Canker Development and Biocontrol Potential of CHV-1 Infected English Isolates of Cryphonectria parasitica Is Dependent on the Virus Concentration and the Compatibility of the Fungal Inoculums
Abstract
1. Introduction
2. Material and Methods
2.1. Viral and Fungal Strains
2.2. Preservation of Virus-Infected Fungal Strains and Assessment of Their Viral Load
2.3. Inoculation of Sweet Chestnut Seedlings and Branch Segments, and Fungal Re-Isolation
2.4. Direct One-Step Reverse Transcription PCR and Comparison with Other Endpoint and Real-Time Virus Detection Methods
2.5. Statistical Analyses
3. Results
3.1. Transmissions
3.2. Preservations
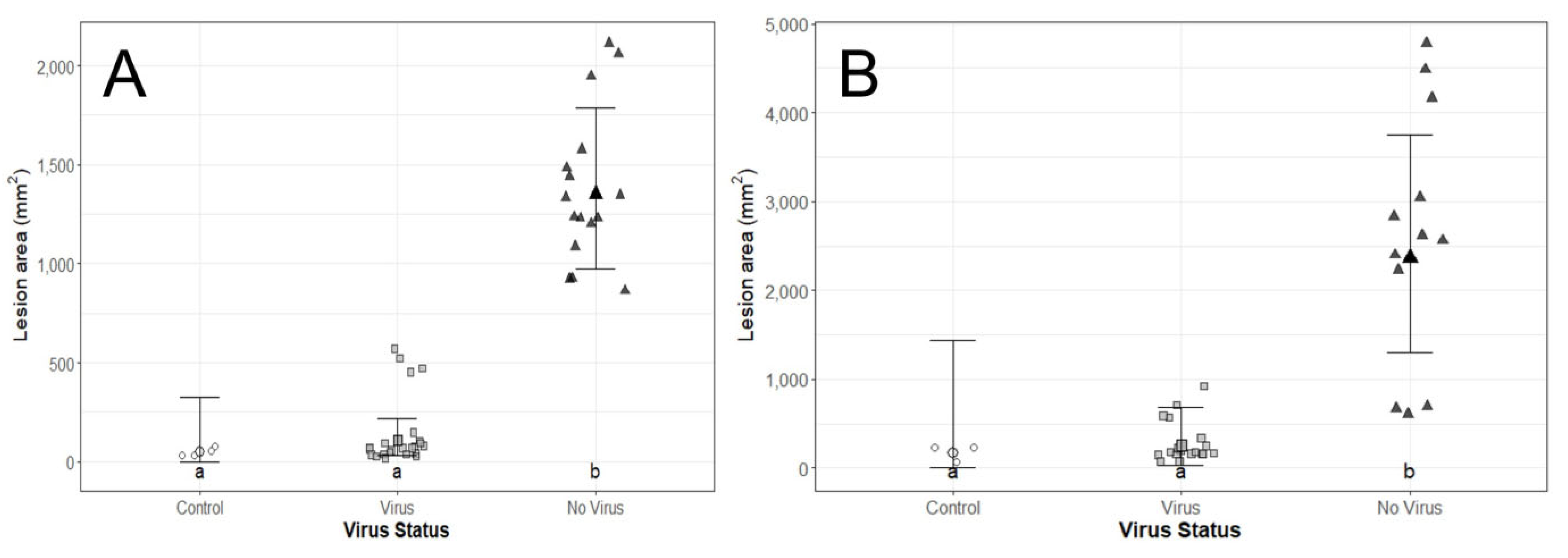
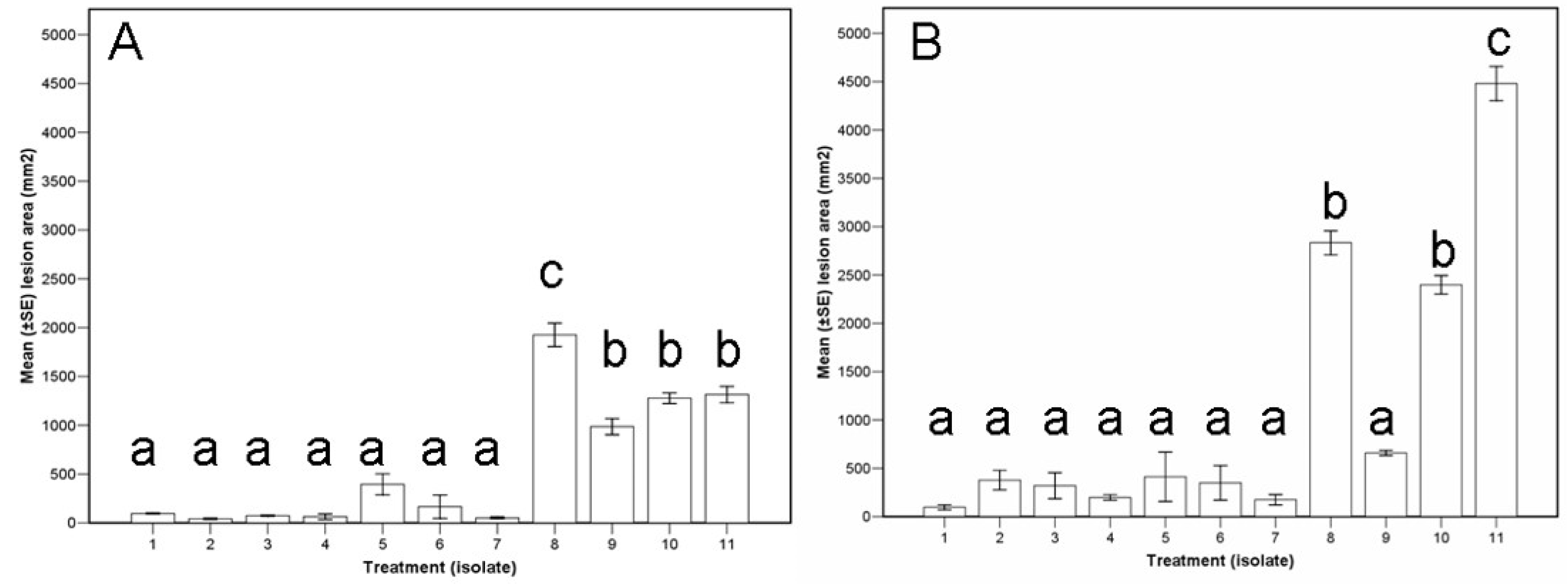
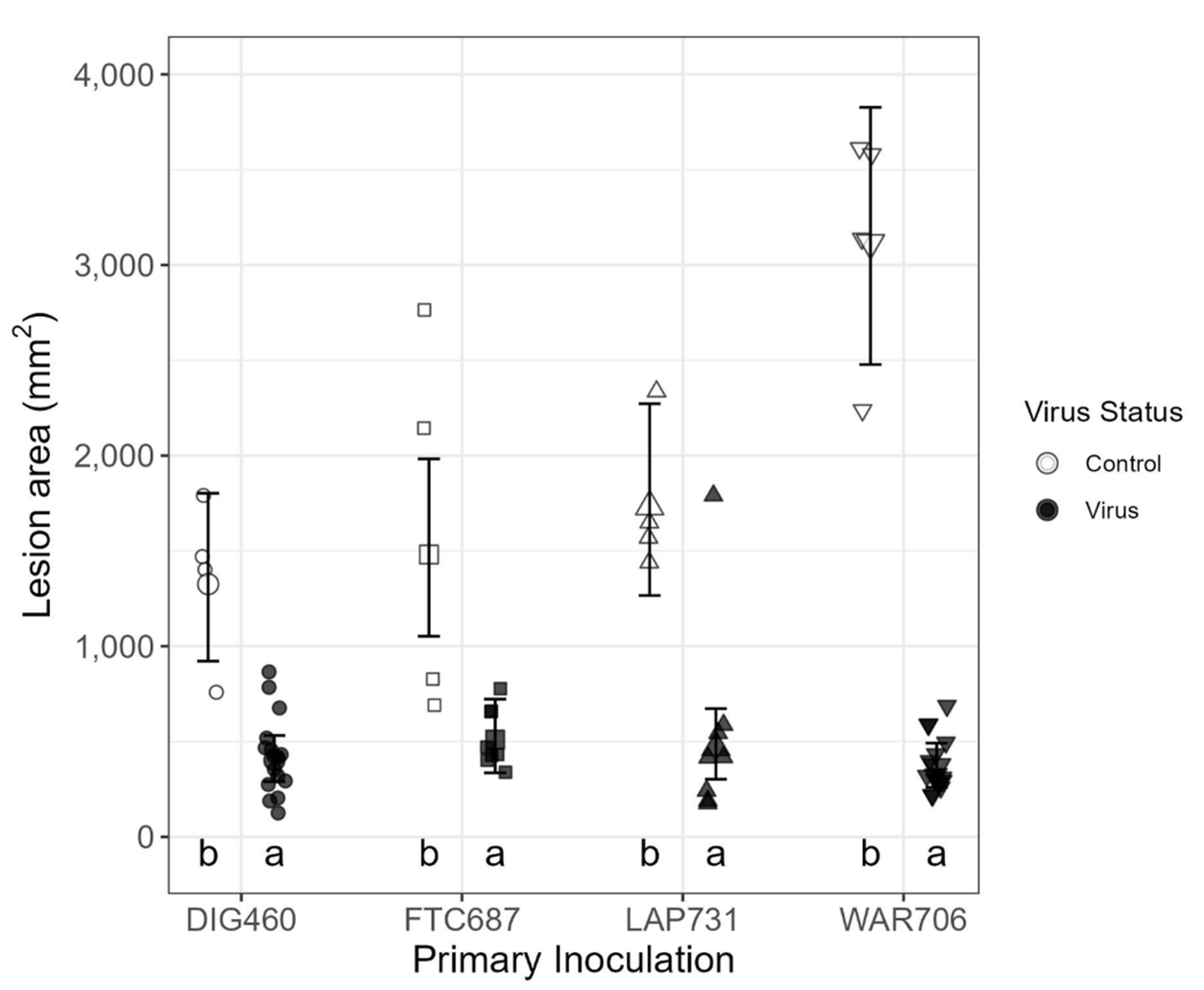
3.3. Assay I, Pathogenicity Test
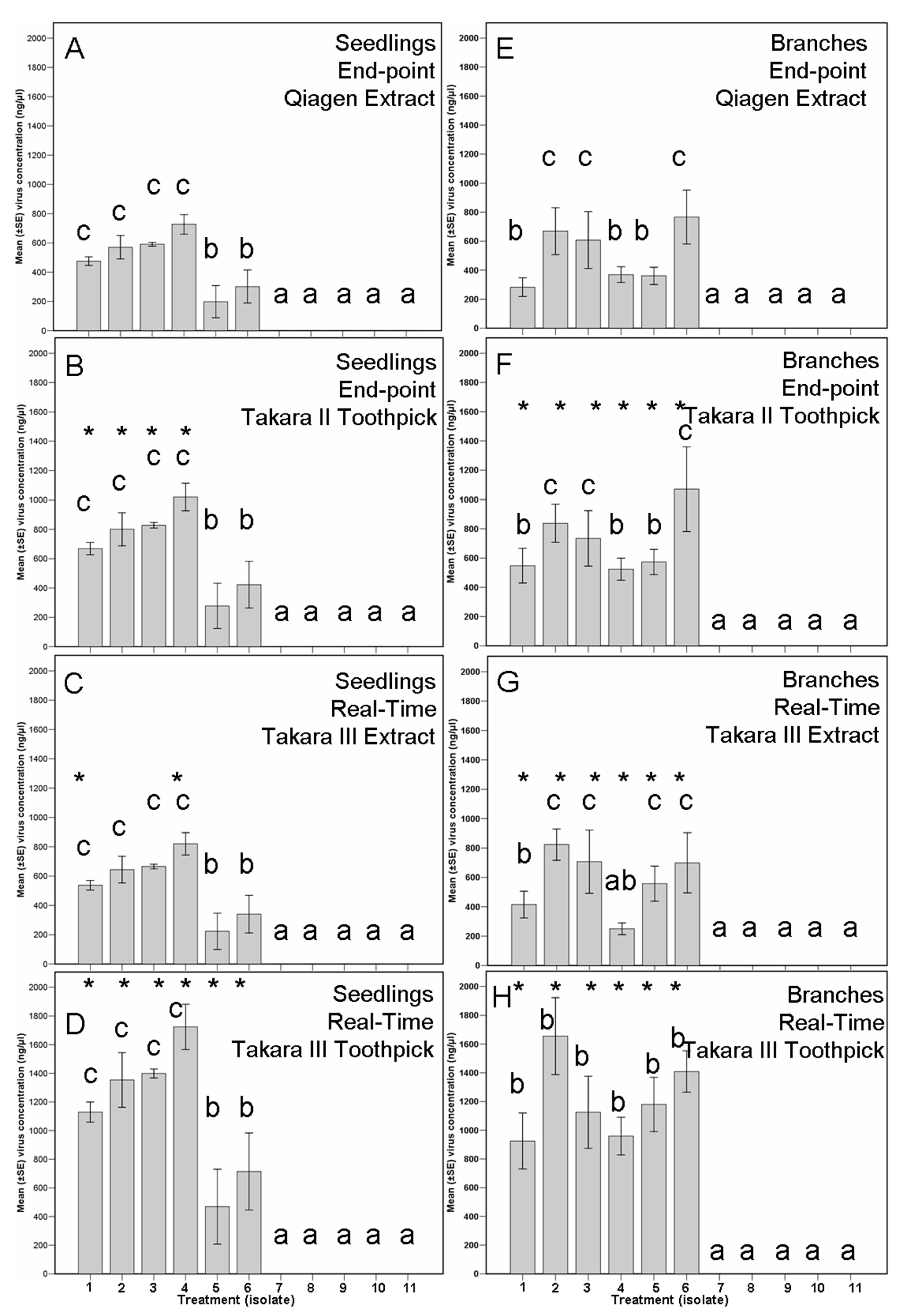
3.4. Assay II, Biocontrol Potential
3.5. New Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.-C.; Dynek, J.N.; Hillman, B.I.; Milgroom, M.G. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res. 2007, 111, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Robin, C.; Heiniger, U. Chestnut blight in Europe: Diversity of Cryphonectria parasitica, hypovirulence and biocontrol. For. Snow Land. Res. 2001, 76, 361–367. [Google Scholar]
- Hunter, G.; Wylder, B.; Jones, B.; Webber, J.F. First finding of Cryphonectria parasitica causing chestnut blight on Castanea sativa trees in England. New Dis. Rep. 2013, 27, 1. [Google Scholar] [CrossRef]
- Forestry Commission. Sweet Chestnut Blight (Cryphonectria parasitica). 2018. Available online: https://www.forestresearch.gov.uk/tools-and-resources/fthr/pest-and-disease-resources/sweet-chestnut-blight-cryphonectria-parasitica/ (accessed on 29 June 2022).
- Pérez-Sierra, A.; Romon-Ochoa, P.; Gorton, C.; Lewis, A.; Rees, H.; van der Linde, S.; Webber, J. High vegetative compatibility diversity of Cryphonectria parasitica infecting sweet chestnut (Castanea sativa) in Britain indicates multiple pathogen introductions. Plant Pathol. 2019, 68, 727–737. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Kranjec Orlovic, J.; Gorton, C.; Lewis, A.; van der Linde, S.; Pérez-Sierra, A. New detections of chestnut blight in Great Britain during 2019–2020 reveal high Cryphonectria parasitica diversity and limited spread of the disease. Plant Pathol. 2021, 71, 793–804. [Google Scholar] [CrossRef]
- Hillman, B.I.; Suzuki, N. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 2004, 63, 423–472. [Google Scholar]
- Fahima, T.; Kazmierczak, P.; Hansen, D.R.; Pfeiffer, P.; van Alfen, N.K. Membrane-associated replication of an un-encapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 1993, 195, 81–89. [Google Scholar] [CrossRef]
- Mlinarec, J.; Jezic, M.; Cosic, J.; Curkovic-Perica, M. Multilocus PCR assay reveals high diversity of vegetative compatibility types in populations of Cryphonectria parasitica in Croatia. Plant Pathol. 2018, 67, 741–749. [Google Scholar] [CrossRef]
- Cortesi, P.; McCulloch, C.E.; Song, H.Y.; Lin, H.Q.; Milgroom, M.G. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 2001, 159, 107–118. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Romon-Ochoa, P.; Gorton, C.; Lewis, A.; van der Linde, S.; Webber, J.; Pérez-Sierra, A. Hypovirulent effect of the Cryphonectria Hypovirus 1 in British isolates of Cryphonectria parasitica. Pest Manag. Sci. 2020, 76, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Gobbin, D.; Hoegger, P.J.; Heiniger, U.; Rigling, D. Sequence variation and evolution of Cryphonectria hypovirus 1 (CHV-1) in Europe. Virus Res. 2003, 97, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Jamal, A.; Kondo, H.; Suzuki, N. SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. USA 2017, 114, 3499–3506. [Google Scholar] [CrossRef]
- Suzuki, N.; Cornejo, C.; Aulia, A.; Shahi, S.; Hillman, B.I.; Rigling, D. In-tree behavior of diverse viruses harbored in the chestnut blight fungus, Cryphonectria parasitica. J. Virol. 2021, 95, e01962-20. [Google Scholar] [CrossRef] [PubMed]
- Urayama, S.; Katoh, Y.; Fukuhara, T.; Arie, T.; Moriyama, H.; Teraoka, T. Rapid detection of Magnaporthe oryzae chrysovirus 1-A from fungal colonies on agar plates and lesions of rice blast. J. Gen. Plant Pathol. 2015, 81, 97–102. [Google Scholar] [CrossRef]
- Aulia, A.; Andika, I.B.; Kondo, H.; Hillman, B.I.; Suzuki, N. A symptomless hypovirus, CHV4, facilitates stable infection of the chestnut blight fungus by a coinfecting reovirus likely through suppression of antiviral RNA silencing. Virology 2019, 533, 99–107. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 25 July 2022).
- Length, R.V. Emmeans: Estimated Marginal Means, aka Least-Squares Means. R Package Version 1.7.2. 2022. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 25 July 2022).
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using ime4. J. Statis. Soft. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Statis. Soft. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 2nd ed.; Sage: Thousand Oaks, CA, USA, 2011; Available online: http://socserv.socsci.mcmaster.ca/jfox/Books/Companion (accessed on 25 July 2022).
- Peever, T.L.; Liu, Y.-C.; Wang, K.; Hillman, B.I.; Foglia, R.; Milgroom, M.G. Incidence and diversity of double-stranded RNAs occurring in the chestnut blight fungus, Cryphonectria parasitica, in China and Japan. Phytopathology 1998, 88, 811–817. [Google Scholar] [CrossRef]
- Park, S.-M.; Kim, J.-M.; Chung, H.-J.; Lim, J.-Y.; Kwon, B.-R.; Lim, J.-G.; Kim, J.-A.; Kim, M.-J.; Cha, B.-J.; Lee, S.-H.; et al. Occurrence of diverse dsRNA in a Korean population of the chestnut blight fungus Cryphonectria parasitica. Phytopathology 2008, 112, 1220–1226. [Google Scholar]
- Rigling, D.; Borst, N.; Cornejo, C.; Supatashvili, A.; Prospero, S. Genetic and phenotypic characterization of Cryphonectria hypovirus 1 from Eurasian Georgia. Viruses 2018, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Allemann, C.; Hoegger, P.; Heiniger, U.; Rigling, D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 1999, 8, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Bryner, S.F.; Rigling, D. Hypovirus virulence and vegetative incompatibility in populations of the chestnut blight fungus. Phytopathology 2012, 102, 1161–1167. [Google Scholar] [CrossRef]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Krajacic, M.; Curkovic-Perica, M. Chestnut blight fungus in Croatia: Diversity of vegetative compatibility types and genetic variability of associated Cryphonectria hypovirus 1. Plant Pathol. 2008, 57, 1086–1096. [Google Scholar] [CrossRef]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Curkovic-Perica, M. Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol. 2011, 60, 752–761. [Google Scholar] [CrossRef]
- Sotiroski, K.; Milgroom, M.G.; Rigling, D.; Heiniger, U. Occurrence of Cryphonectria hypovirus 1 in the chestnut blight fungus in Macedonia. For. Pathol. 2006, 36, 136–143. [Google Scholar] [CrossRef]
- Akilli, S.; Serce, C.U.; Katircioglu, Y.Z.; Maden, S.; Rigling, D. Characterization of hypovirulent isolates of the chestnut blight fungus, Cryphonectria parasitica from the Marmara and Black Sea regions of Turkey. Eur. J. Plant Pathol. 2013, 135, 323–334. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Heiniger, U.; Holdenrieder, O.; Rigling, D. Differential transfer and dissemination of hypovirus and nuclear and mitochondrial genomes of a hypovirus-infected Cryphonectria parasitica strain after introduction into a natural population. Appl. Environ. Microb. 2003, 69, 3767–3771. [Google Scholar] [CrossRef]
- Feau, N.; Dutech, C.; Brusini, J.; Rigling, D.; Robin, C. Multiple introductions and recombination in Cryphonectria hypovirus 1: Perspective of a sustainable biological control of chestnut blight. Evol. Appl. 2014, 9, 580–596. [Google Scholar] [CrossRef]
- Zamora, P.; Martin, A.B.; Rigling, D.; Diez, J.J. Diversity of Cryphonectria parasitica in western Spain and identification of hypovirus-infected isolates. For. Pathol. 2012, 42, 412–419. [Google Scholar] [CrossRef]
- Peters, F.S.; Busskamp, J.; Prospero, S.; Rigling, D.; Metzler, B. Genetic diversification of the chestnut blight fungus Cryphonectria parasitica and its associated hypovirus in Germany. Fungal Biol. 2014, 118, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Hogan, E.P.; Griffin, G.J. Spread of Cryphonectria hypovirus 1 into 45 vegetative compatibility types of Cryphonectria parasitica on grafted American chestnut trees. For. Pathol. 2002, 32, 73–85. [Google Scholar] [CrossRef]
- Double, M.L.; Nuss, D.L.; Rittenour, W.R.; Holásková, I.; Short, D.P.G.; Kasson, M.T.; MacDonald, W.L. Long-term field study of transgenic hypovirulent strains of Cryphonectria parasitica in a forest setting. For. Pathol. 2017, 47, e12367. [Google Scholar] [CrossRef]
- Double, M.L.; Jarosz, A.M.; Fulbright, D.W.; Davelos-Baines, A.; MacDonald, W.L. Evaluation of two decades of Cryphonectria parasitica hypovirus introduction in an American chestnut stand in Wisconsin. Phytopathology 2018, 108, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Stauder, C.M.; Nuss, D.L.; Zhang, D.-X.; Double, M.L.; MacDonald, W.L.; Metheny, A.M.; Kasson, M.T. Enhanced hypovirus transmission by engineered super donor strains of the chestnut blight fungus, Cryphonectria parasitica, into a natural population of strains exhibiting diverse vegetative compatibility genotypes. Virology 2019, 528, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bryner, S.F.; Rigling, D.; Brunner, P.C. Invasion history and demographic pattern of Cryphonectria hypovirus 1 across European populations of the chestnut blight fungus. Ecol. Evol. 2012, 2, 3227–3241. [Google Scholar] [CrossRef]
- Zamora, P.; Martin, A.B.; San Martin, R.; Martinez-Alvarez, P.; Diez, J.J. Control of chestnut blight by the use of hypovirulent strains of the fungus Cryphonectria parasitica in northwestern Spain. Biol. Conserv. 2014, 79, 58–66. [Google Scholar] [CrossRef]
- Krstin, L.; Katanić, Z.; Ježić, M.; Poljak, I.; Nuskern, L.; Matković, I.; Idžojtić, M.; Ćurković-Perica, M. Biological control of chestnut blight in Croatia: An interaction between host sweet chestnut, its pathogen Cryphonectria parasitica and the biocontrol agent Cryphonectria hypovirus 1. Pest Manag. Sci. 2016, 73, 582–589. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Milgroom, M.G. Correlation between hypovirus transmission and the number of vegetative incompatibility (Vic) genes different among isolates from a natural population of Cryphonectria parasitica. Phytopathology 1996, 86, 79–86. [Google Scholar] [CrossRef]
- Brusini, J.; Robin, C. Mycovirus transmission revisited by in situ pairings of vegetatively incompatible isolates of Cryphonectria parasitica. J. Virol. Methods 2013, 187, 435–442. [Google Scholar] [CrossRef] [PubMed]

| Treatment Number | Fungal Strain | Description | VCG | Mating Type | Virus Strain |
|---|---|---|---|---|---|
| 1 | FTC687 | Virus-infected strain, transmitted from SDA540 M2273 | EU10 (2122-11) | MAT-2 | E-5 |
| 2 | WAR706 | Virus-infected strain, transmitted from WAP125 M2273 | EU9 (2111-11) | MAT-2 | E-5 |
| 3 | POWP709 | Virus-infected strain, transmitted from WAP125 M2273 | EU9 (2111-11) | MAT-2 | E-5 |
| 4 | FTC687 | Virus-infected strain, transmitted from SDA540 M2357 | EU10 (2122-11) | MAT-2 | L-18 |
| 5 | WAR706 | Virus-infected strain, transmitted from WAP125 M2357 | EU9 (2111-11) | MAT-2 | L-18 |
| 6 | POWP709 | Virus-infected strain, transmitted from WAP125 M2357 | EU9 (2111-11) | MAT-2 | L-18 |
| 7 | PDA CONTROL | Not Applicable (N/A) | N/A | N/A | N/A |
| 8 | LAP731 | Standard virus-free strain | EU10 (2122-11) | MAT-2 | N/A |
| 9 | FTC687 VIRUS-FREE | Standard virus-free strain | EU10 (2122-11) | MAT-2 | N/A |
| 10 | WAR706 VIRUS-FREE | Standard virus-free strain | EU9 (2111-11) | MAT-2 | N/A |
| 11 | DIG460 | Standard virus-free strain | EU9 (2111-11) | MAT-2 | N/A |
| PROBE SPECIFIC FOR CHV-1 | Tm * °C | GC % | ΔG Kcal/mol |
|---|---|---|---|
| CHV1-F: 5′-TGAGGAACGTCAACTTCG-3′ | 53.8 | 50.0 | 23.2 |
| CHV1-R: 5′-TTGTGACGACGGAAATAATC-3′ | 54.3 | 40.0 | 24.10 |
| HVEP1 Fluo: 5′-56-FAM/TGACACGGAAGCTGAGTGTC/3BHQ1/-3′ | 60.5 | 55.0 | 26.70 |
| PROBE FOR INTERNAL CONTROL TARGETING ACTIN mRNA & DNA | |||
| CpActinCF1: 5′-CCATGGTATCATGATTGGTATG-3′ | 58.4 | 41 | 25.0 |
| CpActinCR1: 5′-TACCGCAGAGTCAGGATA-3′ | 53.8 | 50 | 22.4 |
| CpActinCP1: 5′-56-JOE/TCATCACCAACATACGAGTCCTTCTG/3BHQ1/-3′ | 66.2 | 46 | 33.6 |
| Treatment Number | Strain | Before Preservation | After Glycerol Preservation | After Disks Preservation |
|---|---|---|---|---|
| 1 | FTC687 | 234.37 | 293.60 (±6.30) a | 240.30 (±140.96) a |
| 2 | WAR706 | 407.09 | 371.84 (±47.94) a | 240.98 (±295.33) a |
| 3 | POWP709 | 371.42 | 364.70 (±44.69) a | 230.96 (±314.99) a |
| 4 | FTC687 | 612.83 | 382.92 (±47.33) a | 236.83 (±199.30) a |
| 5 | WAR706 | 458.04 | 425.86 (±99.05) a | 137.64 (±185.07) a |
| 6 | POWP709 | 343.02 | 412,37 (±75.27) a | 144.00 (±195.52) a |
| Mean total | 404.46 | 375.21 (±62.98) a | 199.12 (±175.95) a |
| ASSAY II USING SEEDLINGS | End-Point PCRs | Real-Time PCRs | Virus Copy Number | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor Number | Virus Strain (0 None, 1 E-5, 2 L-18) | Lesion Area (mm2) | Original Virus Concentration (ng/µL) | VCG Compatibility (0 None, 1 Yes) | Qiagen Extract (ng/µL) | Takara Dye Toothpick (ng/µL) | Takara III Extract (ng/µL) | Takara III Toothpick (ng/µL) | |||
| Donor number | Pearson Correlation | 1 | |||||||||
| Sig. (2-tailed) | |||||||||||
| Virus strain (0 None, 1 E-5, 2 L-18) | Pearson Correlation | −0.255 | 1 | ||||||||
| Sig. (2-tailed) | 0.042 | ||||||||||
| Lesion area (mm2) | Pearson Correlation | 0.579 | −0.635 | 1 | |||||||
| Sig. (2-tailed) | 5.277E-7 | 1.785E-8 | |||||||||
| Original virus concentration (ng/µL) | Pearson Correlation | −0.768 | 0.701 | −0.779 | 1 | ||||||
| Sig. (2-tailed) | 1.238E-13 | 1.081E-10 | 3.427E-14 | ||||||||
| VCG compatibility (0 None, 1 Yes) | Pearson Correlation | −0.716 | 0.832 | −0.793 | 0.963 | 1 | |||||
| Sig. (2-tailed) | 3.010E-11 | 1.631E-17 | 5.926E-15 | 6.315E-37 | |||||||
| Qiagen Extract (ng/µL) | Pearson Correlation | −0.633 | 0.690 | −0.766 | 0.829 | 0.852 | 1 | ||||
| Sig. (2-tailed) | 1.936E-8 | 2.796E-10 | 1.706E-13 | 2.564E-17 | 4.482E-19 | ||||||
| Takara Dye Toothpick (ng/µL) | Pearson Correlation | −0.634 | 0.690 | −0.766 | 0.829 | 0.852 | 1.000 | 1 | |||
| Sig. (2-tailed) | 1.925E-8 | 2.817E-10 | 1.700E-13 | 2.535E-17 | 4.447E-19 | 1.64E-159 | |||||
| Takara III Extract (ng/µL) | Pearson Correlation | −0.635 | 0.689 | −0.765 | 0.830 | 0.852 | 1.000 | 1.000 | 1 | ||
| Sig. (2-tailed) | 1.721E-8 | 3.192E-10 | 1.773E-13 | 2.469E-17 | 4.779E-19 | 4.645E-129 | 3.422E-127 | ||||
| Takara III Toothpick (ng/µL) | Pearson Correlation | −0.633 | 0.690 | −0.766 | 0.829 | 0.852 | 1.000 | 1.000 | 1.000 | 1 | |
| Sig. (2-tailed) | 1.957E-8 | 2.800E-10 | 1.707E-13 | 2.589E-17 | 4.488E-19 | 3.336E-154 | 1.198E-167 | 8.542E-125 | |||
| Virus copy number | Pearson Correlation | −0.271 | 0.575 | −0.543 | 0.535 | 0.583 | 0.775 | 0.774 | 0.773 | 0.775 | 1 |
| Sig. (2-tailed) | 0.030 | 6.540E-7 | 3.644E-6 | 5.281E-6 | 4.220E-7 | 5.990E-14 | 6.310E-14 | 7.26E-14 | 6.015E-14 | ||
| Negative correlation is significant at the 0.05 level (2-tailed). | |||||||||||
| Positive correlation is significant at the 0.05 level (2-tailed). | |||||||||||
| N | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | |
| ASSAY II USINGBRANCHES | End-Point PCRs | Real-Time PCRs | Virus Copy Number | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor Number | Virus Strain (0 None, 1 E-5, 2 L-18) | Lesion Area (mm2) | Original Virus Concentration (ng/µL) | VCG Compatibility (0 None, 1 Yes, 2 No) | Qiagen Extract (ng/µL) | Takara Dye Toothpick (ng/µL) | Takara III Extract (ng/µL) | Takara III Toothpick (ng/µL) | |||
| Donor number | Pearson Correlation | 1 | |||||||||
| Sig. (2-tailed) | 1 | ||||||||||
| Virus strain (0 None, 1 E-5, 2 L-18) | Pearson Correlation | 6.74337E-18 | 1 | ||||||||
| Sig. (2-tailed) | 1 | ||||||||||
| Lesion area (mm2) | Pearson Correlation | 0.203 | −0.032 | 1 | |||||||
| Sig. (2-tailed) | 1.562E-2 | 0.706 | |||||||||
| Original virus concentration (ng/µL) | Pearson Correlation | −0.238 | 0.897 | −0.096 | 1 | ||||||
| Sig. (2-tailed) | 5E-3 | 1E-4 | 0.257 | ||||||||
| VCG compatibility (0 None, 1 Yes, 2 No) | Pearson Correlation | −0.459 | 0.562 | 0.156 | 0.675 | 1 | |||||
| Sig. (2-tailed) | 1.14525E-08 | 4.73908E-13 | 6.539E-2 | 1E-4 | |||||||
| Qiagen Extract (ng/µL) | Pearson Correlation | −0.199 | −0.018 | −0.871 | 0.072 | −0.134 | 1 | ||||
| Sig. (2-tailed) | 1.789E-2 | 0.828 | 1.65686E-44 | 0.395 | 0.113 | ||||||
| Takara Dye Toothpick (ng/µL) | Pearson Correlation | −0.199 | −0.018 | −0.871 | 0.072 | −0.134 | 0.999 | 1 | |||
| Sig. (2-tailed) | 1.788E-2 | 0.828 | 1.64435E-44 | 0.395 | 0.113 | 1E-4 | |||||
| Takara III Extract (ng/µL) | Pearson Correlation | −0.199 | −0.018 | −0.871 | 0.072 | −0.134 | 0.999 | 0.999 | 1 | ||
| Sig. (2-tailed) | 1.788E-2 | 0.828 | 1.73168E-44 | 0.395 | 0.113 | 1E-4 | 1E-4 | ||||
| Takara III Toothpick (ng/µL) | Pearson Correlation | −0.199 | −0.018 | −0.871 | 0.072 | −0.134 | 0.999 | 0.999 | 0.999 | 1 | |
| Sig. (2-tailed) | 1.788E-2 | 0.827 | 1.74169E-44 | 0.395 | 0.113 | 1E-4 | 1E-4 | 1E-4 | |||
| Virus copy number | Pearson Correlation | −0.129 | −0.051 | −0.383 | 0.004 | −0.056 | 0.659 | 0.659 | 0.659 | 0.659 | 1 |
| Sig. (2-tailed) | 0.128 | 0.549 | 1E-4 | 0.964 | 0.510 | 8.397E-19 | 8.639E-19 | 8.326E-19 | 8.519E-19 | ||
| Negative correlation is significant at the 0.05 level (2-tailed). | |||||||||||
| Positive correlation is significant at the 0.05 level (2-tailed). | |||||||||||
| N | 140 | 140 | 140 | 140 | 140 | 140 | 140 | 140 | 140 | 140 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romon-Ochoa, P.; Forster, J.; Chitty, R.; Gorton, C.; Lewis, A.; Eacock, A.; Kupper, Q.; Rigling, D.; Pérez-Sierra, A. Canker Development and Biocontrol Potential of CHV-1 Infected English Isolates of Cryphonectria parasitica Is Dependent on the Virus Concentration and the Compatibility of the Fungal Inoculums. Viruses 2022, 14, 2678. https://doi.org/10.3390/v14122678
Romon-Ochoa P, Forster J, Chitty R, Gorton C, Lewis A, Eacock A, Kupper Q, Rigling D, Pérez-Sierra A. Canker Development and Biocontrol Potential of CHV-1 Infected English Isolates of Cryphonectria parasitica Is Dependent on the Virus Concentration and the Compatibility of the Fungal Inoculums. Viruses. 2022; 14(12):2678. https://doi.org/10.3390/v14122678
Chicago/Turabian StyleRomon-Ochoa, Pedro, Jack Forster, Ruth Chitty, Caroline Gorton, Alex Lewis, Amy Eacock, Quirin Kupper, Daniel Rigling, and Ana Pérez-Sierra. 2022. "Canker Development and Biocontrol Potential of CHV-1 Infected English Isolates of Cryphonectria parasitica Is Dependent on the Virus Concentration and the Compatibility of the Fungal Inoculums" Viruses 14, no. 12: 2678. https://doi.org/10.3390/v14122678
APA StyleRomon-Ochoa, P., Forster, J., Chitty, R., Gorton, C., Lewis, A., Eacock, A., Kupper, Q., Rigling, D., & Pérez-Sierra, A. (2022). Canker Development and Biocontrol Potential of CHV-1 Infected English Isolates of Cryphonectria parasitica Is Dependent on the Virus Concentration and the Compatibility of the Fungal Inoculums. Viruses, 14(12), 2678. https://doi.org/10.3390/v14122678






