Effect of BIO-PLYTM, a Platelet-Rich Plasma Derived Biologic on PRRSV-2-Infected Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Macrophage Isolation
2.3. Viruses
2.4. BIO-PLYTM Production
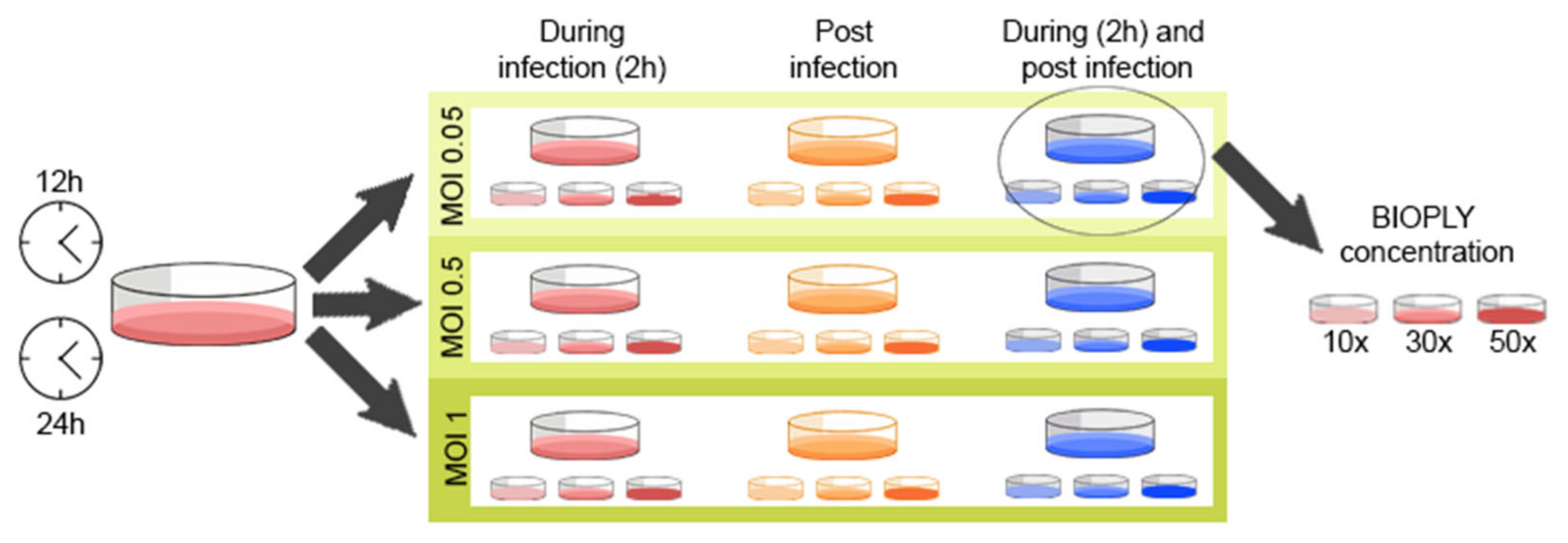
2.5. Experimental Layout
2.6. TCID50 Assay
2.7. RT-qPCR
2.8. Cytokine Production
2.9. ROS Production
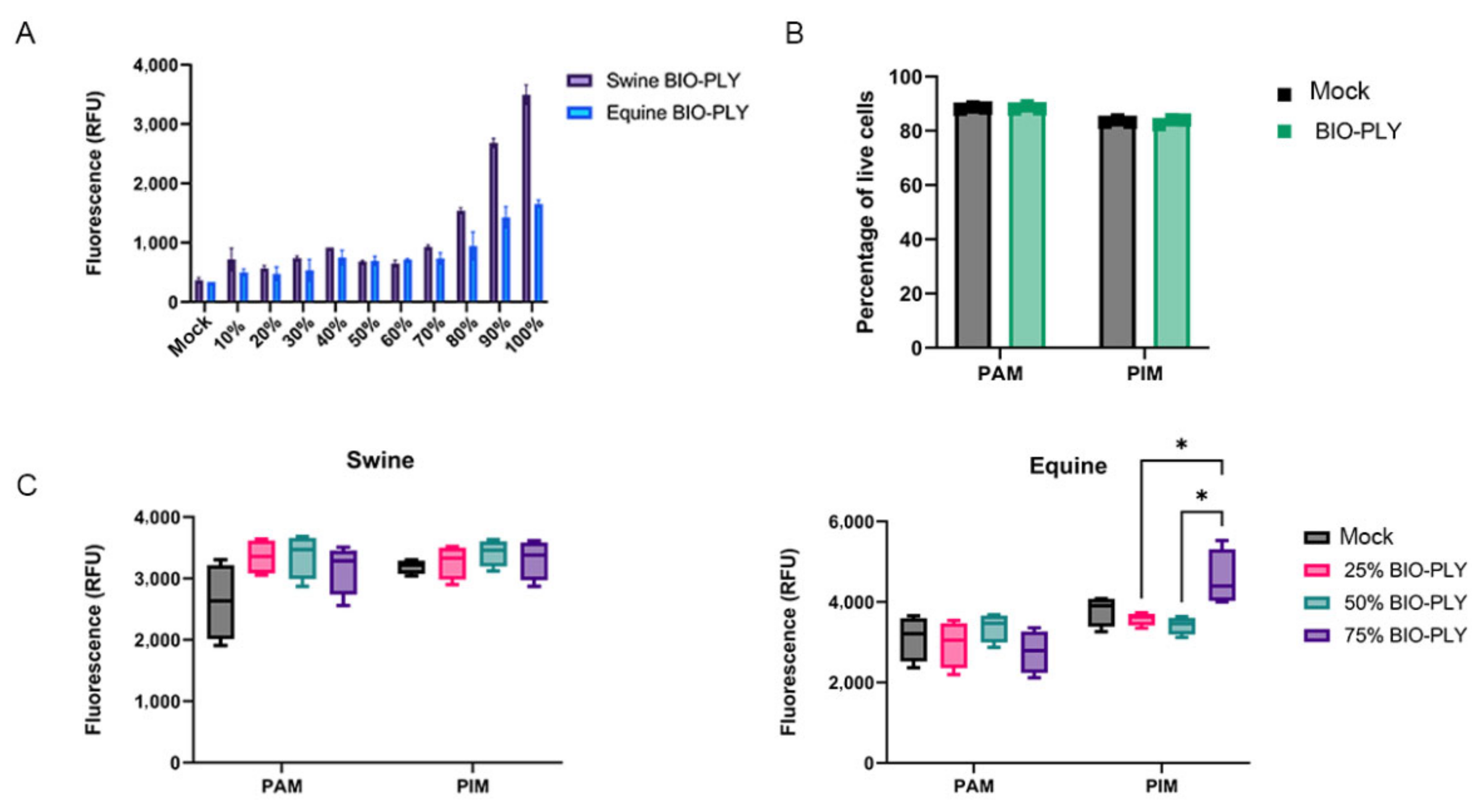
2.10. Cytotoxicity
2.11. Flow Cytometry
3. Results
3.1. Antiviral Capacity of BIO-PLYTM
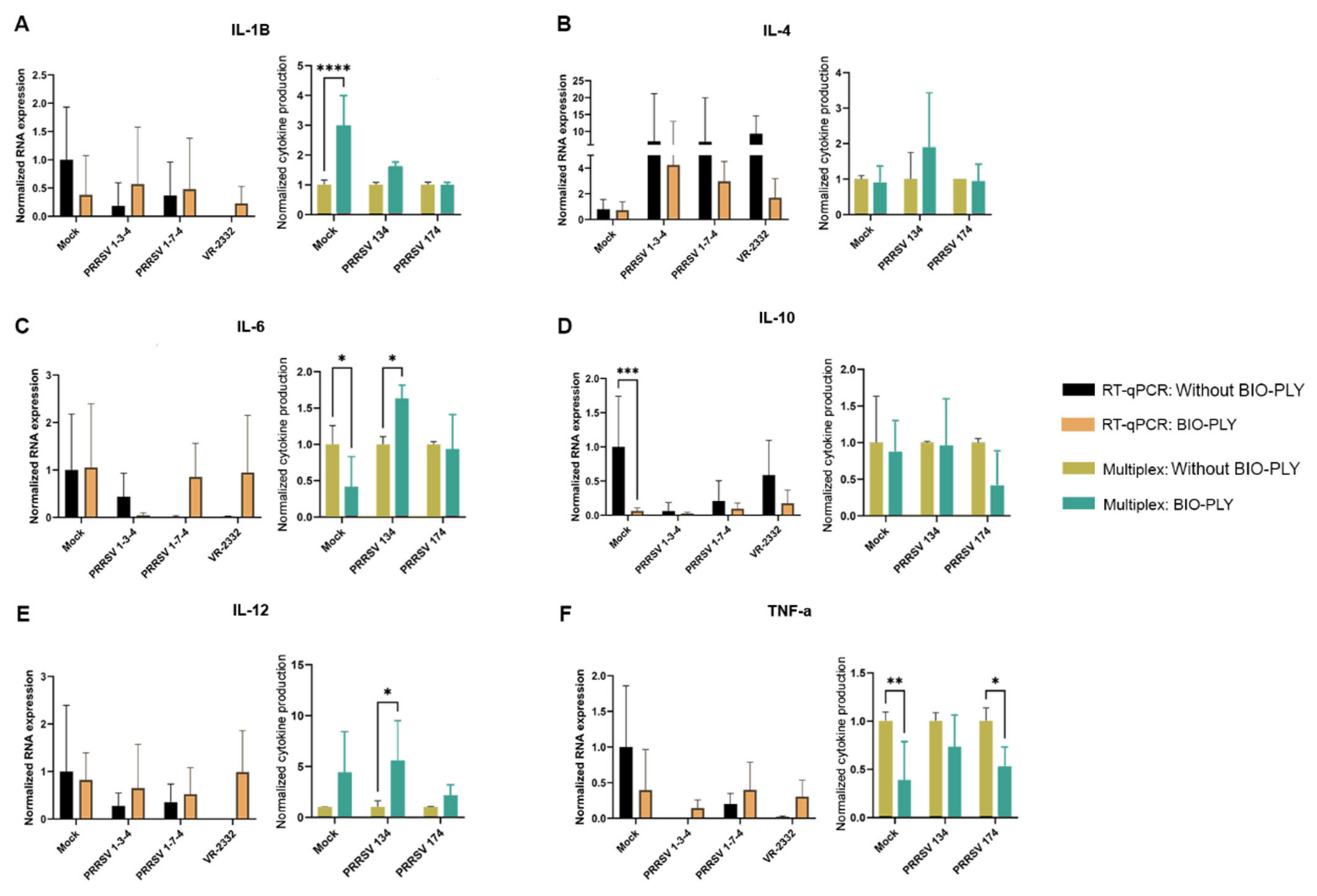
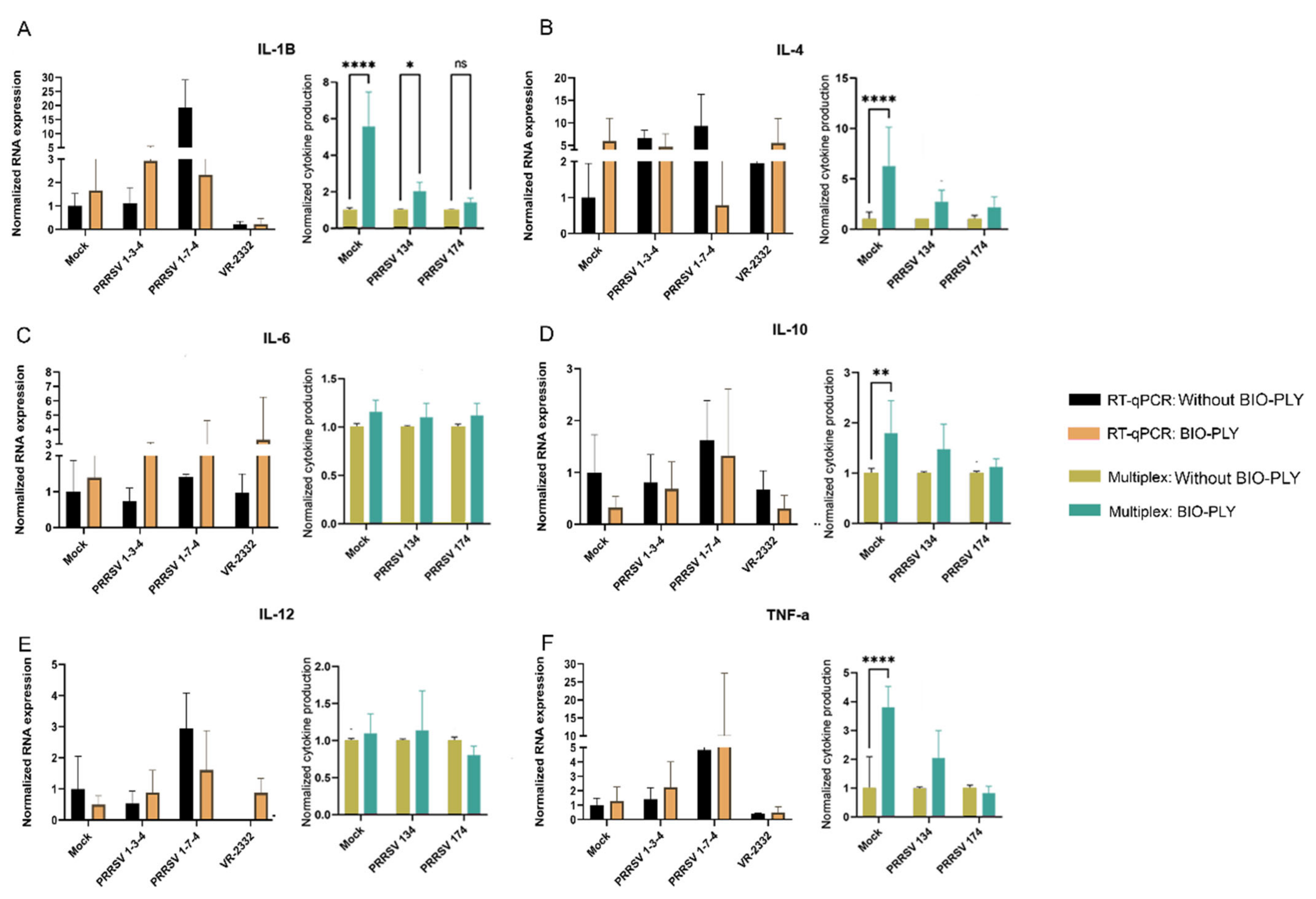
3.2. Cytokine Modulation
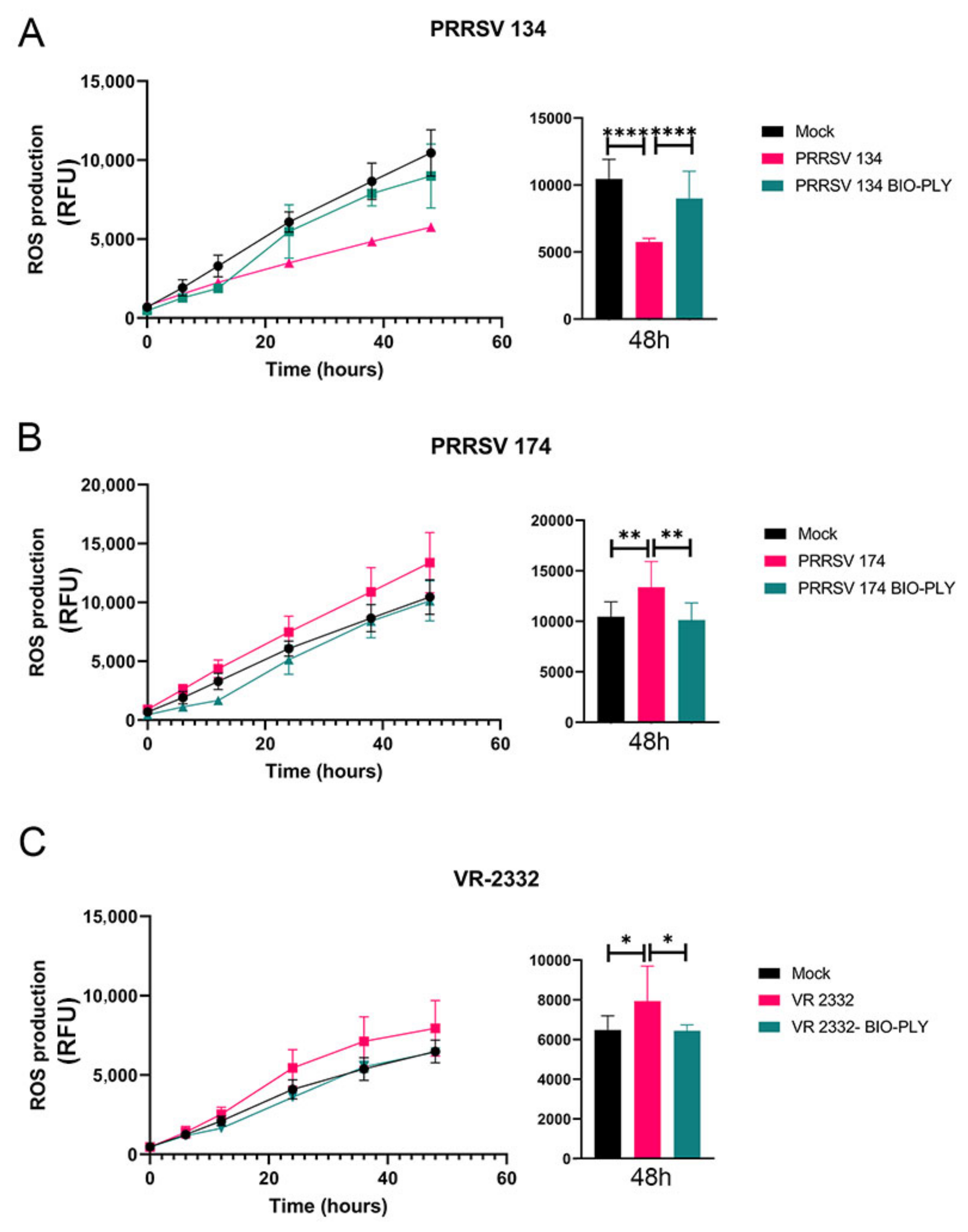
3.3. Reactive Oxygen Species (ROS) Production
3.4. BIO-PLYTM Cytotoxicity and Cell Viability
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA. Pork 2019 Export Highlights; U.S. Department of Agriculture: Washington, DC, USA, 2019. [Google Scholar]
- Holtkamp, D.J.; Kliebenstein, J.B.; Zimmerman, J.J.; Neumann, E.; Rotto, H.; Yoder, T.K.; Wang, C.; Yeske, P.; Mowrer, C.L.; Haley, C. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 2013, 21, 72–84. [Google Scholar]
- Fablet, C.; Marois-Crehan, C.; Grasland, B.; Simon, G.; Rose, N. Factors associated with herd-level PRRSV infection and age-time to seroconversion in farrow-to-finish herds. Vet. Microbiol. 2016, 192, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Straw, B.E.; Zimmerman, J.J.; D’Allaire, S.; Taylor, D.J. Diseases of Swine; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Jara, M.; Rasmussen, D.A.; Corzo, C.A.; Machado, G. Porcine reproductive and respiratory syndrome virus dissemination across pig production systems in the United States. Transbound. Emerg. Dis. 2021, 68, 667–683. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.G.; Dee, S.A. Porcine reproductive and respiratory syndrome virus. Theriogenology 2006, 66, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Corzo, C.A.; Mondaca, E.; Wayne, S.; Torremorell, M.; Dee, S.; Davies, P.; Morrison, R.B. Control and elimination of porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 185–192. [Google Scholar] [CrossRef]
- MSHMP. PRRS Charts. 2020. Available online: https://www.vetmed.umn.edu/centers-programs/swine-program/outreach-leman-mshmp/mshmp/mshmp-prrs-figures (accessed on 26 October 2022).
- Arjin, C.; Hongsibsong, S.; Pringproa, K.; Seel-Audom, M.; Ruksiriwanich, W.; Sutan, K.; Sommano, S.R.; Sringarm, K. Effect of ethanolic Caesalpinia sappan fraction on in vitro antiviral activity against porcine reproductive and respiratory syndrome virus. Vet. Sci. 2021, 8, 106. [Google Scholar] [CrossRef]
- Wang, X.; Dong, W.; Zhang, X.; Zhu, Z.; Chen, Y.; Liu, X.; Guo, C. Antiviral mechanism of tea polyphenols against porcine reproductive and respiratory syndrome virus. Pathogens 2021, 10, 202. [Google Scholar] [CrossRef]
- Graham, S.P.; Cheong, Y.-K.; Furniss, S.; Nixon, E.; Smith, J.A.; Yang, X.; Fruengel, R.; Hussain, S.; Tchorzewska, M.A.; La Ragione, R.M. Antiviral Efficacy of Metal and Metal Oxide Nanoparticles against the Porcine Reproductive and Respiratory Syndrome Virus. Nanomaterials 2021, 11, 2120. [Google Scholar] [CrossRef]
- Pattnaik, S.; Chaudhury, B.; Mohapatra, M. Exploration of Inorganic Materials with Antiviral Properties. In COVID-19 Pandemic; Springer: Berlin/Heidelberg, Germany, 2022; pp. 53–74. [Google Scholar]
- Savard, C.; Pinilla, V.; Provost, C.; Segura, M.; Gagnon, C.A.; Chorfi, Y. In vitro effect of deoxynivalenol (DON) mycotoxin on porcine reproductive and respiratory syndrome virus replication. Food Chem. Toxicol. 2014, 65, 219–226. [Google Scholar] [CrossRef]
- Tang, Y.-Q.; Yeaman, M.R.; Selsted, M.E. Antimicrobial peptides from human platelets. Infect. Immun. 2002, 70, 6524–6533. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Bayer, A.S. Antimicrobial peptides from platelets. Drug Resist. Updates 1999, 2, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Hu, Z.; Zhang, Y.; Wang, P.; Li, S.; Huang, S.; Dong, Y.; Xu, H.; Rong, Y.; Zhu, W. Clinical Studies on Platelet-Rich Plasma Therapy for Chronic Cutaneous Ulcers: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Adv. Wound Care 2022, 11, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Dos-Santos, G.; Hurdle, M.F.B.; Clendenen, S.R.; Eldrige, J.S.; Qu, W. Autologous Platelet-Rich Plasma Applications in Chronic Pain Medicine: Establishing a Framework for Future Research-A Narrative Review. Pain Physician 2022, 25, 15–27. [Google Scholar]
- Evish Kamrava, M.; Aaron Calodney, M.; Kent Remley, M.; Ramsin Benyamin, M.; Sheldon Jordan, M. Safety and Efficacy of Platelet Rich Plasma for Treatment of Lumbar Discogenic Pain: A Prospective, Multicenter, Randomized, Double-blind Study. Pain Physician 2022, 25, 29–34. [Google Scholar]
- El-Kadiry, A.E.-H.; Lumbao, C.; Salame, N.; Rafei, M.; Shammaa, R. Bone marrow aspirate concentrate versus platelet-rich plasma for treating knee osteoarthritis: A one-year non-randomized retrospective comparative study. BMC Musculoskelet. Disord. 2022, 23, 23. [Google Scholar] [CrossRef]
- Jamal, M.; Hurley, E.; Asad, H.; Asad, A.; Taneja, T. The role of Platelet Rich Plasma and other orthobiologics in bone healing and fracture management: A systematic review. J. Clin. Orthop. Trauma 2022, 25, 101759. [Google Scholar] [CrossRef]
- Alio, J.L.; Rodriguez, A.E.; Abdelghany, A.A.; Oliveira, R.F. Autologous Platelet-Rich Plasma Eye Drops for the Treatment of Post-LASIK Chronic Ocular Surface Syndrome. J. Ophthalmol. 2017, 2017, 2457620. [Google Scholar] [CrossRef]
- Farghali, H.A.; AbdElKader, N.A.; AbuBakr, H.O.; Ramadan, E.S.; Khattab, M.S.; Salem, N.Y.; Emam, I.A. Corneal ulcer in dogs and cats: Novel clinical application of regenerative therapy using subconjunctival injection of autologous platelet-rich plasma. Front. Vet. Sci. 2021, 8, 641265. [Google Scholar] [CrossRef]
- Kamiya, K.; Takahashi, M.; Shoji, N. Effect of Platelet-Rich Plasma on Corneal Epithelial Healing after Phototherapeutic Keratectomy: An Intraindividual Contralateral Randomized Study. BioMed Res. Int. 2021, 2021, 5752248. [Google Scholar] [CrossRef]
- Berndt, S.; Turzi, A.; Modarressi, A. Production of autologous platelet-rich plasma for boosting in vitro human fibroblast expansion. JoVE (J. Vis. Exp.) 2021, 2021, e60816. [Google Scholar] [CrossRef]
- Del Amo, C.; Perez-Valle, A.; Atilano, L.; Andia, I. Unraveling the Signaling Secretome of Platelet-Rich Plasma: Towards a Better Understanding of Its Therapeutic Potential in Knee Osteoarthritis. J. Clin. Med. 2022, 11, 473. [Google Scholar] [CrossRef] [PubMed]
- Riewruja, K.; Phakham, S.; Sompolpong, P.; Reantragoon, R.; Tanavalee, A.; Ngarmukos, S.; Udomsinprasert, W.; Suantawee, T.; Dechsupa, S.; Honsawek, S. Cytokine profiling and intra-articular injection of autologous platelet-rich plasma in knee osteoarthritis. Int. J. Mol. Sci. 2022, 23, 890. [Google Scholar] [CrossRef] [PubMed]
- Gilbertie, J.M.; Schaer, T.P.; Engiles, J.B.; Seiler, G.S.; Deddens, B.L.; Schubert, A.G.; Jacob, M.E.; Stefanovski, D.; Ruthel, G.; Hickok, N.J.; et al. A Platelet-Rich Plasma-Derived Biologic Clears Staphylococcus aureus Biofilms While Mitigating Cartilage Degeneration and Joint Inflammation in a Clinically Relevant Large Animal Infectious Arthritis Model. Front. Cell. Infect. Microbiol. 2022, 12, 895022. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Wang, S.; Yang, J.; Wu, T.; Fei, J. Application of platelet-rich plasma in traumatic bone infections. Expert Rev. Anti-Infect. Ther. 2021, 19, 867–875. [Google Scholar] [CrossRef]
- Metsemakers, W.; Kuehl, R.; Moriarty, T.; Richards, R.; Verhofstad, M.; Borens, O.; Kates, S.; Morgenstern, M. Infection after fracture fixation: Current surgical and microbiological concepts. Injury 2018, 49, 511–522. [Google Scholar] [CrossRef]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Monteiro, F.J.; Ferraz, M.P. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter 2012, 2, 176–194. [Google Scholar] [CrossRef]
- Gilbertie, J.M.; Schaer, T.P.; Schubert, A.G.; Jacob, M.E.; Menegatti, S.; Ashton Lavoie, R.; Schnabel, L.V. Platelet-rich plasma lysate displays antibiofilm properties and restores antimicrobial activity against synovial fluid biofilms in vitro. J. Orthop. Res. 2020, 38, 1365–1374. [Google Scholar] [CrossRef]
- Gilbertie, J.M.; Schnabel, L.V.; Schaer, T.P. Platelet-Rich Plasma Lysate Compositions and Related Methods. U.S. Patent 20210128626, 6 May 2021. [Google Scholar]
- López, C.; Álvarez, M.E.; Carmona, J.U. Temporal bacteriostatic effect and growth factor loss in equine platelet components and plasma cultured with methicillin-sensitive and methicillin-resistant Staphylococcus aureus: A comparative in vitro study. Vet. Med. Int. 2014, 2014, 525826. [Google Scholar] [CrossRef]
- Moojen, D.J.F.; Everts, P.A.; Schure, R.M.; Overdevest, E.P.; van Zundert, A.; Knape, J.T.; Castelein, R.M.; Creemers, L.B.; Dhert, W.J. Antimicrobial activity of platelet-leukocyte gel against Staphylococcus aureus. J. Orthop. Res. 2008, 26, 404–410. [Google Scholar] [CrossRef]
- Karina, K.; Christoffel, L.M.; Novariani, R.; Rosidah, S.; Rosadi, I.; Rosliana, I.; Sobariah, S.; Fatkhurohman, N.; Hertati, Y.; Puspitaningrum, N. Potential breakthrough for treating severe Coronavirus Disease-2019 (COVID-19) patients in intensive care unit: The use of autologous activated platelet-rich plasma in serial cases of Indonesian patients. Sapporo Med. J. 2021, 55, 1–12. [Google Scholar]
- Karina, K.; Rosliana, I.; Rosadi, I.; Sobariah, S.; Christoffel, L.M.; Novariani, R.; Rosidah, S.; Fatkhurohman, N.; Hertati, Y.; Puspitaningrum, N. Phase I/II Clinical Trial of Autologous Activated Platelet-Rich Plasma (aaPRP) in the Treatment of Severe Coronavirus Disease 2019 (COVID-19) Patients. Int. J. Inflamm. 2021, 2021, 5531873. [Google Scholar] [CrossRef] [PubMed]
- Tyrnenopoulou, P.; Diakakis, N.; Karayannopoulou, M.; Savvas, I.; Koliakos, G. Evaluation of intra-articular injection of autologous platelet lysate (PL) in horses with osteoarthritis of the distal interphalangeal joint. Vet. Q. 2016, 36, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Liesenborghs, L.; Verhamme, P.; Vanassche, T. Staphylococcus aureus, master manipulator of the human hemostatic system. J. Thromb. Haemost. 2018, 16, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, C.E.; López, C.; Álvarez, M.E.; Samudio, I.J.; Prades, M.; Carmona, J.U. Effects of the breed, sex and age on cellular content and growth factor release from equine pure-platelet rich plasma and pure-platelet rich gel. BMC Vet. Res. 2013, 9, 29. [Google Scholar] [CrossRef]
- Rinnovati, R.; Romagnoli, N.; Gentilini, F.; Lambertini, C.; Spadari, A. The influence of environmental variables on platelet concentration in horse platelet-rich plasma. Acta Vet. Scand. 2015, 58, 45. [Google Scholar] [CrossRef]
- Bordet, E.; Blanc, F.; Tiret, M.; Crisci, E.; Bouguyon, E.; Renson, P.; Maisonnasse, P.; Bourge, M.; Leplat, J.-J.; Giuffra, E. Porcine reproductive and respiratory syndrome virus type 1.3 Lena triggers conventional dendritic cells 1 activation and T helper 1 immune response without infecting dendritic cells. Front. Immunol. 2018, 9, 2299. [Google Scholar] [CrossRef]
- Maisonnasse, P.; Bouguyon, E.; Piton, G.; Ezquerra, A.; Urien, C.; Deloizy, C.; Bourge, M.; Leplat, J.; Simon, G.; Chevalier, C. The respiratory DC/macrophage network at steady-state and upon influenza infection in the swine biomedical model. Mucosal Immunol. 2016, 9, 835–849. [Google Scholar] [CrossRef]
- Inaba, K.; Inaba, M.; Deguchi, M.; Hagi, K.; Yasumizu, R.; Ikehara, S.; Muramatsu, S.; Steinman, R.M. Granulocytes, macrophages, and dendritic cells arise from a common major histocompatibility complex class II-negative progenitor in mouse bone marrow. Proc. Natl. Acad. Sci. USA 1993, 90, 3038–3042. [Google Scholar] [CrossRef]
- Saade, G.; Ménard, D.; Hervet, C.; Renson, P.; Hue, E.; Zhu, J.; Dubreil, L.; Paillot, R.; Pronost, S.; Bourry, O. Porcine reproductive and respiratory syndrome virus interferes with Swine influenza A virus infection of epithelial cells. Vaccines 2020, 8, 508. [Google Scholar] [CrossRef]
- Kick, A.R.; Amaral, A.F.; Cortes, L.M.; Fogle, J.E.; Crisci, E.; Almond, G.W.; Käser, T. The T-Cell Response to Type 2 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). Viruses 2019, 11, 796. [Google Scholar] [CrossRef] [PubMed]
- Gilbertie, J.M. Bacterial Behavior in Synovial Fluid and Development of a Novel Therapeutic to Combat Infectious Arthritis; North Carolina State University: Raleigh, NC, USA, 2019. [Google Scholar]
- Frias-De-Diego, A.; Crisci, E. Use of Crystal Violet to Improve Visual Cytopathic Effect-based Reading for Viral Titration using TCID50 Assays. J. Vis. Exp. Jove 2022. [Google Scholar] [CrossRef] [PubMed]
- Spear, A.; Faaberg, K.S. Development of a genome copy specific RT-qPCR assay for divergent strains of type 2 porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2015, 218, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yu, S.; Tamhane, A.; Causey, Z.L.; Steg, A.; Danila, M.I.; Reynolds, R.J.; Wang, J.; Wanzeck, K.C.; Tang, Q. Simple regression for correcting ΔCt bias in RT-qPCR low-density array data normalization. BMC Genom. 2015, 16, 82. [Google Scholar] [CrossRef]
- Nelli, R.K.; Dunham, S.P.; Kuchipudi, S.V.; White, G.A.; Baquero-Perez, B.; Chang, P.; Ghaemmaghami, A.; Brookes, S.M.; Brown, I.H.; Chang, K.-C. Mammalian innate resistance to highly pathogenic avian influenza H5N1 virus infection is mediated through reduced proinflammation and infectious virus release. J. Virol. 2012, 86, 9201–9210. [Google Scholar] [CrossRef]
- Delgado-Ortega, M.; Olivier, M.; Sizaret, P.-Y.; Simon, G.; Meurens, F. Newborn pig trachea cell line cultured in air-liquid interface conditions allows a partial in vitro representation of the porcine upper airway tissue. BMC Cell Biol. 2014, 15, 14. [Google Scholar] [CrossRef]
- Thanawongnuwech, R.; Halbur, P.G.; Thacker, E.L. The role of pulmonary intravascular macrophages in porcine reproductive and respiratory syndrome virus infection. Anim. Health Res. Rev. 2000, 1, 95–102. [Google Scholar] [CrossRef]
- Kausar, S.; Said Khan, F.; Ishaq Mujeeb Ur Rehman, M.; Akram, M.; Riaz, M.; Rasool, G.; Hamid Khan, A.; Saleem, I.; Shamim, S.; Malik, A. A review: Mechanism of action of antiviral drugs. Int. J. Immunopathol. Pharmacol. 2021, 35, 20587384211002621. [Google Scholar] [CrossRef]
- Balfour, H.H., Jr. Resistance of Herpes Simplex to Acyclovir; Ann Intern Med American College of Physicians: Philadelphia, PA, USA, 1983; Volume 98, pp. 404–406. [Google Scholar]
- Fyfe, J.; Keller, P.; Furman, P.; Miller, R.; Elion, G. Thymidine kinase from herpes simplex virus phosphorylates the new antiviral compound, 9-(2-hydroxyethoxymethyl) guanine. J. Biol. Chem. 1978, 253, 8721–8727. [Google Scholar] [CrossRef]
- Derse, D.; Cheng, Y.-C.; Furman, P.; St Clair, M.; Elion, G. Inhibition of purified human and herpes simplex virus-induced DNA polymerases by 9-(2-hydroxyethoxymethyl) guanine triphosphate. Effects on primer-template function. J. Biol. Chem. 1981, 256, 11447–11451. [Google Scholar] [CrossRef]
- Furman, P.A.; St Clair, M.; Spector, T. Acyclovir triphosphate is a suicide inactivator of the herpes simplex virus DNA polymerase. J. Biol. Chem. 1984, 259, 9575–9579. [Google Scholar] [CrossRef] [PubMed]
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A critical review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Jomah, S.; Asdaq, S.M.B.; Al-Yamani, M.J. Clinical efficacy of antivirals against novel coronavirus (COVID-19): A review. J. Infect. Public Health 2020, 13, 1187–1195. [Google Scholar] [CrossRef]
- Bellinger, F.P.; Raman, A.V.; Reeves, M.A.; Berry, M.J. Regulation and function of selenoproteins in human disease. Biochem. J. 2009, 422, 11–22. [Google Scholar] [CrossRef]
- Lee, S.-M.; Kleiboeker, S.B. Porcine reproductive and respiratory syndrome virus induces apoptosis through a mitochondria-mediated pathway. Virology 2007, 365, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Suarez, P.; Diaz-Guerra, M.; Prieto, C.; Esteban, M.; Castro, J.M.; Nieto, A.; Ortín, J. Open reading frame 5 of porcine reproductive and respiratory syndrome virus as a cause of virus-induced apoptosis. J. Virol. 1996, 70, 2876–2882. [Google Scholar] [CrossRef]


| Cytokine | Primer | Sequence | Reference |
|---|---|---|---|
| IL-1β | Forward | 5′-TGC CAA CGT GCA GTC TAT GG-3′ | [51] |
| Reverse | 5′-TGG GCC AGC CAG CAC TAG-3′ | ||
| IL-4 | Forward | 5′-GCC GGG CCT CGA CTG T-3′ | [42] |
| Reverse | 5′-TCC GCT CAG GAG GCT CTT C-3′ | ||
| IL-6 | Forward | 5′-CTG CTT CTG GTG ATG GCT ACT G-3′ | [42] |
| Reverse | 5′-GGC ATC ACC TTT GGC ATC TT-3′ | ||
| IL-10 | Forward | 5′-GAG CCA ACT GCA GCT TCC A-3′ | [42] |
| Reverse | 5′-TCA GGA CAA ATA GCC CAC TAG CTT-3’ | ||
| IL-12 | Forward | 5′-GGA GCA CCC CAC ATT CCT ACT-3′ | [42] |
| Reverse | 5′-TTC TCT TTT GTT CTT GCC CTG AA-3′ | ||
| TNF-α | Forward | 5′-TGG TGG TGC CGA CAG ATG -3′ | [42] |
| Reverse | 5′-CAG CCT TGG CCC CTG AA -3′ | ||
| RPS24 | Forward | 5′-TTT GCC AGC ACC AAC GTT G-3′ | [42] |
| Reverse | 5′-AAG GAA CGC AAG AAC AGA ATG AA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frias-De-Diego, A.; Gilbertie, J.M.; Scholle, F.; Dejarnette, S.; Crisci, E. Effect of BIO-PLYTM, a Platelet-Rich Plasma Derived Biologic on PRRSV-2-Infected Macrophages. Viruses 2022, 14, 2666. https://doi.org/10.3390/v14122666
Frias-De-Diego A, Gilbertie JM, Scholle F, Dejarnette S, Crisci E. Effect of BIO-PLYTM, a Platelet-Rich Plasma Derived Biologic on PRRSV-2-Infected Macrophages. Viruses. 2022; 14(12):2666. https://doi.org/10.3390/v14122666
Chicago/Turabian StyleFrias-De-Diego, Alba, Jessica M. Gilbertie, Frank Scholle, Sarah Dejarnette, and Elisa Crisci. 2022. "Effect of BIO-PLYTM, a Platelet-Rich Plasma Derived Biologic on PRRSV-2-Infected Macrophages" Viruses 14, no. 12: 2666. https://doi.org/10.3390/v14122666
APA StyleFrias-De-Diego, A., Gilbertie, J. M., Scholle, F., Dejarnette, S., & Crisci, E. (2022). Effect of BIO-PLYTM, a Platelet-Rich Plasma Derived Biologic on PRRSV-2-Infected Macrophages. Viruses, 14(12), 2666. https://doi.org/10.3390/v14122666






