.
Initially, the PHASTER program was able to predict a total of 180 putative intact prophages (172 integrated and 8 plasmid forms) from 123 B. subtilis strains. In the genome of one strain (B. subtilis QB928), the PHASTER program identified two Ips with different scores from the same region, the reason for which is not clear. Thus, only one, the one with the highest score, was considered for further analysis. Overall, a total of 179 Ips (171 integrated and 8 plasmid forms) were predicted by PHASTER, and the details are provided in Table S1.
Out of 171 integrated IPs predicted by PHASTER, 162 were re-identified by the second prophage prediction program, PhiSpy, which showed some degree of deviation in the exact locations of the start and end coordinates. Seven of the integrated IPs were not identified by PhiSpy and were hence excluded from the downstream analysis of the IPs. The remaining two integrated IPs predicted by PHASTER from the B. subtilis NCD-2 chromosome could not be checked by PhiSpy because its genome was not annotated. However, following manual inspection, most of the genes of these IPs were annotated to be phage proteins; thus, they were kept for further analysis. Taken together, the further downstream analysis was carried out by using 172 IPs, i.e., 164 integrated (162 verified by PhiSpy and 2 PhiSpy unverified) and 8 plasmid IPs (Table S3).
3.2. Characterization of Intact Prophages in B. subtilis
Summary statistics were carried out for the genomic features of 172 IPs (Table 1). The mean genome length, genome proportion, protein number, and GC percent were found to be 55.02 kb, 1.86%, 71.33, and 41.63%, respectively. The attachment-site show-up and the number of tRNAs encoded by the IPs generated by the PHASTER program were evaluated. While the program identified the attachment sites for 56.39% (n = 97) of the IPs, 43.60% (n = 75) of the IPs did not show attachment sites. tRNA was only encoded in 8.14% (n = 14) of the IPs (Table S3).

Table 1.
Genomic features of 171 intact prophages in B. subtilis predicted by PHASTER.
The abundance of IPs in each host genome was found to be 1 to 5, while their genomic proportion ranged from 0.01 to 7.18% compared to the genomes of their hosts. A total of 82 (66.66%) of the strains had one IP per genome (accounting <1.5% of host genome), while 27 (21.95%) of the strains had two IPs with genome proportions ranging from 1.5% to 4.5%. Nine (7.32%) strains harbored three IPs with a high genomic proportion ranging from 4.5 to 7.5% (Figure 3a). Generally, we observed that the genomic proportion of the prophages increased as the genome abundance per strain increased, as predicted.
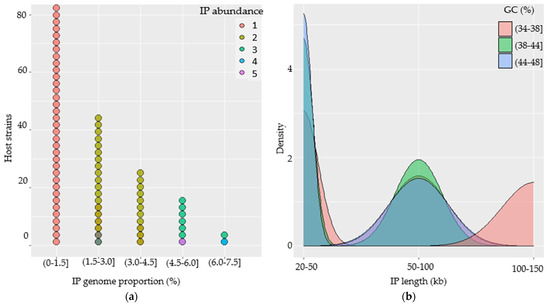
Figure 3.
Characterization of intact prophages (IPs) in the genomes of B. subtilis strains. (a) Dotplot illustrating IP abundance and genome proportion per genome of their host and their associations. (b) Density plot showing the distribution and association of IPs in terms of their genome size and GC-content.
The IPs were plotted against their length and GC-contents to analyze the features and associated genome distribution. (a) The IP genomes had genome lengths spanning from 20 to 150 kb with GC-contents ranging from 34 to 48%. (b) Nearly two-thirds of the IPs (n = 118) had genomes that were less than 50 kb long and broader GC-content distributions. (c) Some of the IPs (n = 28) had genomes that were 50 to 100 kb long with significantly broader GC-content distributions ranging from 34 to 48%. (d) Others (n = 26) IPs contain genomes that extended from 100 to 150 kb and that had a relatively narrow GC-content of 34 to 38% (Figure 3b).
The study into the relationship between the genome length of the IPs and their GC-contents revealed a slight negative association. When IPs have shorter genomes, they tend to have a higher GC-content and vice versa. The majority of the prophages with genome lengths that are less than 50 kb appeared to have a relatively higher GC-content (38 to 48%), while those with lengths greater than 100 kb tend to have lower GC-content (34 to 38%).
3.3. Diversity of B. subtilis 172 Intact Prophages in B. subtilis
Both nucleotide and translated amino acid sequence comparisons were carried out with the Ips at genome-scale using Gegenees software. A comparison heatmap revealed the presence of prophage clusters with identical/nearly identical and highly or less similarity scores. Following BLASTN comparison, the majority of the IPs (n = 153) could be assigned into 13 main clusters and were put into blocks. Identical or nearly identical and highly similar IPs (score > 70%) are depicted in green and t in black rectangles. Some IPs are similar (score > 40% and depicted in yellow and put in blue rectangles), while others are dissimilar (score <40% and depicted in red) (Figure 4a). Moreover, some of the IPs showed no similarity and existed as singletons.
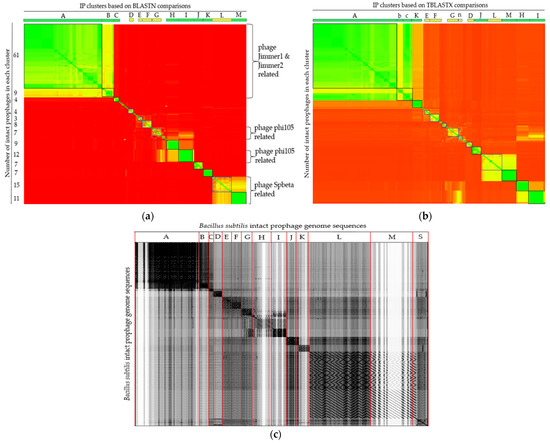
Figure 4.
Comparison of 172 IP genome sequences predicted in B. subtilis (a,b) Gegenees heatplot generated using BLASTN and TBLASTX, respectively, with 200/100 settings. Heatplot colors indicate similarity ranging from nearly identical (green) to similar (yellow) or dissimilar (red). The corresponding numbers of group members of each cluster are indicated on the y-axis. Labeled boxes at the top represent IP clusters. (c) Dotplot alignment of IP sequences generated by Gepard.
Some members of the IPs of the cluster depicted in green (e.g., clusters H, I, and K) have similarity scores as high as >99%, implying their ubiquity, where their sequences are shared by numerous strains, rendering them to colonize a diversity of strains. Dissimilarity among the IPs might imply their distinctness across the strains. The details of the similarity scores used to build the heatmap and the members of each cluster are presented in Table S4.
The TBLASTX-translated IP comparison analysis generated similar clusters to that of the nucleotide comparison method, but with reshuffling of few IPs from one cluster to another. Cluster labels that do not coincide with the BLASTN comparison in Figure 4a are shown by small letters in Figure 4b. Members of cluster ‘B’ in BLASTN comparison joined cluster ‘A’ in the TBLASTX comparison, while some members of cluster ‘A’ left their prior cluster and restructured into a new cluster (assigned as cluster ‘c’) in TBLASTX comparison mode (Figure 4b, Table S5).
All 172 IPs sequences were concatenated into a single file in the same order as in the heatmap similarity matrix (Table S4), with the exception of the singletons, which were concatenated to be at the end. Gepard with a sliding window of 10 nucleotides was used to generate a dotplot alignment, as shown in Figure 4c.
Looking into the annotations of the IPs, Bacillus phage SPbeta (NC_001884), Bacillus phage phi105 (NC_004167), Brevibacillus phage Jimmer2 (NC_041976), and Brevibacillus phage Jimmer1 (NC_029104) were among the topmost common phages that showed homology among the BLAST hits. The majority of the Ips that have the same most common phages hits were clustered together in the Gegenees analysis. To see the relationships at the genomic level, the genomes of the IPs were further compared based on the most common phages that they had in common as well as the B. subtilis temperate phage phi3T (KY030782.1) and the skin element defective prophage containing the B. subtilis 48 kb region found in NCBI (D32216.1), as seen in Table S6.
3.4. Analysis of Insertion Sites of B. subtilis Intact Prophages
The insertion sites of the integrated IPs (n = 161) were analyzed to demonstrate the presence of common and distinct insertion sites as well as the 5′ and 3′ end genes of the prophages. Considering the position of both the 5′ and 3′ end coordinates, 138 of the IPs showed an intergenic site insertion, while five were intragenic. There were 18 other IPs that showed intragenic site insertion in their 5′ end coordinates only, and 10 others showed intragenic insertion at their 3′ end coordinates only. The DinB family- and sporulation-related proteins encoding the genes were among the frequently flanking genes of the prophage region on the left side. The spoIISC gene that encodes three-component toxin–antitoxin–antitoxin system antitoxin SpoIISC was the most frequent gene lanking the prophage regions from the right side. Most IPs of the same cluster tend to have the same or similar preferential insertion sites. Yet, different insertion sites were also observed in some IPs of the same cluster, suggesting their richness in diversity (Table S7). Moreover, in the case of intergenic-inserted Ips, the 5′ end coordinate may be positioned at the 5′ end or downstream of the first prophage’s gene. Some prophages show intragenic insertions into RNAs, in which the prophages’ start and end coordinates are inserted within the tRNA sequences, including tRNA-Arg, tRNA-Val, tRNA-Thr, and rRNA-23S ribosomal RNA. Most IP members of a cluster prefer to have the same genes at their 5′ and 3′ ends (Table 2).

Table 2.
Genes present at 5′ and 3′ integrated intact prophage ends.
3.5. Phylogenomic Analysis of 172 B. subtilis Intact Prophages
Phylogenomic trees of Ips were constructed following the NJ method in SplitsTree4. The result showed that the Ips could be assigned into 14 clusters. Some of the clusters have many Ips while others have only a few. The phylogenomic tree analysis also showed the presence of singletons, indicating the diversity of the IPs (Figure 5).
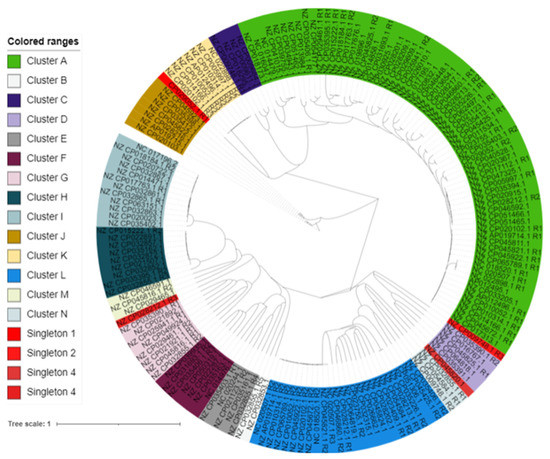
Figure 5.
Phylogenomic relationships of B. subtilis IPs. The distances of the IP sequence were calculated using Gegenees. Then, the tree was constructed by SplitsTree4 and visualized by iTOL. Clusters are coded by different colors, and singletons are in red.
3.6. Functional Annotation Analysis of Putative Proteins Encoded by B. subtilis Intact Prophages
Functional annotation analysis was carried out for 50 heterogeneous cluster-representative putative Ips that were selected based on their phylogenomic relationships. The analysis primarily focused on the gene products of the prophages in order to look into the plausible roles of the prophages in their host properties. The exercise demonstrated that the prophage-encoded putative proteins included various enzymes, such as proteases and peptidases, lipases, catalase, hydrolases, oxidases, and transferases. The prophages also encode putative sporulation-, bacteriocin-, immunity-, and arsenite and antibiotics resistance-related proteins. Some of the proteins were linked to transporters, while other products were associated with toxin–antitoxin and apoptotic control systems (Figure 6).
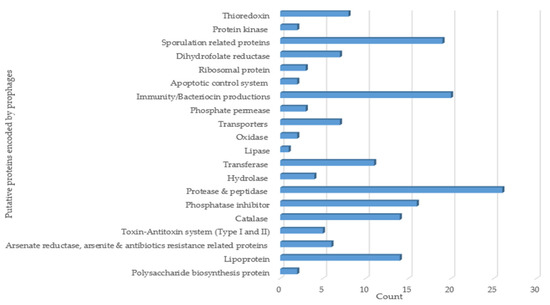
Figure 6.
Functional annotation analysis of putative proteins encoded by B. subtilis prophages.
3.7. Optimization of UV-Light Treatment for Induction of B. subtilis Prophages
The effect of UV-light on the survival of the B. subtilis was optimized before the prophage induction study. UV-light exposure for 1 min at 8, 9, and 10 cm distances inactivated 65 to 80% of B. subtilis. The deactivation level increased from 75 to 87% upon increasing the exposure time by a factor of 1 (Figure 7). A linear relationship was observed between the survival rate and UV-light exposure distance for up to 2 min. As the exposure time increased to 2 min, the exposure distance tended to have less impact, suggesting the greater effect of the exposure time compared to the exposure distance on the survival of B. subtilis. Prophage induction increased as the viability of the host decreased [7]. It has also been documented that a survival rate of about 11% is suitable for meaningful growth comparisons [36]. In our case, since exposing the bacteria for 2 min at a 9 cm distance yielded an ~15% survival rate, that exposure setting (for 2 min from 9 cm) was adopted for prophage induction.
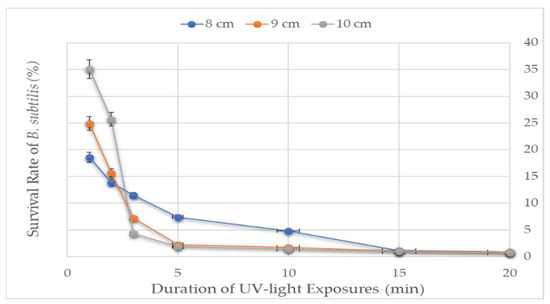
Figure 7.
The survival rate of B. subtilis to UV-light treatment. UV-light tolerance of the B. subtilis was measured at three distances between the sample and the UV-lamp for seven varying exposure times.
3.8. Examination of B. subtilis Supernatants for the Presence of Infective Prophages and Phage Isolation
Supernatants prepared from 91 UV-light-treated B. subtilis strains were dotted on 91 B. subtilis indicator strains using the spot assay method to assess the presence of induced infective prophages. The dotting assay profiles of all of the supernatants are presented in Table S2. A given supernatant can display clear lysis zones in multiple or single indicators. Clear lysis-forming supernatants were considered for the next phage isolation steps. Some supernatants displayed clear zones in multi-indicators, providing an opportunity to consider alternative indicator hosts for phage isolation trials, given that clear lysis showing supernatants remains unresponsive to plaque formation tests or fails to produce sufficient phage titers. Infective phages were isolated from clear lysis-forming supernatants.
Some supernatants formed turbid lysis but not clear lysis. Ten (10) turbid lysis-forming supernatants and two (2) turbid and clear lysis-forming supernatants were considered for further phage isolation tests using indicators on which they showed turbid lysis to assess whether or not these supernatants are effective in producing functional phages. All of these supernatants failed to support plaque formation except the two (2) that form both turbid and clear lysis. Moreover, the UV-light-treated supernatants that formed the plaques were considered for comparisons against their corresponding supernatants that had been prepared without UV-light treatment. Some of the supernatants prepared from non-UV-light-treated B. subtilis strains were able to form a clear zone similar to the treated ones, suggesting the spontaneous induction of the prophages (data not shown).
3.9. Inducibility Potential of B. subtilis Prophages
Supernatants were prepared from 91 B. subtilis strains following UV-light treatment to assess the inducibility potential of the prophages. The spot assay test showed that 26.37% (n = 24) of the supernatants formed clear lysis, while 48.4% (n = 44) formed only turbid lysis zones. Likewise, the plaque-forming assay showed that 20.88% (n = 19) of the supernatants formed plaques. In conclusion, infective phages were isolated from 18.68% (n = 17) of the clear lysis-forming supernatants (Figure 8).
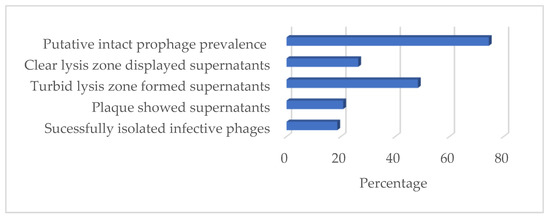
Figure 8.
Prevalence of in silico-predicted prophages and UV-light inducible of prophages in B. subtilis.
3.10. A Comparison of In Silico Predictions with UV-Light Induced Prophages
When searching for the B. subtilis strains to be used in both the in silico and induction studies, we were able to identify five strains (Table 3). The in silico prophage analysis showed that four of the strains contained at least one integrated IP, while one contained IPs in both their integrated and plasmid forms. The UV-light induction experiment showed that while all of the supernatants of the strains formed turbid lysis zones, only three display clear lysis zones tested on 91 indicator strains. During the plaque-forming assay, only one supernatant (NBRC 13719) showed plaques. The supernatants from the SRCM102751 and ATCC 6633 strains were able to form a clear lysis zone in multiple indicator strains but failed to form plaques during the plaque-forming assay.

Table 3.
The prevalence of UV-light inducible prophages in 5 B. subtilis.
3.11. Large-Scale Propagation Conditions and Host Range Analysis
Seventeen (17) B. subtilis phages were isolated and their respective host, and their propagation conditions are presented in the Supplementary Materials (Table S8). Most of the phages were propagated in large-scale liquid media at 37 °C. Few phages were propagated in solid media at 30 °C.
Host range analyses of the 17 phages were carried out in 50 indicators. Twenty-eight (28) indicators on which the phages showed clear lysis or no lysis were sorted for host range lysis pattern comparison (Table 4). Two groups of phages (green and blue) showed similar lysis patterns in all of the indicators, while the rest showed distinctly different lysis patterns. The member phages of the different groups appeared to have a narrower host range, while those that showed distinctly different lysis patterns tended to have broader host ranges. The phages with similar lysis patterns were further analyzed using relative efficiency of plating (EOP). However, they did not show satisfactory score variations to make any generalizations (data not shown).

Table 4.
Phage host range analysis of B. subtilis phages on 28 B. subtilis indicator strains.
Furthermore, the host range of the phages was investigated using 19 purposively selected non-B. subtilis bacterial species. These include Gram-positive bacteria, namely five B. licheniformis and B. cereus, three Lactobacillus brevis, and one Staphylococcus aureus, Listeria monocytogenes, B. thurengenesi, and B. mycoides and Gram-negative bacteria including one Escherichia coli and Cronobacter sakazakii. The phages showed no lysis on the non-B. subtilis strains, except for the B. licheniformis strains, on which some of the phages showed clear or turbid lysis (data not shown).
4. Discussion
The present in silico prophage study uncovered a high prevalence of IPs in a large collection of B. subtilis genomes. The study identified and examined 172 IPs from 123 sequenced B. subtilis strains using two prophage prediction programs, namely PHASTER and PhiSpy. Of the sequenced 164 B. subtilis strains included in this study, 75.00% of them contain IPs (Figure 8)—mostly in integrated form (93.50%). A total of 4.07% of the strains have both IP forms, while 2.44% only have prophages in their plasmid form, expanding the diversity of B. subtilis prophages. Previous studies documented that most temperate phages perpetuate their genomes through integration into the chromosomes of their hosts and that some maintain lysogeny via plasmid formation [37]. Bacillus phage vB_BtS-B83 *NC_048762.1) [38] and vB_BceS-IEBH [NC_011167.1] [39] are examples of experimentally proven plasmid prophages in B. thuringiensis host.
Prophages are particularly prominent in pathogenic bacteria [7,40]. Virulence factor-coding genes of pathogenic bacteria are often associated with prophages [41]. Previous in silico prophage prevalence and diversity analyses have mainly been focused on various pathogens. In one study using PHASTER, the prevalence of full-length or putatively full-length prophages in Gram-positive pathogenic Pneumococcus (n = 482) was 45.00%, and the total identified full-length prophages were 286 [42]. Studies using the same method reported prophage prevalence of up to 64.90% in Gram-negative Pectobacteriaceae phytopathogen (n = 57), with 37 intact prophages being identified [13], and up to 81.00% in Gram-positive B. thuringiensis (n = 61), in which 135 putative complete prophages were identified [9]. Likewise, studies using the multiplex PCR method showed 87.00% prophage prevalence in Gram-positive S. aureus pathogens [43] and 80.00% in Gram-negative Flavobacterium psychrophilum pathogens [44].
Relevant information on the prevalence of prophages in non-pathogenic bacterial species is limited. Thus, a higher or comparable prevalence of B. subtilis prophages with that of the aforementioned pathogens is interesting and of high value. Previously, it was suggested that lysogeny is frequent in fast growers, as they provide more resources for virion production [7]. Thus, it is possible that as fast-growing bacteria, in B. subtilis infection and lysognization across strains by temperate phages takes place frequently, resulting in a high prevalence of prophages.
High genetic diversity was noticed among the predicted IPs. Comparative genomics of the IPs revealed that the prophages demonstrated diverse but structured relationships, with most of them being grouped into 13 clusters and four distinct singletons. The presence of highly similar prophages in different B. subtilis strains could imply that such prophages may have high infective power, whereas the less similar ones and singletons may have lower infection ability. However, it was not easy to make generalizations because the number of hosts used for prophage identification or the number of related phages used for homology searches may not be comprehensive enough.
Detailed analysis of the gene annotations indicated by PHASTER showed that Bacillus phage SPbeta, Bacillus phage phi105, Brevibacillus phage Jimmer2, and Brevibacillus phage Jimmer 1 are the most common phages to 28 (16.28%), 25 (14.53%), 35 (20.35%), and 17 (9.88%) IPs. Upon the comparison of these IPs and phages, significant similarities were only able to be observed among some of the IP with the SPbeta and phi3T phages but not the others, supporting the high diversity of the IPs. Most of the Spbeta-related IPs were grouped into two clusters, L and M. Only about half of the phi105-related IPs could be assigned into cluster I, with the rest being distributed into multiple clusters. The observation that some of the Ips have the most common phages from different host species may indicate the limitation of the information provided by B. subtilis phages.
According to the current International Committee on Taxonomy of Viruses (ICTV) classification scheme, the Bacillus phage SPbeta and phi105 are the only two B. subtilis temperate phages classified to the genus Spbetavirus and Spizizenvirus, respectively, under the family Siphoviridae. Based on the ICTV criteria, to classify phages into the same genus, they should share >50% DNA sequence identity [45]. Our findings showed that only 30.81% (cluster L and M members) of the identified IPs shared significant sequence similarity with the Bacillus phage SPbeta and phi105, meaning that they may belong to the same respective genus. The majority of the other species showed no relatedness to either SPbeta or phi105, suggesting the rich diversity of the prophages in B. subtilis strains. It is important to note that the diversity of the IPs and their insertion site analysis results can vary with different prediction platforms. In fact, the two prophage prediction tools used in this study mostly showed variation in terms of the same IP end coordinates.
We used a large collection of B. subtilis strains (n = 91) to study the UV-light inducibility potential of B. subtilis prophages and 18.68% of them produced infective phages that can form clear plaques and be isolated. Hinting at the inducibility prevalence of the prophages in B. subtilis, five strains were studied in both in silico and inducibility studies. All of the strains appeared to have at least one IP in the in silico analysis, but following UV-light induction treatment, only one (20%) could produce plaques.
The inducibility potential of the prophages observed in this study is significantly lower than the prevalence of the IPs (75%), which are commonly identified from two prophage-predicting programs in the in silico analysis. During the induction study, some supernatants a showing clear lysis zone failed to form plaques and few plaques remained unresponsive in the subsequent phage isolation steps, indicating that some prophages might still be induced but somehow defective to further their lifestyles. A similar observation was reported in a study with Staphylococcus suis, where cells were lysed after the addition of mitomycin C but failed to form plaque in all of the tested indicators [41]. In addition, the gradual loss in the ability of the prophages to form plaque or phage particles and subsequently to lyse cells is documented [46]. This loss in ability occurs because of the inactivation of point mutations, genome rearrangements, modular exchanges, invasion by further mobile DNA elements, and massive DNA deletions [47]. Exploring the prevalence of inducible prophages in B. subtilis would require additional prophage inducibility studies using various inducing agents and a large collection of sequenced B. subtilis strains.
High IP prevalence in B. subtilis genomes and the inducibility potential traits observed in this study leads us to deduce that prophages may have considerable effects on their hosts. The functional annotation of 50 representative intact prophage genomes resulted in the identification of putative proteins such as bacteriocins, transporters, enzymes (hydrolases, catalases, phosphatases, lipases, to name a few), and others associated with immunity, sporulation, arsenate-, arsenite-, and bleomycin resistances as well as polysaccharide biosynthesis (Figure 6).
Some of these may affect B. subtilis in a desirable way from an industry point of view. For example, immunity-related proteins could be involved in defending B. subtilis from foreign phage infection [48]. Bacteriocin-related proteins could assist B. subtilis in having competitive advantages [14].Resistance-associated proteins, such as the arsenite resistance protein, may enhance host survival. A previous study involving the inactivation of arsenate and arsenite encoding genes in a skin element, a defective B. subtilis prophage, showed that they confer resistance to arsenate and arsenite [49].
On the other hand, prophages may also have undesirable effects. One such effect is the risk of interfering with the B. subtilis-mediated fermentation processes. Phage infections of starter cultures are serious risks in the food industry [50]. Moreover, agents that favor prophage inductions are common in food industries where B. subtilis may be used for various purposes. For example, UV-light, an established prophage inducer [16], is used for sterilization in food industries [51]. Likewise, some foods such as soy sauce, which are predominantly fermented by B. subtilis [52,53], are reported as being prophage inducers [54]. Desirable or not, it needs to be experimentally proven whether any of these putative genes play a role in the bacterial property and thus require further studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v14030483/s1, Table S1: Bacillus subtilis strain genomes used in prophage prediction; Table S2: Dotting assay profiles of UV-light treated B. subtilis supernatants; Table S3: Intact prohages predicted in Bacillus subtilis strains two-prophage finding programs, PHASTER and PhiSpy; Table S4: Bacillus subtilis intact prophage genome sequence similarity scores (BLASTN); Table S5: Bacillus subtilis intact prophage genome sequence similarity scores (TBLASTX); Table S6: Intact prophages and topmost common phages similarity scores; Table S7: Bacillus subtilis prophages insertion sites; Table S8: Large-scale propagation conditions of Bacillus subtilis phages.
Author Contributions
Conceptualization, K.-P.K. and H.B.A.; methodology, investigation, and resources, K.-P.K., H.B.A., Y.C., W.H., J.-W.L., H.S.K. and M.S.S.; software, H.B.A.; validation, K.-P.K., D.S. and J.-H.J.; formal analysis and data curation, H.B.A.; original draft preparation and enriching the review, H.B.A.; validation, visualization, review and editing, H.B.A., K.-P.K., D.S., D.B.S. and J.-H.J.; funding acquisition, supervision and project administration, K.-P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT), grant number 2021R1A2C2008022.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset that supports the central findings of this study are contained within the Supplementary Materials.
Acknowledgments
We thank Korean Government (Korean Government Scholarship Program (KGSP)), the National Research Foundation of Korea (NRF), and Jeonbuk National University (JBNU).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sharma, S.; Chatterjee, S.; Datta, S.; Prasad, R.; Dubey, D.; Prasad, R.K.; Vairale, M.G. Bacteriophages and its applications: An overview. Folia Microbiol. 2017, 62, 17–55. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colavecchio, A.; D’Souza, Y.; Tompkins, E.; Jeukens, J.; Freschi, L.; Emond-Rheault, J.-G.; Kukavica-Ibrulj, I.; Boyle, B.; Bekal, S.; Tamber, S. Prophage integrase typing is a useful indicator of genomic diversity in Salmonella enterica. Front. Microbiol. 2017, 8, 1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piligrimova, E.G.; Kazantseva, O.A.; Kazantsev, A.N.; Nikulin, N.A.; Skorynina, A.V.; Koposova, O.N.; Shadrin, A.M. Putative plasmid prophages of Bacillus cereus sensu lato may hold the key to undiscovered phage diversity. Sci. Rep. 2021, 11, 7611. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Davidson, A.R. When a virus is not a parasite: The beneficial effects of prophages on bacterial fitness. J. Microbiol. 2014, 52, 235–242. [Google Scholar] [CrossRef]
- Touchon, M.; Bernheim, A.; Rocha, E.P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef] [Green Version]
- Casjens, S.R. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 2005, 8, 451–458. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, Y.; Yuan, Y.; Gao, M. Prevalence and diversity analysis of candidate prophages to provide an understanding on their roles in Bacillus thuringiensis. Viruses 2019, 11, 388. [Google Scholar] [CrossRef] [Green Version]
- Beres, S.B.; Sylva, G.L.; Barbian, K.D.; Lei, B.; Hoff, J.S.; Mammarella, N.D.; Liu, M.-Y.; Smoot, J.C.; Porcella, S.F.; Parkins, L.D. Genome sequence of a serotype M3 strain of group A Streptococcus: Phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 2002, 99, 10078–10083. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.-G.; Ohtsubo, E.; Nakayama, K.; Murata, T. Complete genome sequence of enterohemorrhagic Eschelichia coli O157: H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javan, R.R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czajkowski, R. May the phage be with you? Prophage-like elements in the genomes of Soft Rot Pectobacteriaceae: Pectobacterium spp. and Dickeya spp. Front. Microbiol. 2019, 10, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanda, A.M.; Thormann, K.; Frunzke, J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 2015, 197, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Wilcox, R.M.; Fuhrman, J.A. Bacterial viruses in coastal seawater: Lytic rather than lysogenic production. Mar. Ecol.-Prog. Ser. 1994, 114, 35. [Google Scholar] [CrossRef]
- Jiang, S.C.; Paul, J.H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 1996, 142, 27–38. [Google Scholar] [CrossRef]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 173. [Google Scholar] [CrossRef]
- Errington, J.; van der Aart, L.T. Microbe Profile: Bacillus subtilis: Model organism for cellular development, and industrial workhorse. Microbiology 2020, 166, 425–427. [Google Scholar] [CrossRef]
- Kohm, K.; Floccari, V.A.; Lutz, V.T.; Nordmann, B.; Mittelstädt, C.; Poehlein, A.; Dragos, A.; Commichau, F.M.; Hertel, R. The Bacillus phage SPβ and its relatives: A temperate phage model system reveals new strains, species, prophage integration loci, conserved proteins and lysogeny management components. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kohm, K.; Hertel, R. The life cycle of SPβ and related phages. Arch. Virol. 2021, 166, 2119–2130. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Arndt, D.; Marcu, A.; Liang, Y.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief Bioinform. 2017, 44, W16–W21. [Google Scholar] [CrossRef]
- Ågren, J.; Sundström, A.; Håfström, T.; Segerman, B. Gegenees: Fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS ONE 2012, 7, e39107. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Estimating Phylogenetic Trees and Networks Using SplitsTree4. Manuscr. Prep. Softw. 2005. Available online: www.splitstree.org (accessed on 15 October 2021).
- Krumsiek, J.; Arnold, R.; Rattei, T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007, 23, 1026–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandara, N.; Jo, J.; Ryu, S.; Kim, K.P. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol. 2012, 31, 9–16. [Google Scholar] [CrossRef]
- Djurdjevic-Milosevic, D.; Solaja, M.; Topalic-Trivunovic, L.N.; Stijepic, M.; Glusac, J. The survival of Escherichia coli upon exposure to irradiation with non-coherent polychromatic polarized light. Vet. Med. 2011, 56, 520–527. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-P.; Born, Y.; Lurz, R.; Eichenseher, F.; Zimmer, M.; Loessner, M.J.; Klumpp, J. Inducible Clostridium perfringens bacteriophages ΦS9 and ΦS63: Different genome structures and a fully functional sigK intervening element. Bacteriophage 2012, 2, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar]
- Ghosh, K.; Kang, H.S.; Hyun, W.B.; Kim, K.-P. High prevalence of Bacillus subtilis-infecting bacteriophages in soybean-based fermented foods and its detrimental effects on the process and quality of Cheonggukjang. Food Microbiol. 2018, 76, 196–203. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. In Bacteriophages; Springer: Berlin/Heidelberg, Germany, 2009; pp. 141–149. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- R Core Team. R Version 4.0.3 (Bunny-Wunnies Freak Out): The R Project for Statistical Computing. 2020. Available online: https://www.r-project.org/ (accessed on 2 February 2022).
- RStudio. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 2 February 2022).
- Abedi-Moghaddam, N.; Bulic, A.; Herderson, L.; Lam, E. Survival of Escherichia coli to UV irradiation during exponential and stationary phases of growth. J. Exp. Microbiol. Immunol 2004, 5, 44–49. [Google Scholar]
- Tan, J.A.; Christie, G.E. Lysogeny. eLS 2017, 1–11. [Google Scholar] [CrossRef]
- Piligrimova, E.G.; Kazantseva, O.A.; Nikulin, N.A.; Shadrin, A.M. Bacillus phage vB_BtS_b83 previously designated as a plasmid may represent a new siphoviridae genus. Viruses 2019, 11, 624. [Google Scholar] [CrossRef] [Green Version]
- Smeesters, P.R.; Drèze, P.-A.; Bousbata, S.; Parikka, K.J.; Timmery, S.; Hu, X.; Perez-Morga, D.; Deghorain, M.; Toussaint, A.; Mahillon, J. Characterization of a novel temperate phage originating from a cereulide-producing Bacillus cereus strain. Res. Microbiol. 2011, 162, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Fournous, G.; Brüssow, H. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 2004, 53, 9–18. [Google Scholar] [CrossRef]
- Tang, F.; Bossers, A.; Harders, F.; Lu, C.; Smith, H. Comparative genomic analysis of twelve Streptococcus suis (pro) phages. Genomics 2013, 101, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Brueggemann, A.B.; Harrold, C.L.; Javan, R.R.; Van Tonder, A.J.; McDonnell, A.J.; Edwards, B.A. Pneumococcal prophages are diverse, but not without structure or history. Sci. Rep. 2017, 7, 42976. [Google Scholar] [CrossRef] [Green Version]
- Goerke, C.; Pantucek, R.; Holtfreter, S.; Schulte, B.; Zink, M.; Grumann, D.; Bröker, B.M.; Doskar, J.; Wolz, C. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 2009, 191, 3462–3468. [Google Scholar] [CrossRef] [Green Version]
- Adriaenssens, E.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [Green Version]
- Castillo, D.; Espejo, R.; Middelboe, M. Genomic structure of bacteriophage 6H and its distribution as prophage in Flavobacterium psychrophilum strains. FEMS Microbiol. Lett. 2014, 351, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef] [Green Version]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexeeva, S.; Liu, Y.; Zhu, J.; Kaczorowska, J.; Kouwen, T.R.; Abee, T.; Smid, E.J. Genomics of tailless bacteriophages in a complex lactic acid bacteria starter culture. Int. Dairy J. 2021, 114, 104900. [Google Scholar] [CrossRef]
- Sato, T.; Kobayashi, Y. The ars operon in the skin element of bacillus subtilis confers resistance to arsenate and arsenite. J. Bacteriol. 1998, 180, 1655–1661. [Google Scholar] [PubMed]
- del Rio, B.; Binetti, A.G.; Martin, M.; Fernandez, M.; Magadan, A.; Alvarez, M. Multiplex PCR for the detection and identification of dairy bacteriophages in milk. Food Microbiol. 2007, 24, 75–81. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.; Sauceda-Gálvez, J.N.; Codina-Torrella, I.; Hernández-Herrero, M.; Gervilla, R.; Roig-Sagués, A.X. Evaluation of Continuous UVC Treatments and its Combination with UHPH on Spores of Bacillus subtilis in Whole and Skim Milk. Foods 2019, 8, 539. [Google Scholar] [CrossRef] [Green Version]
- Det-Udom, R.; Gilbert, C.; Liu, L.; Prakitchaiwattana, C.; Ellis, T.; Ledesma-Amaro, R. Towards semi-synthetic microbial communities: Enhancing soy sauce fermentation properties in B. subtilis co-cultures. Microb. Cell Factories 2019, 18, 101. [Google Scholar] [CrossRef]
- Choi, H.J.; Shin, D.; Shin, M.; Yun, B.; Kang, M.; Yang, H.-J.; Jeong, D.-Y.; Kim, Y.; Oh, S. Comparative Genomic and Functional Evaluations of Bacillus subtilis Newly Isolated from Korean Traditional Fermented Foods. Foods 2020, 9, 1805. [Google Scholar] [CrossRef]
- Willner, D.; Furlan, M.; Schmieder, R.; Grasis, J.A.; Pride, D.T.; Relman, D.A.; Angly, F.E.; McDole, T.; Mariella, R.P.; Rohwer, F. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc. Natl. Acad. Sci. USA 2011, 108, 4547–4553. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).