Species Fowl aviadenovirus B Consists of a Single Serotype despite Genetic Distance of FAdV-5 Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. FAdV-B Isolates
2.2. Sequencing and Genome Assembly
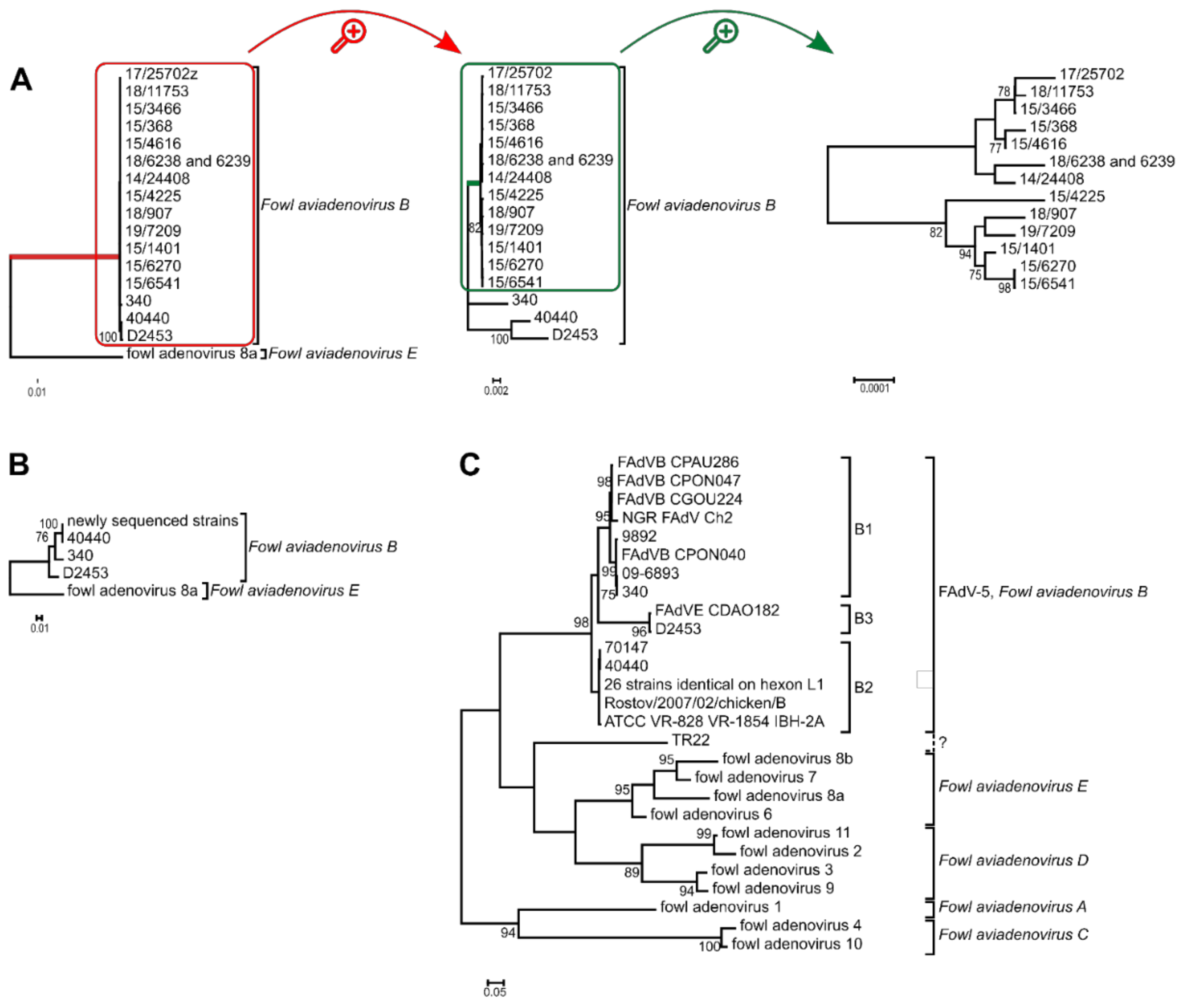
2.3. Phylogenetic Analysis
| Strain Name (GenBank acc.no.) | Country, Year of Origin | Type of Sample | Clinical Signs, Pathologies | Other Bacterial/Viral Findings |
|---|---|---|---|---|
| 14-24408 (OK283042) | Austria a, 2014 | Organ pool (liver, intestine, caecal tonsils) | Birds dead on farm | Viral: none |
| 15-368 (OK283043) | Austria b, 2015 | Organ pool (liver, intestine, caecal tonsils) | Broilers, dead on farm | Viral: none |
| 15-1401 (OK283044) | Austria c, 2015 | Organ pool (liver, intestine, caecal tonsils) | Birds dead on farm | Viral: none |
| 15-3466 (OK283045) | Austria d, 2015 | Organ pool (liver, intestine, caecal tonsils) | Broilers, dead on farm No lesions recorded | FAdV-1 |
| 15-4225 (OK283051) | France, 2015 | Pancreas | 21-day-old chickens with severe tenosynovitis (2–3% incidence) | Tendons: Reovirus |
| 15-4616 (OK283046) | Austria e, 2015 | Organ pool (liver, intestine, caecal tonsils) | Broilers, dead on farm Liver swollen, perihepatitis Pericarditis Ascites | E. coli, Staphylococcus spp. Reovirus |
| 15-6270 (OK283047) | Austria f, 2015 | Organ pool (liver, intestine, caecal tonsils) | Broilers, dead on farm Liver swollen, necroses Ascites Intestinal dilatation, foamy contents | Proteus spp. in all organs |
| 15-6541 (OK283048) | Austria g, 2015 | Organ pool (liver, intestine, caecal tonsils) | Broilers, dead on farm | E. coli, Staphylococcus spp. |
| 17-25702 (OK283049) | Hungary a, 2017 | Caecal tonsils | Broiler breeders, dead on farm | Viral: none |
| 18-907 (OK283050) | Hungary b, 2018 | Caecal tonsils | Broiler breeders, dead on farm | Viral: none |
| 18-6238 (OK283052) | Germany, 2018 | Cell culture supernatant (material of origin unknown) | n.r. | n.c. |
| 18-6239 (OK283053) | ||||
| 18-11753 (OK283054) | Hungary a, 2018 | Caecal tonsils | Broiler breeders, dead on farm | n.c. |
| 19-7207 (OK283055) | Hungary c, 2018 | Cloacal swab pool | n.r. | n.c. |
2.4. Cross-Neutralization Testing
| Accession Number | Strain Identifier | Country of Origin | Year of Origin | Taxonomic Lineage (If not FAdV-B) |
|---|---|---|---|---|
| AF508953 | TR22 | Japan | 1960s | |
| KC493646 | 340 | Northern Ireland | 1970s | |
| EF442425 | Rostov/2007/02/chicken/B | Russia | 2007 | Aviadenovirus |
| FN869989 | 08-21472 | Austria | 2008 | |
| FN869990 | 08-8669 | Austria | 2008 | |
| FN869987 | 09-7470-2 | Hungary | 2009 | |
| FN869988 | 09-7473-2 | Hungary | 2009 | |
| FN869991 | 09-6893 | Hungary | 2009 | |
| HQ697592 | K318/09 | South Korea | 2009 | Fowl aviadenovirus D |
| JF304111 | 8844 | Hungary | 2010 | |
| KC750798 | 160 | Hungary | 2011 | |
| KC750799 | 177 | Hungary | 2011 | |
| KP274034 | FAdVB_CGOU224 | Cote d’Ivoire | 2012 | unclassified aviadenovirus |
| KP274035 | FAdVB_CPON047 | Cote d’Ivoire | 2012 | unclassified aviadenovirus |
| KP274036 | FAdVB_CPAU286 | Cote d’Ivoire | 2012 | unclassified aviadenovirus |
| KP274037 | FAdVB_CPON040 | Cote d’Ivoire | 2012 | unclassified aviadenovirus |
| KP274038 | FAdVE_CDAO182 | Cote d’Ivoire | 2012 | unclassified aviadenovirus |
| KP828383 | GB 1643 | Germany | 2012 | |
| MG953228 | 9892 | Hungary | 2013 | |
| MT500572 | D2453 | Ukraine | 2013 | |
| MG953211 | 2255 | Hungary | 2014 | |
| OK283042 | 14/24408 | Austria | 2014 | |
| MG953201 | 40440 | Hungary | 2015 | |
| MG953219 | 45871 | Hungary | 2015 | |
| MG953222 | 5626 | Hungary | 2015 | |
| MG953223 | 70147 | Hungary | 2015 | |
| OK283043 | 15/368 | Austria | 2015 | |
| OK283044 | 15/1401 | Austria | 2015 | |
| OK283045 | 15/3466 | Austria | 2015 | |
| OK283046 | 15/4616 | Austria | 2015 | |
| OK283047 | 15/6270 | Austria | 2015 | |
| OK283048 | 15/6541 | Austria | 2015 | |
| OK283051 | 15/4225 | France | 2015 | |
| MK509019 | NGR_FAdV_Ch2 | Nigeria | 2017 | Fowl aviadenovirus C |
| OK283049 | 17/25702 | Hungary | 2017 | |
| OK283050 | 18/907 | Hungary | 2018 | |
| OK283052 | 18/6238 | Germany | 2018 | |
| OK283053 | 18/6239 | Germany | 2018 | |
| OK283054 | 18/11753 | Hungary | 2018 | |
| OK283055 | 19/7209 | Hungary | 2019 | |
| AF339916 | ATCC VR-828, VR-1854, IBH-2A | n.a. | n.a. | Fowl aviadenovirus D |
| AF508953 | TR22 | Japan | 1960s | |
| KC493646 | 340 | Northern Ireland | 1970s | |
| EF442425 | Rostov/2007/02/chicken/B | Russia | 2007 | Aviadenovirus |
| FN869989 | 08-21472 | Austria | 2008 | |
| FN869990 | 08-8669 | Austria | 2008 |
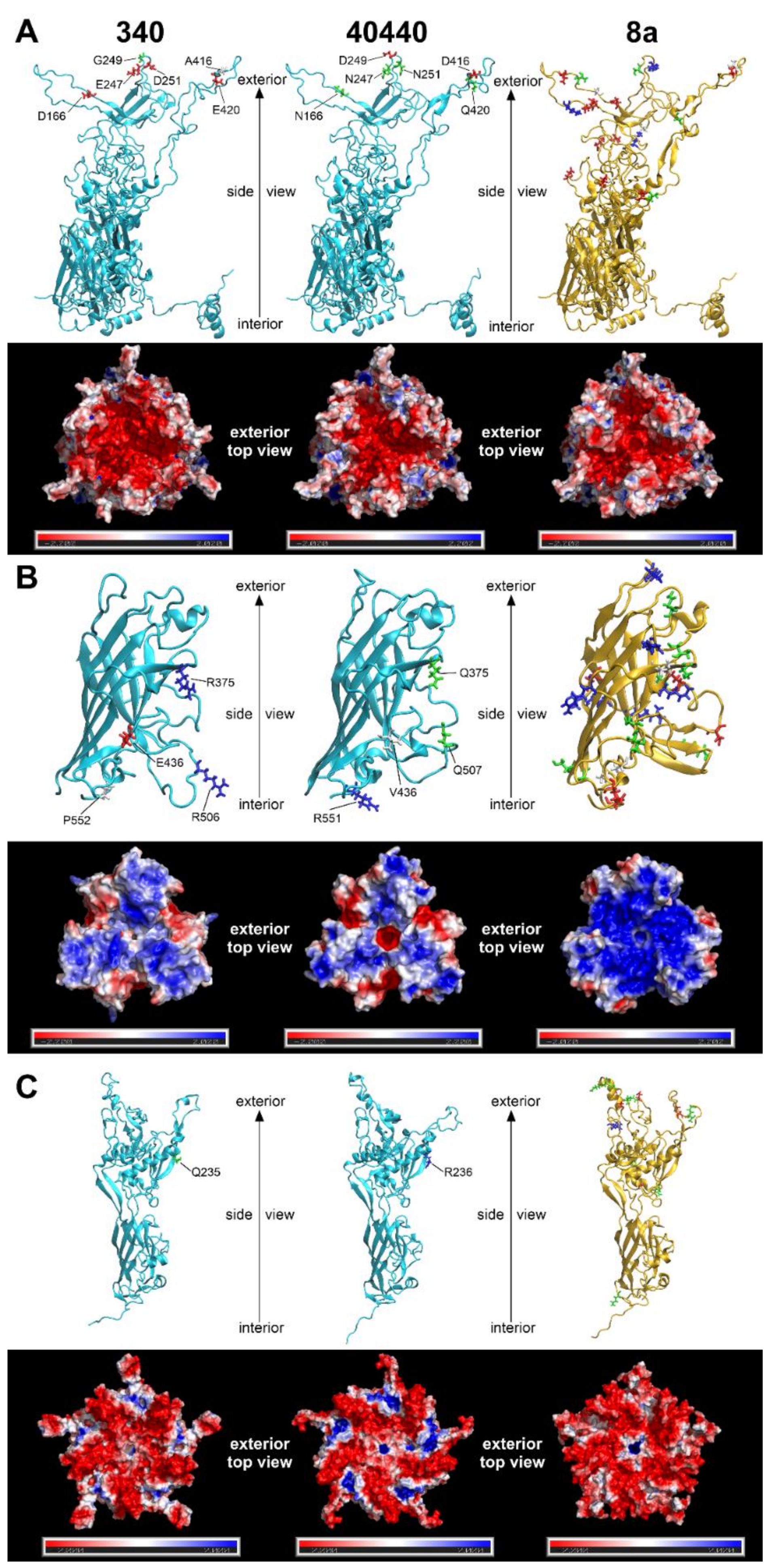
2.5. Modelling of the Major Capsid Proteins
3. Results
3.1. Propagation of FAdV-B Strains
3.2. Genome Sequences
3.3. Phylogenetic Analysis
3.4. Neutralizing Antibodies and Cross-Neutralization of FAdV-B Strains
3.5. Modelling of the Major Capsid Proteins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harrach, B.; Tarján, Z.L.; Benkő, M. Adenoviruses across the animal kingdom: A walk in the zoo. FEBS Lett. 2019, 593, 3660–3673. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; San Martín, C. Adenovirus Structure: What Is New? Int. J. Mol. Sci. 2021, 22, 5240. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.; Dolja, V.; Krupovic, M.; Varsani, A.; Wolf, Y.; Yutin, N.; Zerbin, I.M.; Kuhn, J. Create a Megataxonomic Framework, Filling All Principal Taxonomic Ranks, for DNA viruses Encoding Vertical Jelly Roll-Type Major Capsid Proteins. Available online: https://talk.ictvonline.org/ictv/proposals/2019.003G.zip (accessed on 10 June 2021).
- Harrach, B.; Kaján, G.L. Aviadenovirus. In The Springer Index of Viruses; Tidona, C.A., Darai, G., Eds.; Springer: New York, NY, USA, 2011; pp. 13–28. ISBN 978-0-387-95918-4. [Google Scholar]
- Schachner, A.; Matos, M.; Grafl, B.; Hess, M. Fowl adenovirus-induced diseases and strategies for their control—A review on the current global situation. Avian Pathol. 2018, 47, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Kiss, I.; Homonnay, Z.G.; Mató, T.; Bányai, K.; Palya, V. Research Note: An overview on distribution of fowl adenoviruses. Poult. Sci. 2021, 100, 101052. [Google Scholar] [CrossRef] [PubMed]
- McFerran, J.B.; Clarke, J.K.; Connor, T.J. Serological classification of avian adenoviruses. Arch. Gesamte Virusforsch 1972, 39, 132–139. [Google Scholar] [CrossRef]
- Marek, A.; Kosiol, C.; Harrach, B.; Kaján, G.L.; Schlötterer, C.; Hess, M. The first whole genome sequence of a Fowl adenovirus B strain enables interspecies comparisons within the genus Aviadenovirus. Vet. Microbiol. 2013, 166, 250–256. [Google Scholar] [CrossRef]
- Hess, M. Commensal or pathogen—A challenge to fulfil Koch’s Postulates. Br. Poult. Sci. 2017, 58, 1–12. [Google Scholar] [CrossRef]
- Marek, A.; Günes, A.; Schulz, E.; Hess, M. Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the hexon L1 gene region. J. Virol. Methods 2010, 170, 147–154. [Google Scholar] [CrossRef]
- Kaján, G.L.; Kecskeméti, S.; Harrach, B.; Benkő, M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 2013, 167, 357–363. [Google Scholar] [CrossRef]
- Kaján, G.L.; Affranio, I.; Tóthné Bistyák, A.; Kecskeméti, S.; Benkő, M. An emerging new fowl adenovirus genotype. Heliyon 2019, 5, e01732. [Google Scholar] [CrossRef] [Green Version]
- Homonnay, Z.; Jakab, S.; Bali, K.; Kaszab, E.; Mató, T.; Kiss, I.; Palya, V.; Bányai, K. Genome sequencing of a novel variant of fowl adenovirus B reveals mosaicism in the pattern of homologous recombination events. Arch. Virol. 2021, 166, 1477–1480. [Google Scholar] [CrossRef] [PubMed]
- Meulemans, G.; Boschmans, M.; Berg, T.P.; Decaesstecker, M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001, 30, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In Proceedings of the German conference on bioinformatics, Heidelberg, Germany, 1999; Volume 99, pp. 45–56. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lehwark, P.; Greiner, S. GB2sequin—A file converter preparing custom GenBank files for database submission. Genomics 2019, 111, 759–761. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Schnurr, D. Neutralization. In Clinical Virology Manual; Specter, S.C., Hodinka, R.L., Young, S.A., Wiedbrauk, D.L., Eds.; ASM Press: Washington, DC, USA, 2009; ISBN 978-1-555-81462-5. [Google Scholar]
- Norrby, E. The structural and functional diversity of Adenovirus capsid components. J. Gen. Virol. 1969, 5, 221–236. [Google Scholar] [CrossRef]
- Song, Y.; Dimaio, F.; Wang, R.Y.R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, L. Schrödinger Suite; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual MolecularDynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford-Miksza, L.; Schnurr, D.P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 1996, 70, 1836–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rux, J.J.; Kuser, P.R.; Burnett, R.M. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 2003, 77, 9553–9566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, R.R.; Maxfield, L.F.; Lynch, D.M.; Iampietro, M.J.; Borducchi, E.N.; Barouch, D.H. Adenovirus serotype 5-specific neutralizing antibodies target multiple hexon hypervariable regions. J. Virol. 2012, 86, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Bradley, R.R.; Lynch, D.M.; Iampietro, M.J.; Borducchi, E.N.; Barouch, D.H. Adenovirus Serotype 5 Neutralizing Antibodies Target both Hexon and Fiber following Vaccination and Natural Infection. J. Virol. 2012, 86, 625–629. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Duffy, M.R.; Deng, L.; Dakin, R.S.; Uil, T.; Custers, J.; Kelly, S.M.; McVey, J.H.; Nicklin, S.A.; Baker, A.H. Manipulating adenovirus hexon hypervariable loops dictates immune neutralisation and coagulation factor X-dependent cell interaction in vitro and in vivo. PLoS Pathog. 2015, 11, e1004673. [Google Scholar] [CrossRef] [Green Version]
- Nemerow, G.R.; Stewart, P.L.; Reddy, V.S. Structure of human adenovirus. Curr. Opin. Virol. 2012, 2, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Gall, J.G.; Crystal, R.G.; Falck-Pedersen, E. Construction and characterization of hexon-chimeric adenoviruses: Specification of adenovirus serotype. J. Virol. 1998, 72, 10260–10264. [Google Scholar] [CrossRef] [Green Version]
- Schachner, A.; Gonzalez, G.; Endler, L.; Ito, K.; Hess, M. Fowl Adenovirus (FAdV) recombination with intertypic crossovers in genomes of FAdV-D and FAdV-E, displaying hybrid serological phenotypes. Viruses 2019, 11, 1094. [Google Scholar] [CrossRef] [Green Version]
- Gahéry-Ségard, H.; Farace, F.; Godfrin, D.; Gaston, J.; Lengagne, R.; Tursz, T.; Boulanger, P.; Guillet, J.-G. Immune Response to Recombinant Capsid Proteins of Adenovirus in Humans: Antifiber and Anti-Penton Base Antibodies Have a Synergistic Effect on Neutralizing Activity. J. Virol. 1998, 72, 2388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFerran, J.B.; McCracken, R.M.; Connor, T.J.; Evans, R.T. Isolation of viruses from clinical outbreaks of inclusion body hepatitis. Avian Pathol. 1976, 5, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaján, G.L.; Sameti, S.; Benkő, M. Partial sequence of the DNA-dependent DNA polymerase gene of fowl adenoviruses: A reference panel for a general diagnostic PCR in poultry. Acta Vet. Hung. 2011, 59, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Kaján, G.L. Poultry adenoviruses. In Molecular Detection of Animal Viral Pathogens; Liu, D., Ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 735–746. ISBN 9781498700368. [Google Scholar]
- Gomis, S.; Goodhope, A.R.; Ojkic, A.D.; Willson, P. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 2006, 50, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Ojkić, D.; Martin, E.; Swinton, J.; Vaillancourt, J.P.; Boulianne, M.; Gomis, S. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 2008, 37, 95–100. [Google Scholar] [CrossRef]
- Changjing, L.; Haiying, L.; Dongdong, W.; Jingjing, W.; Youming, W.; Shouchun, W.; Jida, L.; Ping, L.; Jianlin, W.; Shouzhen, X.; et al. Characterization of fowl adenoviruses isolated between 2007 and 2014 in China. Vet. Microbiol. 2016, 197, 62–67. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, Q.; Zhang, G.; Sun, W.; Liu, X.; Xiao, Y.; Shang, Y.; Liu, S. Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transbound. Emerg. Dis. 2018, 65, e121–e126. [Google Scholar] [CrossRef]
- De la Torre, D.; Nuñez, L.F.N.; Santander Parra, S.H.; Astolfi-Ferreira, C.S.; Piantino Ferreira, A.J. Molecular characterization of fowl adenovirus group I in commercial broiler chickens in Brazil. VirusDisease 2018, 29, 83–88. [Google Scholar] [CrossRef]
- Chen, L.; Yin, L.; Zhou, Q.; Peng, P.; Du, Y.; Liu, L.; Zhang, Y.; Xue, C.; Cao, Y. Epidemiological investigation of fowl adenovirus infections in poultry in China during 2015–2018. BMC Vet. Res. 2019, 15, 271. [Google Scholar] [CrossRef] [Green Version]
- Hess, M. Aviadenovirus infections. In Diseases of Poultry; Swayne, D.E., Ed.; Wiley-Blackwell: Oxford, UK, 2013; pp. 290–300. ISBN 978-0-470-95899-5. [Google Scholar]
- McCarthy, T.; Lebeck, M.G.; Capuano, A.W.; Schnurr, D.P.; Gray, G.C. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J. Clin. Virol. 2009, 46, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Kawamura, H.; Shimizu, F.; Tsubahara, H. Avian adenovirus: Its properties and serological classification. Natl. Inst. Anim. Health Q. 1964, 4, 183–193. [Google Scholar]
- Meulemans, G.; Couvreur, B.; Decaesstecker, M.; Boschmans, M.; van den Berg, T.P. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. 2004, 33, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Kaján, G.L.; Kecskeméti, S. First Hungarian isolation of an adenovirus type belonging to species Fowl adenovirus B. Magy. Állatorvosok Lapja 2011, 133, 347–352. [Google Scholar]
- Kleine, A.; Hafez, H.M.; Lüschow, D. Investigations on aviadenoviruses isolated from turkey flocks in Germany. Avian Pathol. 2016, 46, 181–187. [Google Scholar] [CrossRef]
- Niczyporuk, J.; Kozdrun, W.; Czekaj, H.; Piekarska, K.; Stys-Fijol, N. Characterisation of adenovirus strains represented species B and E isolated from broiler chicken flocks in eastern Poland. Heliyon 2021, 7, e06225. [Google Scholar] [CrossRef]
- Pauly, M.; Akoua-Koffi, C.; Buchwald, N.; Schubert, G.; Weiss, S.; Couacy-Hymann, E.; Anoh, A.E.; Mossoun, A.; Calvignac-Spencer, S.; Leendertz, S.A.; et al. Adenovirus in Rural Côte D‘Ivoire: High Diversity and Cross-Species Detection. Ecohealth 2015, 12, 441–452. [Google Scholar] [CrossRef] [Green Version]
- Lim, T.H.; Lee, H.J.; Lee, D.H.; Lee, Y.N.; Park, J.K.; Youn, H.N.; Kim, M.S.; Youn, H.S.; Lee, J.B.; Park, S.Y.; et al. Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis. 2011, 55, 554–560. [Google Scholar] [CrossRef]
- Dar, A.; Gomis, S.; Shirley, I.; Mutwiri, G.; Brownlie, R.; Potter, A.; Gerdts, V.; Tikoo, S.K. Pathotypic and molecular characterization of a fowl adenovirus associated with inclusion body hepatitis in Saskatchewan chickens. Avian Dis. 2012, 56, 73–81. [Google Scholar] [CrossRef]
- Cui, J.; Xu, Y.; Zhou, Z.; Xu, Q.; Wang, J.; Xiao, Y.; Li, Z.; Bi, D. Pathogenicity and Molecular Typing of Fowl Adenovirus-Associated With Hepatitis/Hydropericardium Syndrome in Central China (2015–2018). Front. Vet. Sci. 2020, 7, 190. [Google Scholar] [CrossRef]
- Chitradevi, S.; Sukumar, K.; Suresh, P.; Balasubramaniam, G.A.; Kannan, D. Molecular typing and pathogenicity assessment of fowl adenovirus associated with inclusion body hepatitis in chicken from India. Trop. Anim. Health Prod. 2021, 53, 412. [Google Scholar] [CrossRef]
- Steer, P.A.; O’Rourke, D.; Ghorashi, S.A.; Noormohammadi, A.H. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust. Vet J. 2011, 89, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Niczyporuk, J.S. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch. Virol. 2016, 161, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Niczyporuk, J.S.; Czekaj, H. A comparative pathogenicity analysis of two adenovirus strains, 1/A and 8a/E, isolated from poultry in Poland. Arch. Virol. 2018, 163, 3005–3013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redondo, H.; Fragoso, J.S.; Tahala, M.A.; Bensassi, Y.; Gil, I.; Elbachir, E.; Rodríguez, M.J.; Moreno, J.C.A. Characterization of strain of fowl adenoviruses circulating in Morocco. Poult. Sci. 2018, 97, 4057–4062. [Google Scholar] [CrossRef]
- Oliver-Ferrando, S.; Dolz, R.; Calderón, C.; Valle, R.; Rivas, R.; Pérez, M.; Biarnés, M.; Blanco, A.; Bertran, K.; Ramis, A.; et al. Epidemiological and pathological investigation of fowl aviadenovirus serotypes 8b and 11 isolated from chickens with inclusion body hepatitis in Spain (2011–2013). Avian Pathol. 2017, 46, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Morshed, R.; Hosseini, H.; Langeroudi, A.G.; Fard, M.H.B.; Charkhkar, S. Fowl Adenoviruses D and e Cause Inclusion Body Hepatitis Outbreaks in Broiler and Broiler Breeder Pullet Flocks. Avian Dis. 2017, 61, 205–210. [Google Scholar] [CrossRef]
- McFerran, J.B.; Connor, T.J.; McCracken, R.M. Isolation of adenoviruses and reoviruses from avian species other than domestic fowl. Avian Dis. 1976, 20, 519–524. [Google Scholar] [CrossRef]
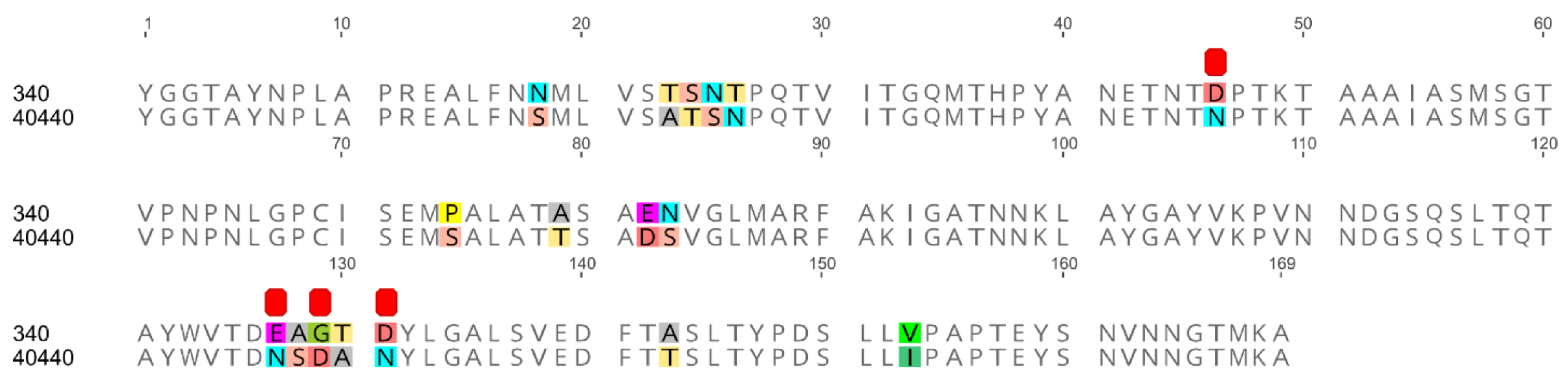
| Analyzed Stretch | Sequence Identity of the Newly Sequenced Strains to the Two Strains: | |
|---|---|---|
| 340 | 40440 | |
| Complete genome (NA) | 98.50–98.54% | 97.97–97.98% |
| DNA polymerase (AA) | 99.46–99.54% | 99.07–99.15% |
| Penton base (AA) | 99.63% | 100.00% |
| Hexon complete (AA) | 97.27% | 100.00% |
| Hexon loop-1 (AA) | 89.94% | 100.00% |
| Fiber (AA) | 92.93% | 96.56% |
| Fiber knob (AA) | 89.80% | 100.00% |
| Concatenate (AA) * | 95.40% | 100.00% |
| Virus (100 TCID50) | Antiserum (Reciprocal Titer) | ||||
|---|---|---|---|---|---|
| 40440 | 15-6541 | 15-4225 | 18-6238 | 340 | |
| 40440 | 640 | 640 | 1280 | 640 | - |
| 15-6541 | 320 | 640 | 1280 | 640 | - |
| 15-4225 | 640 | 640 | 1280 | 640 | - |
| 18-6238 | 320 | 640 | 1280 | 640 | - |
| 340 | 320 | 320 | 320 | 160 | 320 |
| Protein | 340 Versus 40440 | 340 Versus FAdV-8a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Diff. | Charge-Altering Substitutions | Number of Diff. | Charge-Altering Substitutions | |||||||
| Polar-to-Charged a | Nonpolar-to-Charged b | Charge Reversal c | Polar-to-Charged a | Nonpolar-to-Charged b | Charge Reversal c | Insertion/Deletion d | ||||
| Hexon | 26 | 5 | 1 | 0 | 126 | 9 | 9 | 2 | 0 | |
| Fiber knob e | 24 | 2 | 2 | 0 | 113 | 14 | 8 | 1 | 1 | |
| Penton base f | 1 | 1 | 0 | 0 | 63 | 10 | 2 | 0 | 1 | |
| Sum | 51 | 8 | 3 | 0 | 302 | 33 | 19 | 3 | 2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaján, G.L.; Schachner, A.; Gellért, Á.; Hess, M. Species Fowl aviadenovirus B Consists of a Single Serotype despite Genetic Distance of FAdV-5 Isolates. Viruses 2022, 14, 248. https://doi.org/10.3390/v14020248
Kaján GL, Schachner A, Gellért Á, Hess M. Species Fowl aviadenovirus B Consists of a Single Serotype despite Genetic Distance of FAdV-5 Isolates. Viruses. 2022; 14(2):248. https://doi.org/10.3390/v14020248
Chicago/Turabian StyleKaján, Győző L., Anna Schachner, Ákos Gellért, and Michael Hess. 2022. "Species Fowl aviadenovirus B Consists of a Single Serotype despite Genetic Distance of FAdV-5 Isolates" Viruses 14, no. 2: 248. https://doi.org/10.3390/v14020248
APA StyleKaján, G. L., Schachner, A., Gellért, Á., & Hess, M. (2022). Species Fowl aviadenovirus B Consists of a Single Serotype despite Genetic Distance of FAdV-5 Isolates. Viruses, 14(2), 248. https://doi.org/10.3390/v14020248







