Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae) †
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Insects and Viruses
2.2. Viral Content of Soft Scales and Source Sampled
2.3. Recipient Grapevines
2.4. Transmission Experiments
2.5. Virus Detection in Recipient Plants by ELISA
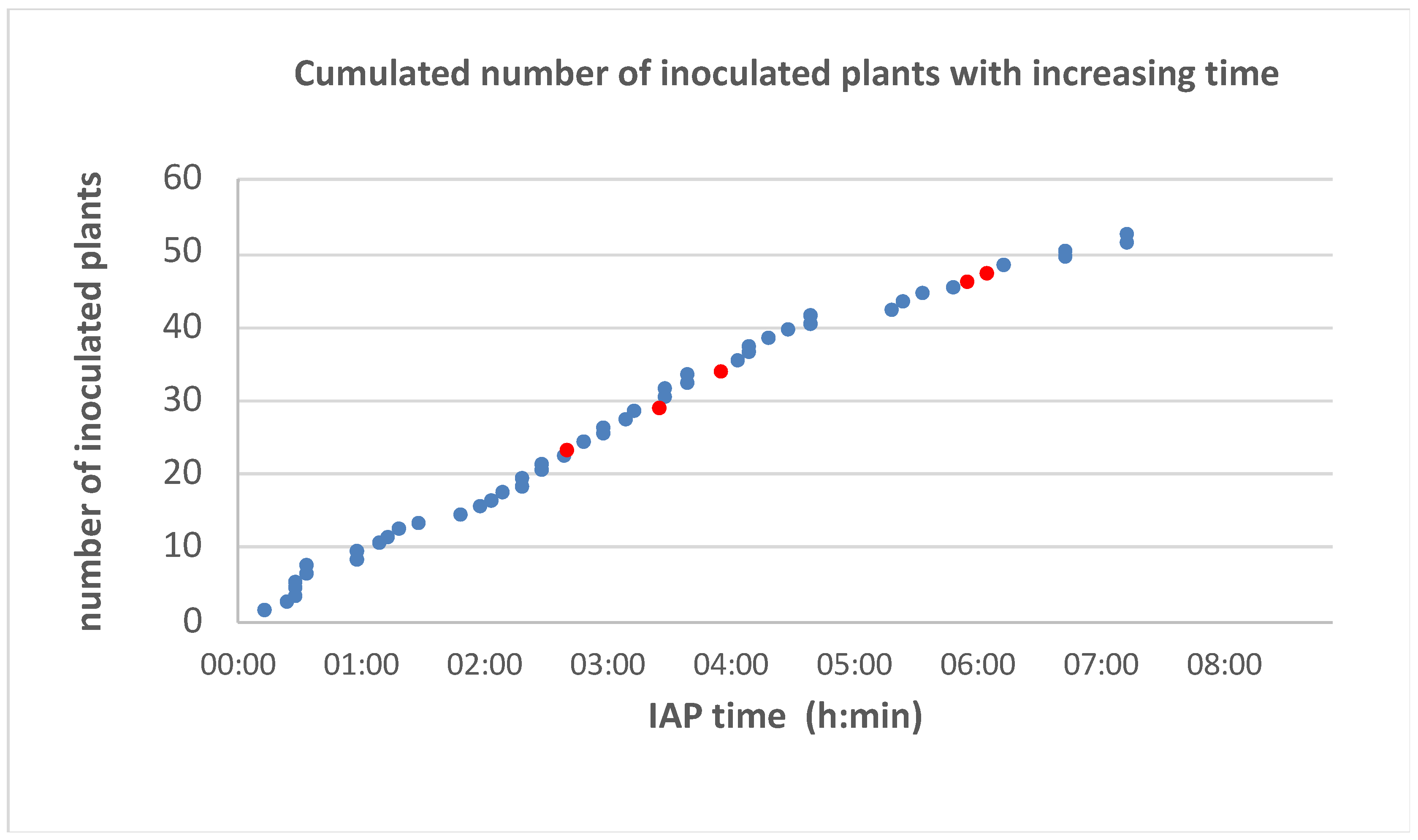
2.6. Study of Minimal Inoculation Access Period
2.7. Statistical Analysis
3. Results
3.1. Virus Detection in Natural Populations of P. corni
3.2. Virus Transmission Experiments by Natural Populations of P. corni
3.3. Time Threshold of Inoculation Access Period (IAP)
4. Discussion
4.1. Virus Transmission Experiments by Natural Populations of P. corni
4.2. Case of GLRaV 3
4.3. Case of GLRaV 2
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atallah, S.S.; Gomez, M.I.; Fuchs, M.F.; Martinson, T.E. Economic impact of grapevine leafroll disease on Vitis vinifera cv. Cabernet Franc in Finger Lakes vineyards of New York. Am. J. Enol. Vitic. 2012, 63, 73–79. [Google Scholar] [CrossRef]
- Abou Ghanem-Sabanadzovic, N.; Sabanadzovic, S.; Gugerli, P.; Rowhani, A. Genome organization; serology and phylogeny of Grapevine leafroll-associated viruses 4 and 6: Taxonomic implications. Virus Res. 2012, 163, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Dolja, V.V.; Daubert, S.; Koonin, E.V.; Rowhani, A. Genomic and biological analysis of Grapevine leafroll-associated virus 7 reveals a possible new genus within the family Closteroviridae. Virus Res. 2012, 163, 302–309. [Google Scholar] [CrossRef]
- Ito, T.; Nakaune, R. Molecular characterization of a novel putative ampelovirus tentatively named grapevine leafroll-associated virus 13. Arch. Virol. 2016, 161, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.P. An overview on grapevine viruses, viroids, and the diseases they cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B.Z., Martelli, G.P., Fuchs, M., Golino, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 31–46. [Google Scholar] [CrossRef]
- Herrbach, É.; Le Maguet, J.; Hommay, G. Virus transmission by mealybugs and soft scales (Hemiptera: Coccoidea). In Vector-Mediated Transmission of Plant Pathogens; Brown, J.K., Ed.; American Phytopathological Society Press: St. Paul, MN, USA, 2016; pp. 147–161. [Google Scholar] [CrossRef]
- Herrbach, É.; Alliaume, A.; Prator, C.A.; Daane, K.M.; Cooper, M.L.; Almeida, R.P.P. Vector transmission of grapevine-leafroll associated viruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B.Z., Martelli, G.P., Fuchs, M., Golino, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 483–503. [Google Scholar] [CrossRef]
- Prator, C.A.; Kashiwagi, C.M.; Voncina, D.; Almeida, R.P.P. Infection and colonization of Nicotiana benthamiana by Grapevine leafroll-associated virus 3. Virology 2017, 510, 60–66. [Google Scholar] [CrossRef]
- Engelbrecht, D.J.; Kasdorf, G.G.F. Association of a closterovirus with grapevines indexing positive for grapevine leafroll disease and evidence for its natural spread in grapevine. Phytopathol. Mediterr. 1985, 24, 101–105. [Google Scholar]
- La Notte, P.; Buzkan, N.; Choueiri, E.; Minafra, A.; Martelli, G.P. Acquisition and transmission of grapevine virus A by the mealybug Pseudococcus longispinus. J. Plant Pathol. 1997, 79, 79–85. [Google Scholar]
- Hommay, G.; Komar, V.; Lemaire, O.; Herrbach, É. Grapevine virus A transmission by larvae of Parthenolecanium corni. Eur. J. Plant Pathol. 2008, 121, 185–188. [Google Scholar] [CrossRef]
- Zorloni, A.; Prati, S.; Chiesa, S.; Bianco, P.A. Transmission of Grapevine leafroll associated virus 3 by the soft scale Neopulvinaria innumerabilis. J. Plant Pathol. 2006, 88, S61. [Google Scholar]
- Nakaune, R.; Toda, S.; Mochizuki, M.; Nakano, M. Identification and characterization of a new vitivirus from grapevine. Arch. Virol. 2008, 153, 1827–1832. [Google Scholar] [CrossRef]
- Alliaume, A.; Reinbold, C.; Erhardt, M.; Beuve, M.; Hily, J.M.; Lemaire, O.; Herrbach, É. Virus preparations from the mixed-infected P70 Pinot Noir accession exhibit GLRaV-1/GVA ‘end-to-end’ particles. Arch. Virol. 2018, 163, 3149–3154. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, C.; Segura, A. Some characteristics of the transmission of grapevine leafroll-associated virus 3 by Planococcus citri Risso. Eur. J. Plant Pathol. 1997, 10, 373–378. [Google Scholar] [CrossRef]
- Tsai, C.W.; Chau, J.; Fernandez, L.; Bosco, D.; Daane, K.M.; Almeida, R.P.P. Transmission of Grapevine leafroll-associated virus 3 by the vine mealybug (Planococcus ficus). Phytopathology 2008, 98, 1093–1098. [Google Scholar] [CrossRef]
- Le Maguet, J.; Beuve, M.; Herrbach, É.; Lemaire, O. Transmission of five ampeloviruses and two vitiviruses to grapevine by Phenacoccus aceris (Signoret). Phytopathology 2012, 102, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Krüger, K.; Saccaggi, D.L.; van der Merwe, M.; Kasdorf, G.G.F. Transmission of Grapevine Leafroll-associated Virus 3 (GLRaV-3): Acquisition, inoculation and retention by the mealybugs Planococcus ficus and Pseudococcus longispinus (Hemiptera: Pseudococcidae). S. Afr. J. Enol. Vitic. 2015, 36, 223–230. [Google Scholar] [CrossRef][Green Version]
- Alliaume, A. Biologie de la Vection de l’Ampélovirus GLRaV-1 et du Vitivirus GVA par la Cochenille Phenacoccus aceris. Ph.D. Thesis, Strasbourg University, Strasbourg, France, 2016. [Google Scholar]
- Hommay, G.; Beuve, M.; Herrbach, É. Transmission of grapevine ampelo- and vitiviruses by the Bohemian mealybug Heliococcus bohemicus Šulc (Hemiptera: Pseudococcidae). Viruses 2022, 14, 1430. [Google Scholar] [CrossRef]
- Belli, G.; Fortusini, A.; Casati, P.; Belli, L.; Bianco, P.A.; Prati, S. Transmission of a grapevine leafroll associated closterovirus by the scale insect Pulvinaria vitis L. Riv. Patol. Veg. 1994, 4, 105–108. [Google Scholar]
- Fortusini, A.G.; Scattini, G.; Prati, S.; Cinquanta, S.; Belli, G. Transmission of grapevine leafroll virus 1 (GLRaV-1) and grapevine virus A (GVA) by scale insects. In Proceedings of the 12th Meeting of the ICVG, Lisbon, Portugal, 29 September–2 October 1997; pp. 121–122. [Google Scholar]
- Sforza, R.; Boudon-Padieu, E.; Greif, C. New mealybug species vectoring Grapevine leafroll associated viruses-1 and -3 (GLRaV-1 and -3). Eur. J. Plant Pathol. 2003, 109, 975–981. [Google Scholar] [CrossRef]
- Hommay, G.; Le Maguet, J.; Komar, V.; Lemaire, O.; Herrbach, É. Transmission of Grapevine leaf roll-associated virus-1 and -3 (Ampelovirus) and Grapevine virus A (Vitivirus) by natural populations of soft scales and mealybugs in the north-eastern French vineyard. In Proceedings of the 16th Meeting of the ICVG, Dijon, France, 31 August–4 September 2009; pp. 286–287. [Google Scholar]
- Hommay, G.; Alliaume, A.; Reinbold, C.; Herrbach, É. Transmission of Grapevine leafroll-associated virus-1 (Ampelovirus) and Grapevine virus A (Vitivirus) by the Cottony Grape Scale, Pulvinaria vitis (Hemiptera: Coccidae). Viruses 2021, 13, 2081. [Google Scholar] [CrossRef]
- Mahfoudhi, N.; Digiaro, M.; Dhouibi, M.H. Transmission of grapevine leafroll viruses by Planococcus ficus (Hemiptera: Pseudococcidae) and Ceroplastes rusci (Hemiptera: Coccidae). Plant Dis. 2009, 93, 999–1002. [Google Scholar] [CrossRef]
- Le Maguet, J. Epidémiologie de l’Enroulement Viral de la Vigne dans les Vignobles Français Septentrionaux et Transmission par Cochenilles Vectrices. Ph.D. Thesis, Strasbourg University, Strasbourg, France, 2012; 204p. [Google Scholar]
- Bahder, B.W.; Poojari, S.; Alabi, O.J.; Naidu, R.A.; Walsh, D.B. Pseudococcus maritimus (Hemiptera: Pseudococcidae) and Parthenolecanium corni (Hemiptera: Coccidae) are capable of transmitting Grapevine Leafroll-Associated Virus 3 between Vitis x labruscana and Vitis vinifera. Environ. Entomol. 2013, 42, 1292–1298. [Google Scholar] [CrossRef]
- Krüger, K.; Douglas-Smit, N. Grapevine leafroll-associated virus 3 (GLRaV-3) transmission of by three soft scale insect species (Hemiptera: Coccidae) with notes on biology. Afr. Entomol. 2013, 21, 1–8. [Google Scholar] [CrossRef]
- Zhou, J.S.; Drucker, M.; Ng, J.C.K. Direct and indirect influences of virus–insect vector–plant interactions on non-circulative, semi-persistent virus transmission. Curr. Opin. Virol. 2018, 33, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Canard, M. Recherches sur la morphologie et la biologie de la cochenille Eulecanium corni Bouché (Homoptères-Coccoidea). Ann. Ecol. Nat. Sup. Agron. Toulouse 1958, 6, 185–271. [Google Scholar]
- Kosztarab, M.; Kozár, F. Scale Insects of Central Europe; Series Entomologica; Dr W. Junk Publishers: Dordrecht, The Netherlands, 1988; 456p. [Google Scholar]
- Schmutterer, H. Die Ökologie der Cocciden (Homoptera, Coccoïdea) Frankens. Z. Angew. Entomol. 1952, 33, 544–584. [Google Scholar] [CrossRef]
- Hommay, G.; Wiss, L.; Chadoeuf, J.; Le Maguet, J.; Beuve, M.; Herrbach, É. Gone with the wind: Aerial dispersal of Parthenolecanium corni crawlers in a newly planted grapevine plot. Ann. Appl. Biol. 2019, 174, 372–387. [Google Scholar] [CrossRef]
- Hommay, G.; Wiss, L.; Reinbold, C.; Chadoeuf, J.; Herrbach, É. Spatial distribution patterns of Parthenolecanium corni (Hemiptera, Coccidae), and of the ampelovirus GLRaV-1 and the vitivirus GVA in a commercial vineyard. Viruses 2020, 12, 1447. [Google Scholar] [CrossRef]
- Beuve, M.; Moury, B.; Spilmont, A.S.; Sempé-Ignatovic, L.; Hemmer, C.; Lemaire, O. Viral sanitary status of declining grapevine Syrah clones and genetic diversity of Grapevine Rupestris stem pitting-associated virus. Eur. J. Plant Pathol. 2013, 135, 439–452. [Google Scholar] [CrossRef]
- Greif, C.; Walter, B. The European reference collection of grapevine virus diseases. In Sanitary Selection of the Grapevine; Walter, B., Ed.; Editions INRA: Paris, France, 1997; pp. 171–181. [Google Scholar]
- Pinot Noir. Available online: https://plantgrape.plantnet-project.org/fr/cepage/Pinot%20noir#115 (accessed on 7 September 2022).
- Zimmermann, D.; Bass, P.; Legin, R.; Walter, B. Characterization and serological detection of four closterovirus-like particles associated with leafroll disease on grapevine. J. Phytopathol. 1990, 130, 205–218. [Google Scholar] [CrossRef]
- Bertin, S.; Cavalieri, V.; Gribaudo, I.; Sacco, D.; Marzachi, C.; Bosco, D. Transmission of Grapevine virus A and Grapevine leafroll-associated virus 1 and 3 by Heliococcus bohemicus (Hemiptera: Pseudococcidae) nymphs from plants with mixed infections. J. Econ. Entomol. 2016, 109, 1504–1511. [Google Scholar] [CrossRef]
- Bertin, S.; Pacifico, D.; Cavalieri, V.; Marzachi, C.; Bosco, D. Transmission of Grapevine virus A and Grapevine leafroll-associated viruses 1 and 3 by Planococcus ficus and Planococcus citri fed on mixed-infected plants. Ann. Appl. Biol. 2016, 169, 53–63. [Google Scholar] [CrossRef]
- Almeida, R.P.P.; Daane, K.M.; Bell, V.A.; Blaisdell, G.K.; Cooper, M.L.; Herrbach, É.; Pietersen, G. Ecology and management of grapevine leafroll disease. Front. Microbiol. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.Y.; Ling, K.S.; Goszczynski, D.E.; McFerson, J.R.; Gonsalves, D. Nucleotide sequence and genome organization of grapevine leafroll-associated virus-2 are similar to beet yellows virus, the closterovirus type member. J. Gen. Virol. 1998, 79, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M. Closteroviruses (Closteroviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; Volume 3, pp. 336–347. [Google Scholar] [CrossRef]
- Wistrom, C.M.; Blaisdell, G.K.; Wunderlich, L.R.; Botton, M.; Almeida, R.P.P.; Daane, K.M. No evidence of transmission of grapevine leafroll-associated viruses by phylloxera (Daktulosphaira vitifoliae). Eur. J. Plant Pathol. 2017, 147, 937–941. [Google Scholar] [CrossRef]
- Klaassen, V.A.; Sim, S.T.; Dangl, G.S.; Osman, F.; Al Rwahnih, M.; Rowhani, A.; Golino, D.A. Vitis californica and Vitis californica × Vitis vinifera hybrids are hosts for Grapevine leafroll-associated virus-2 and -3 and Grapevine virus A and B. Plant Dis. 2011, 95, 657–665. [Google Scholar] [CrossRef]
| Locality | Latitude | Longitude | Cultivar | Number | Negative | Positive for | |||||||
| of Vines Tested | GVA | GLRaV 1 | GLRaV 3 | GLRaV 1, GVA | GLRaV 1, 3 | GLRaV 3, GVA | GLRaV 1, 3; GVA | GLRaV 2 with 1 or 3 or GVA | |||||
| Bennwihr | 48°08′17.7″ N | 7°19′06.6″ E | Pinot noir | 142 | 48 (2) | 4 | 61 (12) | 21 (1) | 6 (1) | 2 | |||
| Kientzheim | 48°08′24.8″ N | 7°16′20.6″ E | Riesling | 24 | 2 | 22 (7) | |||||||
| Nothalten | 48°21′31.7″ N | 7°24′39.7″ E | Riesling | 704 | 279 (28) | 1 | 208 (36) | 24 (12) | 99 (17) | 45 (11) | 5 (5) | 33 (10) | 10 (6) |
| Ribeauvillé | 48°11′55.6″ N | 7°20′11.4″ E | Riesling | 21 | 17 | 2 (1) | 1 (1) | 1 | |||||
| Turckheim | 48°05′41.8″ N | 7°16′34.2″ E | Sylvaner | 14 | 2 (1) | 2 (2) | 9 (5) | 1 | |||||
| Ihringen | 48°03′18.7″ N | 7°37′30.0″ E | Kerner | 16 | 4 (2) | 1 | 11 (9) | ||||||
| Virus | Target ORF | Primer | Nucleotide Sequence 5′-3′ | Hybridisation Temperature | Amplicon Length (pb) |
|---|---|---|---|---|---|
| GLRaV 1 | HSP70 | LR1-H70F1 | GTTGGTGAATTCTCCGTTCGT | 56 °C | 402 |
| LR1-H70R1 | ACTTCGCTTGAACGAGTTATAC | ||||
| GLRaV 3 | Polymerase | LR3-POLF1 | ACGTAACGGGGCAGAATATAGT | 56 °C | 282 |
| LR3-POLR1 | TATCAACACCAAGTGTCAAGAGTA | ||||
| GVA | Coat protein | GVA-CPF1 | GGCTACGACCGAAATATGTAC | 56 °C | 524 |
| GVA-CPR1 | AGAAACGATGGGTCATCCATC |
| Viruses in Source Plant | |||||||||
| Soft scales no. and stage | Viruses detected | GLRaV 1 | GLRaV 3 | GLRaV 1, 3 | GLRaV 1, 2, 3 | GLRaV 1, 2, GVA | GLRaV 1, GVA | GLRaV 1, 3, GVA | Total |
| >100 eggs | GLRaV 1 | 0/10 | 0/10 | ||||||
| (n = 10) | GVA | 0/10 | 0/10 | ||||||
| GLRaV 1 | 5/5 | 2/2 | 22/22 | 6/8 | 35/37 | ||||
| ≥25 L1 | GLRaV 3 | 2/2 | 3/8 | 5/10 | |||||
| (47 ± 16, n = 37) | GVA | 14/22 | 3/8 | 17/30 | |||||
| GLRaV 1 | 16/16 | 0/1 | 1/1 | 10/10 | 11/11 | 38/39 | |||
| 1–20 L2 | GLRaV 2 | 0/1 | 1/1 | 1/2 | |||||
| (8 ± 7, n = 43) | GLRaV 3 | 1/4 | 1/1 | 4/11 | 6/16 | ||||
| GVA | 1/1 | 7/10 | 2/11 | 10/22 | |||||
| 8–45 L2 | GLRaV 1 | 8/14 | 1/1 | 7/7 | 3/3 | 19/25 | |||
| overwintering | GLRaV 3 | 3/4 | 1/1 | 1/3 | 5/8 | ||||
| (24 ± 9, n = 29) | GVA | 2/7 | 0/3 | 2/10 | |||||
| 2-11 maturing | GLRaV 1 | 3/5 | 4/7 | 2/2 | 9/14 | ||||
| females | GLRaV 3 | 1/7 | 1/7 | ||||||
| (8 ± 4, n = 14) | GVA | 0/2 | 0/2 | ||||||
| 25–100 L1 | GLRaV 1 | 0/2 | 2/3 | 0/2 | 1/5 | 3/12 | |||
| under adult | GLRaV 3 | 0/3 | 0/5 | 0/8 | |||||
| shield (n = 12) | GVA | 0/2 | 1/5 | 1/7 | |||||
| L2 honeydew (n = 4) | GLRaV-1 | 4/4 | 4/4 | ||||||
| Viruses of source plants | |||||||||
| No. nymphsand stage | Virus transmission | GLRaV 1 | GLRaV 3 | GLRaV 2 (with 1, 3 or GVA) | GLRaV 1, 3 | GLRaV 1, GVA | GLRaV 3, GVA | GLRaV 1, 3, GVA | Total |
| GLRaV 1 | 4/22 | 2/4 | 2/20 | 12/26 | 7/22 | 27/94 | |||
| GLRaV 2 | 0/4 | 0/4 | |||||||
| 100 L1 | GLRaV 3 | 0/10 | 0/3 | 0/20 | 0/6 | 0/22 | 0/61 | ||
| GVA | 0/2 | 9/26 | 0/6 | 6/22 | 15/56 | ||||
| Test plants | 22 | 10 | 4 | 20 | 26 | 6 | 22 | 110 | |
| GLRaV 1 | 29/68 | 4/5 | 12/56 | 25/55 | 26/47 | 96/231 | |||
| GLRaV 2 | 0/5 | 0/5 | |||||||
| 50–100 L2 | GLRaV 3 | 0/42 | 0/3 | 0/56 | 0/15 | 0/47 | 0/163 | ||
| GVA | 2/3 | 21/55 | 1/15 | 19/47 | 43/120 | ||||
| Test plants | 68 | 42 | 5 | 56 | 55 | 15 | 47 | 288 | |
| 7–50 L2 * | GLRaV 1 | 0/10 | 0/10 | ||||||
| 50–100 L1–L2 | Healthy sources | 0/49 | |||||||
| 3–70 L2 * | Healthy sources | 0/4 | |||||||
| Viruses in Source plants | |||||||
| Locality | Cultivar | GLRaV 1 | GLRaV 3 | GLRaV 1, GVA | GLRaV 1, 3 | GLRaV 3, GVA | GLRaV 1, 3, GVA |
| Bennwihr | Pinot noir | 0/14 | 0/1 | 1/1 | |||
| Ihringen | Kerner | 0/12 | |||||
| Kientzheim | Riesling | 7/10 | |||||
| Turckheim | Sylvaner | 1/2 | 6/9 | ||||
| Nothalten | Riesling | 33/88 | 0/17 | 27/65 | 16/78 | 1/21 | 32/68 |
| Ribeauvillé | Riesling | 0/1 | 1/2 | ||||
| Total transmission rates | 34/90 | 0/44 | 41/86 | 16/79 | 1/21 | 33/69 | |
| GLRaV 1 % transmission | 38% | - | 48% | 20% | - | 48% | |
| Virus % transmission | 38% | 0% | 48% | 20% | 5% | 48% | |
| Recipient | Transmission rates | |
| cultivar | GLRaV 1 | GVA |
| Pinot noir P 114 | 35% (18/52) | 21% (11/52) |
| Pinot noir P 115 | 28% (64/229) | 12% (27/229) |
| Pinot noir | 44% (31/71) | 24% (17/71) |
| Pinot blanc | 32% (9/28) | 14% (4/28) |
| Muscat Ottonel | 5% (1/19) | 0% (0/19) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hommay, G.; Beuve, M.; Herrbach, E. Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae). Viruses 2022, 14, 2679. https://doi.org/10.3390/v14122679
Hommay G, Beuve M, Herrbach E. Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae). Viruses. 2022; 14(12):2679. https://doi.org/10.3390/v14122679
Chicago/Turabian StyleHommay, Gérard, Monique Beuve, and Etienne Herrbach. 2022. "Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae)" Viruses 14, no. 12: 2679. https://doi.org/10.3390/v14122679
APA StyleHommay, G., Beuve, M., & Herrbach, E. (2022). Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae). Viruses, 14(12), 2679. https://doi.org/10.3390/v14122679






