Characterization of HIV-1 Transmission Clusters Inferred from the Brazilian Nationwide Genotyping Service Database
Abstract
1. Introduction
2. Materials and Methods
2.1. The RENAGENO Dataset, Quality Control and Ethical Approval
2.2. HIV-1 Group M Subtyping
2.3. Phylogenetic Analysis
2.4. Clusters Identification
2.5. Statistical Analysis
3. Results
3.1. Clusters’ Frequency and Size Description
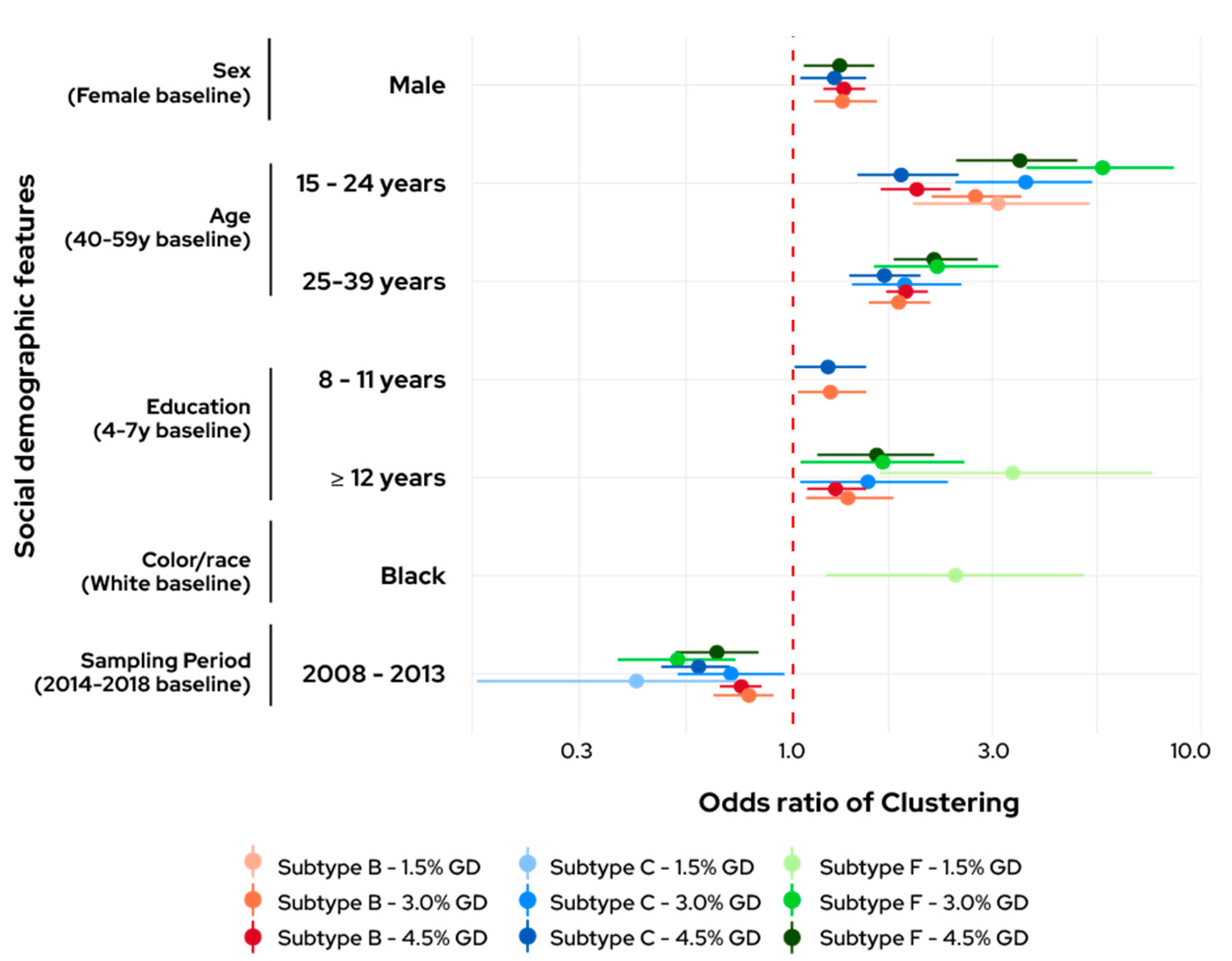
3.2. Clustering Odds
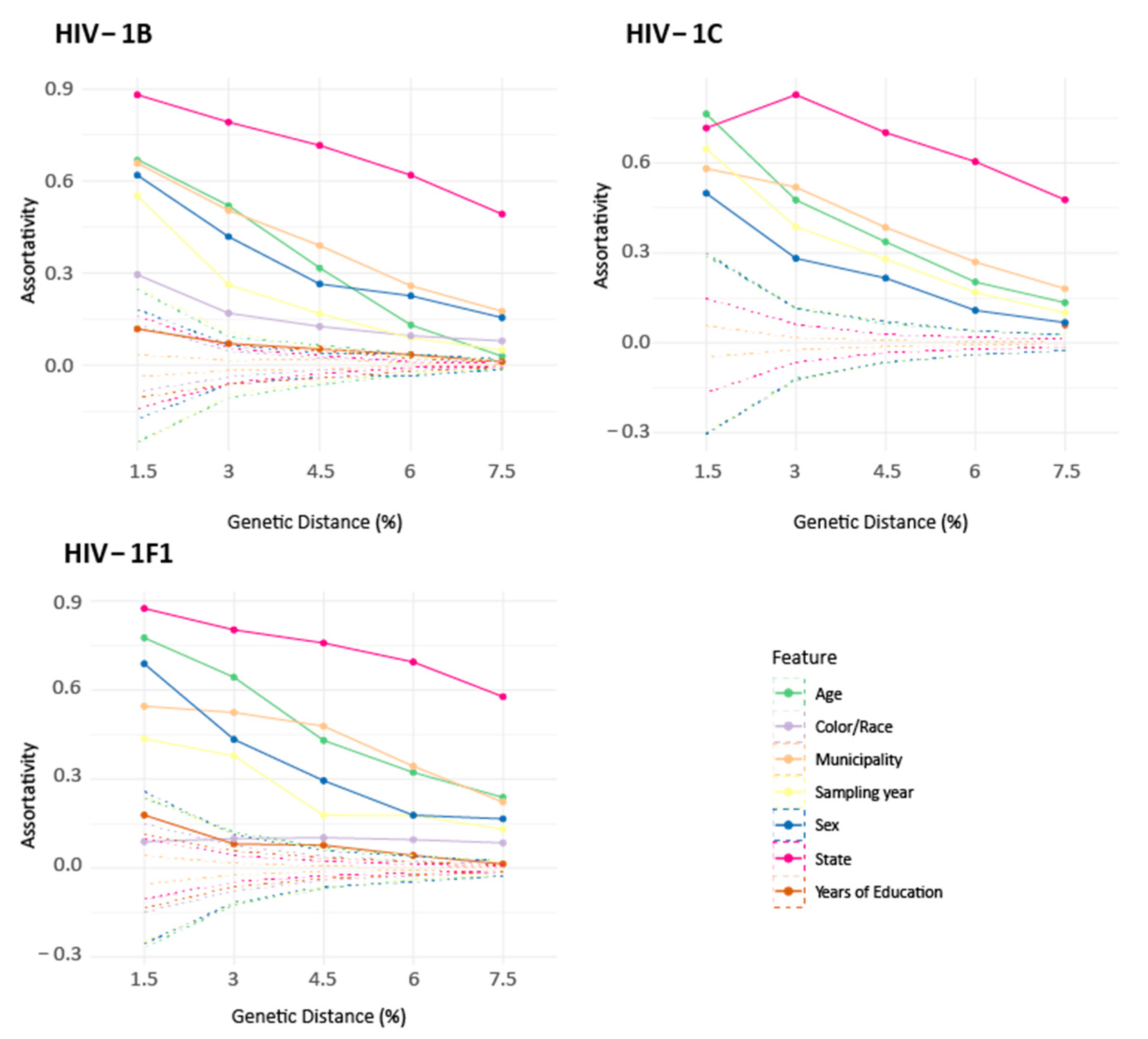
3.3. Assortativity Analysis of the Networks
3.4. Spatial Distribution of the Clusters
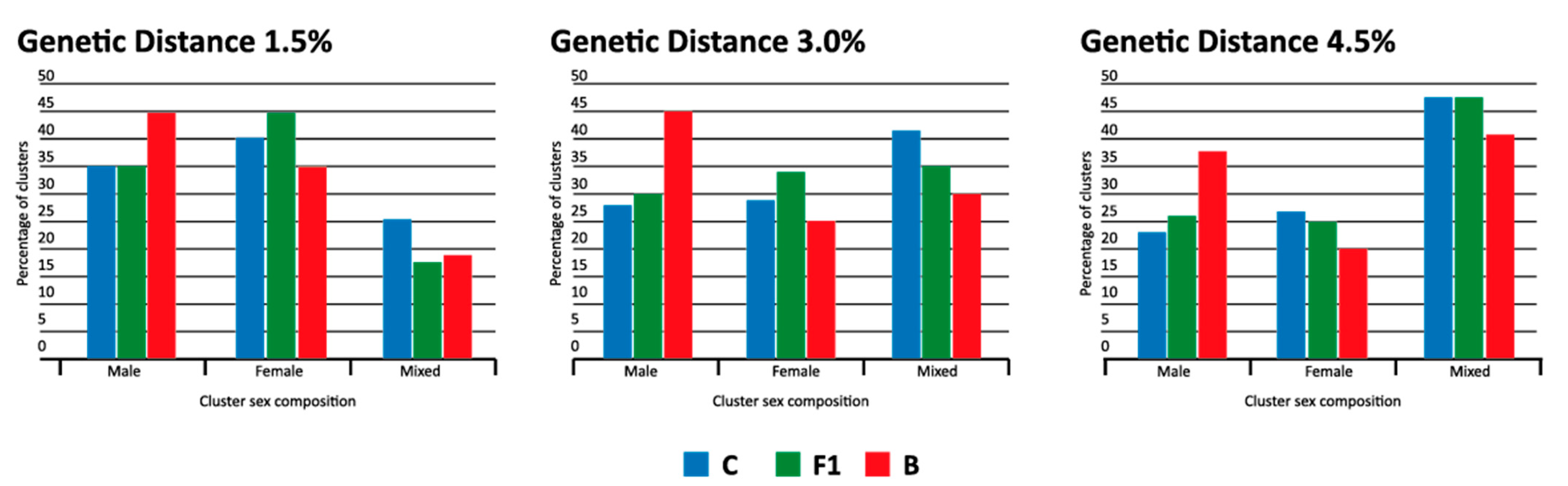
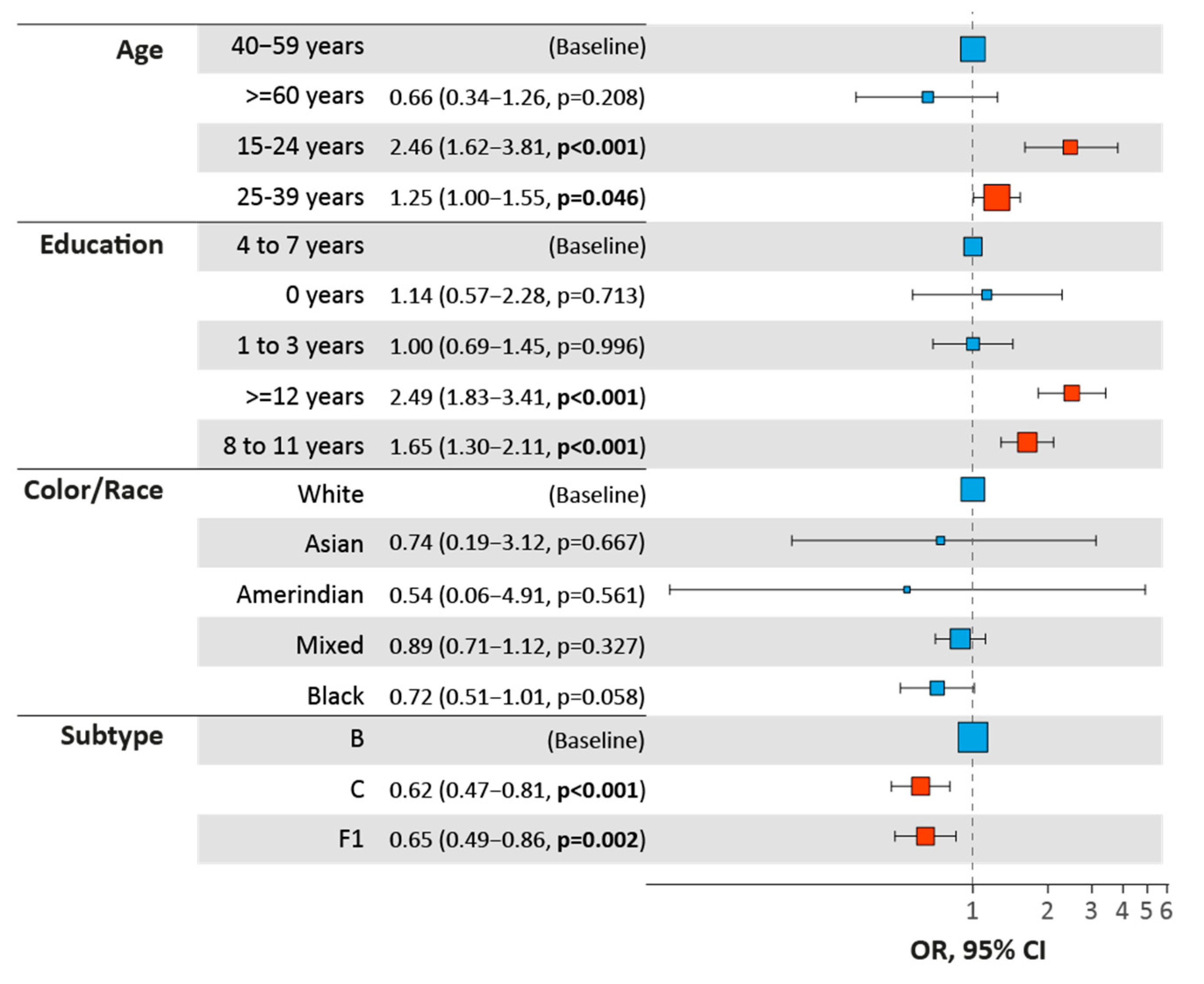
3.5. Male Clusters Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tebit, D.M.; Arts, E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 2011, 11, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Brazilian Ministry of Health. AIDS epidemiological bulletin July 2020-June 2021. Brazilian Ministry of Health, Brasília, DF, Brazil. 2021. Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/especiais/2021/boletim-epidemiologico-especial-hiv-aids-2021.pdf/view (accessed on 18 September 2022).
- UNAIDS. Data 2019. Program. HIV/AIDS 1–476 (2019). 2019. Available online: https://www.unaids.org/en/resources/documents/2019/2019-UNAIDS-data (accessed on 18 September 2022).
- Gräf, T.; Bello, G.; Andrade, P.; Arantes, I.; Pereira, J.M.; da Silva, A.B.P.; Veiga, R.V.; Mariani, D.; Boullosa, L.T.; Arruda, M.B.; et al. HIV-1 molecular diversity in Brazil unveiled by 10 years of sampling by the national genotyping network. Sci. Rep. 2021, 11, 15842. [Google Scholar] [CrossRef] [PubMed]
- Arruda, M.B.; Boullosa, L.T.; Cardoso, C.C.; da Costa, C.M.; Brites, C.; de Lima, S.T.; Kaminski, H.T.; Aleixo, A.W.; Esposito, A.O.; Cavalcanti, A.M.; et al. Brazilian network for HIV Drug Resistance Surveillance (HIV-BresNet): A survey of treatment-naive individuals. J. Int. AIDS Soc. 2018, 21, e25032. [Google Scholar] [CrossRef] [PubMed]
- Preston, B.D.; Poiesz, B.J.; Loeb, L.A. Fidelity of HIV-1 Reverse Transcriptase. Science 1988, 242, 1168–1171. [Google Scholar] [CrossRef]
- Gräf, T.; Delatorre, E.; Bello, G. Phylogenetics applied to the human immunodeficiency virus type 1 (HIV-1): From the cross-species transmissions to the contact network inferences. Memórias Do Inst. Oswaldo Cruz 2020, 115, e190461. [Google Scholar] [CrossRef]
- Lewis, F.; Hughes, G.J.; Rambaut, A.; Pozniak, A.; Brown, A.J.L. Episodic Sexual Transmission of HIV Revealed by Molecular Phylodynamics. PLoS Med. 2008, 5, 392–402. [Google Scholar] [CrossRef]
- Rose, R.; Lamers, S.L.; Dollar, J.J.; Grabowski, M.K.; Hodcroft, E.B.; Ragonnet-Cronin, M.; Wertheim, J.O.; Redd, A.D.; German, D.; Laeyendecker, O. Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res. Hum. Retrovir. 2017, 33, 211–218. [Google Scholar] [CrossRef]
- Brenner, B.G.; Roger, M.; Routy, J.; Moisi, D.; Ntemgwa, M.; Matte, C.; Baril, J.; Thomas, R.; Rouleau, D.; Bruneau, J.; et al. High Rates of Forward Transmission Events after Acute/Early HIV-1 Infection. J. Infect. Dis. 2007, 195, 951–959. [Google Scholar] [CrossRef]
- Drescher, S.M.; von Wyl, V.; Yang, W.-L.; Böni, J.; Yerly, S.; Shah, C.; Aubert, V.; Klimkait, T.; Taffé, P.; Furrer, H.; et al. Treatment-Naive Individuals Are the Major Source of Transmitted HIV-1 Drug Resistance in Men Who Have Sex With Men in the Swiss HIV Cohort Study. Clin. Infect. Dis. 2014, 58, 285–294. [Google Scholar] [CrossRef]
- Chin, B.S.; Chaillon, A.; Mehta, S.R.; Wertheim, J.O.; Kim, G.; Shin, H.-S.; Smith, D.M. Molecular epidemiology identifies HIV transmission networks associated with younger age and heterosexual exposure among Korean individuals. J. Med Virol. 2016, 88, 1832–1835. [Google Scholar] [CrossRef]
- Poon, A.F.Y.; Joy, J.B.; Woods, C.K.; Shurgold, S.; Colley, G.; Brumme, C.J.; Hogg, R.S.; Montaner, J.S.G.; Harrigan, P.R. The Impact of Clinical, Demographic and Risk Factors on Rates of HIV Transmission: A Population-based Phylogenetic Analysis in British Columbia, Canada. J. Infect. Dis. 2014, 211, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Kouyos, R.D.; von Wyl, V.; Yerly, S.; Böni, J.; Taffé, P.; Shah, C.; Börgisser, P.; Klimkait, T.; Weber, R.; Hirschel, B.; et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J. Infect Dis. 2010, 201, 1488–1497. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Hu, Y.W.; Morris, S.R.; Sheng, Z.; Poortinga, K.; O Wertheim, J. HIV transmission networks among transgender women in Los Angeles County, CA, USA: A phylogenetic analysis of surveillance data. Lancet HIV 2019, 6, e164–e172. [Google Scholar] [CrossRef]
- Ratmann, O.; van Sighem, A.; Bezemer, D.; Gavryushkina, A.; Jurriaans, S.; Wensing, A.; de Wolf, F.; Reiss, P.; Fraser, C.; Cohort, A.O. Sources of HIV infection among men having sex with men and implications for prevention. Sci. Transl. Med. 2016, 8, 320ra2. [Google Scholar] [CrossRef]
- Poon, A.F.Y.; Gustafson, R.; Daly, P.; Zerr, L.; Demlow, S.E.; Wong, J.; Woods, C.K.; Hogg, R.S.; Krajden, M.; Moore, D.; et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: An implementation case study. Lancet HIV 2016, 3, e231–e238. [Google Scholar] [CrossRef] [PubMed]
- Dennis, A.M.; Herbeck, J.T.; Brown, A.L.; Kellam, P.; de Oliveira, T.; Pillay, D.; Fraser, C.; Cohen, M.S. Phylogenetic Studies of Transmission Dynamics in Generalized HIV Epidemics. JAIDS J. Acquir. Immune Defic. Syndr. 2014, 67, 181–195. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.; Kharsany, A.B.M.; Gräf, T.; Cawood, C.; Khanyile, D.; Grobler, A.; Puren, A.; Maduraiet, S.; Karim, Q.A.; Karimet, S.S.A. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: A community-wide phylogenetic study. Lancet HIV 2017, 4, e41–e50. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, M.K.; Lessler, J.; Redd, A.D.; Kagaayi, J.; Laeyendecker, O.; Ndyanabo, A.; Nelson, M.I.; Cummings, D.A.T.; Bwanika, J.B.; Mueller, A.C.; et al. The Role of Viral Introductions in Sustaining Community-Based HIV Epidemics in Rural Uganda: Evidence from Spatial Clustering, Phylogenetics, and Egocentric Transmission Models. PLoS Med. 2014, 11, e1001610. [Google Scholar] [CrossRef]
- Junqueira, D.M.; De Medeiros, R.M.; Gräf, T.; Almeida, S.E.D.M. Short-Term Dynamic and Local Epidemiological Trends in the South American HIV-1B Epidemic. PLoS ONE 2016, 11, e0156712. [Google Scholar] [CrossRef]
- Tanaka, T.S.O.; Leite, T.F.; Freitas, S.Z.; Cesar, G.A.; de Rezende, G.R.; Lindenberg, A.D.S.C.; Guimarães, M.L.; Motta-Castro, A.R.C. HIV-1 Molecular Epidemiology, Transmission Clusters and Transmitted Drug Resistance Mutations in Central Brazil. Front. Microbiol. 2019, 10, 20. [Google Scholar] [CrossRef]
- Struck, D.; Lawyer, G.; Ternes, A.-M.; Schmit, J.-C.; Bercoff, D.P. COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014, 42, e144. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Peña, A.-C.; Faria, N.R.; Imbrechts, S.; Libin, P.; Abecasis, A.B.; Deforche, K.; Gómez-López, A.; Camacho, R.J.; de Oliveira, T.; Vandamme, A.-M. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect. Genet. Evol. 2013, 19, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Wayne, P.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Evironments Workshop (GCE) New Orleans, New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Berger, S.A.; Krompass, D.; Stamatakis, A. Performance, Accuracy, and Web Server for Evolutionary Placement of Short Sequence Reads under Maximum Likelihood. Syst. Biol. 2011, 60, 291–302. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Cabello, M.; Romero, H.; Bello, G. Multiple introductions and onward transmission of non-pandemic HIV-1 subtype B strains in North America and Europe. Sci. Rep. 2016, 6, 33971. [Google Scholar] [CrossRef][Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Bennett, D.E.; Camacho, R.J.; Otelea, D.; Kuritzkes, D.R.; Fleury, H.; Kiuchi, M.; Heneine, W.; Kantor, R.; Jordan, M.R.; Schapiro, J.M.; et al. Drug Resistance Mutations for Surveillance of Transmitted HIV-1 Drug-Resistance: 2009 Update. PLoS ONE 2009, 4, e4724. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Lemoine, F.; Entfellner, J.-B.D.; Wilkinson, E.; Correia, D.; Felipe, M.D.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Hodcroft, E.; Hué, S.; Fearnhill, E.; Delpech, V.; Brown, A.J.L.; Lycett, S. Automated analysis of phylogenetic clusters. BMC Bioinform. 2013, 14, 317. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Poon, A.F.Y. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol. 2016, 2, vew031. [Google Scholar] [CrossRef]
- McCloskey, R.M.; Poon, A.F.Y. A model-based clustering method to detect infectious disease transmission outbreaks from sequence variation. PLOS Comput. Biol. 2017, 13, e1005868. [Google Scholar] [CrossRef] [PubMed]
- Barido-Sottani, J.; Vaughan, T.G.; Stadler, T. Detection of HIV transmission clusters from phylogenetic trees using a multi-state birth–death model. J. R. Soc. Interface 2018, 15, 20180512. [Google Scholar] [CrossRef] [PubMed]
- Harling, G.; Bärnighausen, T. The role of partners’ educational attainment in the association between HIV and education amongst women in seven sub-Saharan African countries. J. Int. AIDS Soc. 2016, 19, 20038. [Google Scholar] [CrossRef]
- Le Vu, S.; Ratmann, O.; Delpech, V.; Brown, A.E.; Gill, O.N.; Tostevin, A.; Fraser, C.; Volz, E. Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics 2018, 23, 1–10. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Pond, K.S.L.; Forgione, L.A.; Mehta, S.R.; Murrell, B.; Shah, S.; Smith, D.M.; Scheffler, K.; Torian, L.V. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathog. 2017, 13, e1006000. [Google Scholar] [CrossRef]
- Gregson, S.; Nyamukapa, C.A.; Garnett, G.P.; Mason, P.R.; Zhuwau, T.; Caraël, M.; Chandiwana, S.K.; Anderson, R.M. Sexual mixing patterns and sex-diff erentials in teenage exposure to HIV infection in rural Zimbabwe. Lancet 2002, 359, 1896–1903. [Google Scholar] [CrossRef]
- Rothenberg, R.; Muth, S.Q. Large-Network Concepts and Small-Network Characteristics: Fixed and Variable Factors. Sex Transm Dis 2007, 34, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Crooks, N.; Wise, A.; Frazier, T. Addressing sexually transmitted infections in the sociocultural context of black heterosexual relationships in the United States. Soc. Sci. Med. 2020, 263, 113303. [Google Scholar] [CrossRef] [PubMed]
- Mustanski, B.; Birkett, M.; Kuhns, L.M.; Latkin, C.A.; Muth, S.Q. The Role of Geographic and Network Factors in Racial Disparities in HIV Among Young Men Who have Sex with Men: An Egocentric Network Study. AIDS Behav. 2014, 19, 1037–1047. [Google Scholar] [CrossRef] [PubMed]

| Subtype C (n = 5328) | Subtype F1 (n = 4423) | Subtype B (n = 14628) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1.5% GD | 3% GD | 4.5% GD | 1.5% GD | 3% GD | 4.5% GD | 1.5% GD | 3% GD | 4.5% GD | |
| N of cluster | 40 | 194 | 453 | 51 | 182 | 379 | 138 | 637 | 1494 |
| N of clustered individuals | 80 | 409 | 1054 | 105 | 401 | 933 | 276 | 1362 | 3467 |
| Clustering rate | 1.5% | 7.7% | 19.8% | 2.4% | 9.1% | 21.1% | 1.9% | 9.3% | 23.7% |
| Cluster size (% per cluster) | |||||||||
| 2 individuals | 40 (100%) | 178 (91.8%) | 355 (78.4%) | 49 (96.1%) | 155 (85.2%) | 274 (72.3%) | 276 (100%) | 566 (88.9%) | 1174 (78.6%) |
| 3 individuals | 13 (6.7%) | 71 (15.7%) | 1 (2.0%) | 19 (10.4%) | 69 (18.2%) | 60 (9.4%) | 224 (15.0%) | ||
| 4 individuals | 2 (1.0%) | 13 (2.9%) | 1 (2.0%) | 7 (3.9%) | 19 (5.0%) | 7 (1.1%) | 60 (4.0%) | ||
| 5 individuals | 8 (1.8%) | 10 (2.6%) | 2 (0.3%) | 19 (1.3%) | |||||
| 6 individuals | 1 (0.5%) | 3 (0.7%) | 1 (0.6%) | 2 (0.5%) | 2 (0.3%) | 10 (0.7%) | |||
| 7 individuals | 3 (0.7%) | 2 (0.5%) | 4 (0.3%) | ||||||
| 8 individuals | 1 (0.3%) | 3 (0.2%) | |||||||
| 9 individuals | 2 (0.5%) | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrade, P.; Arantes, I.; Tanuri, A.; Bello, G.; Gräf, T. Characterization of HIV-1 Transmission Clusters Inferred from the Brazilian Nationwide Genotyping Service Database. Viruses 2022, 14, 2768. https://doi.org/10.3390/v14122768
Andrade P, Arantes I, Tanuri A, Bello G, Gräf T. Characterization of HIV-1 Transmission Clusters Inferred from the Brazilian Nationwide Genotyping Service Database. Viruses. 2022; 14(12):2768. https://doi.org/10.3390/v14122768
Chicago/Turabian StyleAndrade, Paula, Ighor Arantes, Amilcar Tanuri, Gonzalo Bello, and Tiago Gräf. 2022. "Characterization of HIV-1 Transmission Clusters Inferred from the Brazilian Nationwide Genotyping Service Database" Viruses 14, no. 12: 2768. https://doi.org/10.3390/v14122768
APA StyleAndrade, P., Arantes, I., Tanuri, A., Bello, G., & Gräf, T. (2022). Characterization of HIV-1 Transmission Clusters Inferred from the Brazilian Nationwide Genotyping Service Database. Viruses, 14(12), 2768. https://doi.org/10.3390/v14122768






