Abstract
In nature, viral coinfection is as widespread as viral infection alone. Viral coinfections often cause altered viral pathogenicity, disrupted host defense, and mixed-up clinical symptoms, all of which result in more difficult diagnosis and treatment of a disease. There are three major virus–virus interactions in coinfection cases: viral interference, viral synergy, and viral noninterference. We analyzed virus–virus interactions in both aspects of viruses and hosts and elucidated their possible mechanisms. Finally, we summarized the protocol of viral coinfection studies and key points in the process of virus separation and purification.
1. Introduction
In nature, it is common for multiple pathogens (viruses, bacteria, fungi, and protozoa) to infect the same host simultaneously or successively. This phenomenon is defined as coinfection [1]. Typically, coinfection complicates the symptoms and diagnosis of a disease. In this article, we only focus on viral coinfection in clinics while focusing on virus–virus interactions.
A virus–virus interaction can be observed in five patterns: interference, synergy, noninterference, dependence assistance, and host–parasite relation. The most common virus–virus interaction in coinfection is interference, in which one virus competes to suppress the replication of another [2]. SARS-CoV-2 can extensively inhibit the replication of multiple respiratory viruses [3,4]. A persistent infection of the Old World arenavirus [5], influenza A virus (IAV) [6], or classical swine fever virus (CSFV) [7,8] eliminates the secondary viral infection; this is known as superinfection exclusion [6].
In contrast to interference, coinfection with certain viruses may enhance the replication of other viruses [9], which we define as synergy. For instance, West Nile virus (WNV) and IAV infection each enhance the replication of Culex flavivirus (CxFV) [10] and human parainfluenza virus type 2 (hPIV2) [9], respectively.
If coinfection has no effect on virus replication, it is defined as non-interference [11,12,13]. Noninterference is usually found between viruses with different tissue tropisms. In human or animal viral infections, we can often detect a “passenger virus” that does not cause any symptoms or disease. The relation between a “causative virus” and a “passenger virus” is independent.
Dependence assistance and host–parasite relations are two specific viral relationships. Viruses with an incomplete genome, such as adeno-associated virus, with defective interfering particles, cannot complicate a replication cycle by themselves; instead, they require the assistance of a “helper virus”, such as adeno virus, herpes virus, or another intact virus, in order to finish their life cycle. These represent dependence assistance in viral interactions [14,15,16].
A host–parasite relation exists between virophages and giant viruses [17]. Virophages, such as Sputnik, are parasites of Mimivirus and Mamavirus. Additionally, Sputnik cannot replicate in Acanthamoeba castellanii but grows rapidly in the giant virus producer found in amoebae coinfected with Mimivirus, and Sputnik growth impacts and reduces Mimivirus replication.
Interference, synergy, and noninterference interactions are commonly identified in clinical viral coinfection cases. In this review, we focus on these three viral interactions and summarize their outcomes, mechanisms, and relative studies.
2. Virus–Virus Interaction in Coinfections
Interactions in viral coinfections are primarily caused by changes in virus replication cycles (virus factors) and the replication environment (host factors), as shown in Figure 1.
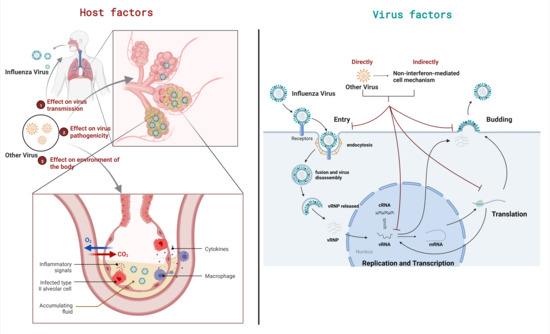
Figure 1.
Two factors leading to the outcomes of viral coinfection. The outcomes of viral coinfection can be mainly attributed to two factors, namely virus factors and host factors. The figure takes IAV coinfection as an example. Host factors in coinfection change the environment of the body, thereby affecting the transmission and pathogenicity of viruses. Virus factors are coinfection changes that affect the intracellular environment and directly or indirectly affect the viral life cycle.
2.1. Viral Interference
The causes of viral interference can be divided into two categories: interferon-mediated and non-interferon-mediated (Figure 2).
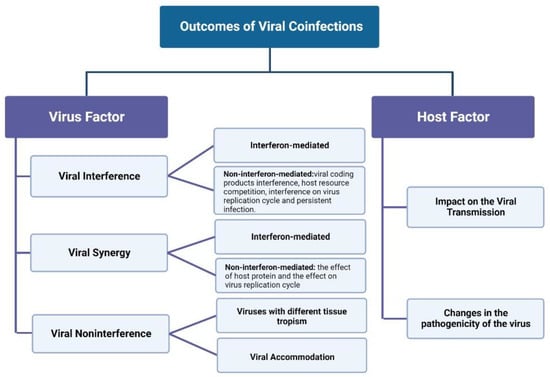
Figure 2.
Outcomes of Viral Coinfections.
Interferon (IFN)-mediated innate immunity is the most common reason for viral interference [18,19]. In vivo studies of coinfection of IAV, respiratory syncytial virus (RSV), and rhinovirus (RV) show that IAV and RSV can interfere with RV replication through type I and type III IFN [20]. In clinical HCV and HIV coinfection, HIV-induced IFNα can reduce the level of HCV viremia [21]. Mouse hepatitis virus strain 1 (MHV-1) inhibits replication of IAV by upregulating IFN-β [22].
IFN induces multiple interferon-stimulating genes (ISGs) and activates multiple innate immunity signaling pathways [23,24,25,26,27,28,29,30,31,32,33]. GB virus C (GBV-C) inhibiting the proliferation of HIV is a typical IFN-mediated viral interference phenomenon. GBV-C promotes the activation of IFN-γ and downstream ISGs expression, as well as the activation/maturation of circulating pDC, which further increases IFN-γ [34]. Additionally, regarding coinfection of RV and IAV/pneumonia virus of mice (PVM), RV significantly inhibits the replication of IAV or PVM. RV induces an increase in Muc5ac gene expression, leading to an increase in IFN-β through the aromatic hydrocarbon receptor (AhR) signal transduction [35,36].
Non-interferon-mediated viral interference, also known as intrinsic interference, is the resistance of viral-infected cells to subsequent viral infections. This is particularly noticeable in foot-and-mouth disease virus (FMDV) coinfection cases, in which the attenuated A24 Cruzeiro strain interferes with the proliferation of homologous and heterologous strains [37]. Another typical intrinsic interference is in the case of Sindbis virus coinfection in viral infected vertebrate cells; the first virus translates non-structural genes to establish homologous exclusion and the genome of the second virus translates only without replication [38,39]. Adam et al. conducted further research based on these findings, and they found that a unipartite non-structural precursor called P123 is necessary to produce viral negative-strand RNA templates. The P123 of the latter virus is rapidly cleaved by the protease of the former virus, resulting in the latter virus being unable to synthesize the negative strand. This explains the phenomenon of intrinsic interference, at least to some extent [40].
The intrinsic interference between unrelated viruses can be found in the case of Newcastle disease virus (NDV) coinfection. Rubella virus can induce an interference state in infected host cells to avoid infection of NDV [41]. There is competition between the coinfected viruses for metabolites, replication sites [42], or a host’s viral replication-required proteins [12,40,43,44,45,46,47,48,49,50,51,52,53,54]. There are some host proteins that play a key role in the life cycle of various viruses, such as tetraspanins. Tetraspanins are transmembrane glycoproteins that are associated with the pathogenesis of non-enveloped viruses (human papillomavirus [HPV]) and enveloped viruses (HIV, Zika virus, IAV and coronavirus) [55]. When coinfections occur among these viruses, tetraspanins serve as the main host protein being explored.
In addition to the contest for host proteins, there are several other interference mediators, including defective interfering particles (DI particles) [56], RNA interferences (RNAi) [57,58,59,60,61], trans-acting viral proteins [62,63,64], and non-specific dsRNAs [65,66].
Virus interference can occur at each step of the virus-replication process, including virus attachment [67,68,69,70,71,72,73,74,75,76,77,78] and entry [54,79,80,81,82], viral genome replication [40,54,83,84,85,86,87,88], viral protein translation and assembly, and progeny virus budding [89]. At the stage of viral attachment, simian immunodeficiency virus (SIV) can significantly inhibit the expression of CD4 glycoprotein on the cell surface, which causes cell resistance to HIV-1 superinfection [78]. At the stage of entry, vesicular stomatitis virus (VSV) inhibits the formation rate of endocytic vesicles and reduces the internalization rate of receptor-binding ligands in order to restrain other viruses from taking over the coated pits [81]. In the viral gene-replication step, the expression of the Borna disease virus (BDV) P, N or X protein makes human cells resistant to superinfection with BDV by selectively blocking the polymerase activity of viruses [84]. In the viral protein translation step, the coinfection of VSV and IAV inhibits the translation of IAV mRNA, which is related to the inhibition of protein synthesis after VSV infection [90]. In the viral assembly and budding stages, Alphabaculovirus-induced actin recombination blocks the assembly and budding of other viruses [89]. Inhibition could happen at multiple steps, as Semliki Forest virus (SFV) infection inhibits the attachment, entry, and budding of subsequent viruses [54].
Viral interference is also often found in persistent infections. Unlike acute infections, in which virus particles are eventually cleared by the immune system or host, viruses stay in infected cells for a long time in persistent infections [1]. Viruses in persistent infections usually reduce their replication level [91,92,93,94,95,96,97,98,99] to keep the infected cell alive. Therefore, the virus in a persistent infection state can resist the influence of other viruses and exist in infected cells for a long time. A good example is the persistent infection of mosquitos by densovirus (DNV) [100]. DNV-infected cells are resistant to dengue virus (DENV) attack, and no CPE appears in these cases [101,102]. Studies on flock house virus (FHV) have shown that host and viral factors are involved in maintaining viral persistence [103,104,105]. Regarding the establishment of persistent infection in vitro, mutations in the viral genome begin to accumulate after several continuous passages [103], indicating that the cellular environment, rather than the virus itself, is essential for the establishment of sustained infection. The continuous replication of viruses could be accomplished by blocking the RNAi response of infected cells. Goic et al. reported that the persistence of FHV in Drosophila melanogaster could be accomplished by regulating RNAi and reverse transcriptase activity [106]. Fragments of different RNA viruses are reversely transcribed at early infection, which results in DNA forms embedded in the retrotransposon sequences. These virus-retrotransposon DNA chimeras trigger cellular RNAi mechanisms that inhibit viral replication. The inhibition of reverse transcriptase by FHV can hinder the emergence of chimeric DNA, thus closing the cell RNAi mechanism and making FHV persist in the cell.
2.2. Viral Synergy
The causes of viral synergy can be divided into two categories: interferon-mediated and non-interferon-mediated (as shown in Figure 2). Interferon-mediated viral promotion is primarily manifested as one virus causing host immunodeficiency; this, in turn, promotes the proliferation of the other viruses. In mouse L cells, coinfection with Vaccinia virus (VV) protects the VSV from IFN inhibition. This is related to the inhibition of IFN-induced dsRNA-dependent protein kinase activity by VV [107]. In coinfections of Hepatitis B virus (HBV) and Hepatitis C virus (HCV), the reduced liver IFN response after HCV clearance can cause HBV reactivation [108]. Another good example of this mechanism is the coinfection between canine parvovirus type 2 (CPV-2) and canine circovirus (CCV); CCV inhibits the activation of the IFN-I promoter by inducing Rep protein expression, thus blocking the subsequent expression of ISGs to promote CPV-2 replication [109]. Additionally, in the coinfection of paramyxovirus 5 wild-type (SV5-WT) and SV5 P/V mutant (rSV5-P/V-CPI−), rSV5-WT can block IFN signaling by inhibiting IRF-3 translocation into the nucleus and degrading STAT1 [110], thus blocking host cytokines involved in antiviral response and those involved in IFN synthesis [111].
Non-interferon-mediated viral synergy could be related to the effect of the host protein or the replication of other viruses. Coinfection between Marek’s disease virus (MDV) and reticuloendotheliosis virus (REV) increases the replication of both viruses in cells [112]. Further studies have shown that host proteins such as IRF7, MX1, TIMP3, and AKT1 may be related to the synergy of MDV and REV. In the coinfection of the Avian leukosis virus subgroup J (ALV-J) and REV, host protein TRIM62 increases replication of the two viruses by regulating the actin cytoskeleton [113].
The effect of coinfection on virus replication cycles is more intuitive and effective. A study by Goto et al. found that hPIV2 infection enhances IAV replication [9] by promoting the fusion of the infected cell’s membrane.
2.3. Viral Noninterference
Noninterference is usually found between viruses with different tissue tropisms. For example, influenza viruses mainly infect the upper respiratory tract and lower respiratory tract, and occasionally infect extrapulmonary tissues, such as the eyes and intestines [114]. HPV infection, however, is mainly distributed on the skin, mouth, nasal cavity and genitals [115]. The tissue tropisms of these two viruses have almost no intersection; therefore, when coinfections between IAV and HPV occur, we generally assume that their relationship is one of noninterference.
A host could be actively and continuously infected by multiple viruses without any obvious signs of disease. This is called viral accommodation. Viral accommodation is usually observed in arthropods [12] and shrimps [102,116,117]. There is little evidence that shrimps or other arthropods have an immune system [118], but exposure to inactivated virions or envelope proteins allows them to acquire short-term resistance to viral attacks [119,120]. In shrimp, viral diseases are the result of virus-induced apoptosis, which is not mediated by the immune system [102,121,122,123]. In viral accommodation, multiple viruses can exist independently and stably in the same cell, and the possibility of gene exchange between them depends on the similarity between the coinfected viruses.
3. Outcome of Viral Coinfections on Host
The outcomes of viral coinfection attributed to the host can be divided into two categories: effects on viral transmission and viral pathogenicity (as shown in Figure 2).
3.1. Effects on Virus Transmission
Inhibition and promotion of virus transmission can both be found in viral coinfection. Coinfection with DENV2 and DENV4 produces a competitive inhibitory effect that reduces the spread of the viruses [124,125,126]. Natural coinfections of RV and IAV occur frequently in humans. RV interferes with IAV transmission by reducing IAV aerosols [127]. Regarding the promotion of virus transmission, one study found that coinfections of CxFV and WNV promoted WNV transmission [10]. Coinfections of Chikungunya virus and Zika virus in mosquitoes leads to an enhancement in transmission of the Zika virus [128]. Therefore, in order to fully understand the effects of viral coinfection on virus transmission, further studies in natural populations are needed.
3.2. Effects on Viral Pathogenicity
Increased pathogenicity of viruses is another common result of viral coinfection. For example, FMDV does not generally kill adult sheep and goats [129]. However, when FMDV is coinfected with peste des petits ruminants virus (PPRV), the mortality rate increases to 50% [130]. The admission rate of coinfection in intensive care units was higher than that of single infection in human viral coinfection cases [131,132,133]. HBV and HCV coinfection causes more severe fibrosis and cirrhosis, as well as higher liver-related mortality, than single infection [134]. Coinfections of guinea pig reovirus and SARS-CoV cause rapid animal death in vivo [135]. Mice coinfected with Autochthonous Group 1 and 2 Brazilian VV showed more severe disease than mice infected with one virus alone [136].
However, not all viral coinfections will aggravate viral pathogenicity, and viral coinfection may not change or even alleviate the symptoms of a disease. Lanjuan Li et al. analyzed the impact of SARS-CoV-2 and IAV on the risk of disease severity in 9498 patients and found no significant association between SARS-CoV-2 and IAV coinfection mortality [137]. Additionally, Xiang et al. suggest that HBV infection does not increase the severity and outcome of COVID-19 [138].
Alleviation of symptoms is mainly reflected in the coinfection of respiratory viruses. RV can reduce the severity of IAV due to a faster reduction in the pulmonary inflammatory response and faster clearance of IAV [22]. Martinez-Roig, A et al. investigated coinfection of respiratory viruses in children and found that the number of viruses detected in nasopharyngeal aspirates was inversely proportional to the number of days of aerobic therapy and hospital stay [139].
Therefore, whether the virulence of a virus in coinfection changes seems to be related to the virus involved in the coinfection.
4. Study of Viral Coinfection
We summarized the study process of virus coinfection, as shown in Figure 3. The detailed methods are described below.
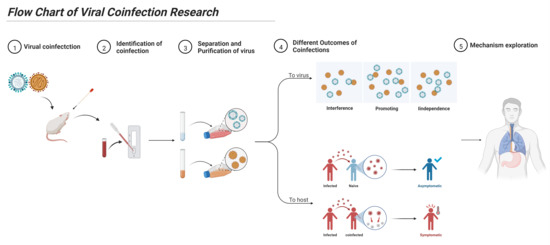
Figure 3.
Flow chart of viral coinfection research. For the study of viral coinfection, it is necessary to correctly identify the occurrence of coinfections, establish a corresponding coinfection virus isolation and detection system, and determine the type of viral coinfection and its effect on a host. Finally, the mechanism of viral coinfection can be explored.
4.1. Identification
The diagnosis of coinfection and the separation of viruses in coinfection samples are the bottlenecks in studies of coinfection [140,141,142]. The identification of coinfections is traceable, and is often accompanied by increased or decreased clinical symptoms [143] and abnormal clinical symptoms (higher mortality, neurological symptoms, immunosuppression, etc.); these cannot be explained by single-pathogen infection [135,141]. Coinfections tend to have similar means of transmission (respiratory tract [144], vector [141], blood [140], etc.) and a similar host tropism. On the other hand, viruses in coinfections are often highly contagious and cross-represented in epidemic areas [142,145]. Therefore, suspected cases of coinfections can be identified from the above aspects.
The diagnosis of viruses is based on serological evidence and viral isolation. However, the sensitivity of serological methods is low and different viruses sometimes cause similar serological responses [146,147]. Virus isolation requires a suitable cell line or animal model, and the presence of multiple viruses may interfere with the replication of the target virus [148,149].
The development and application of PCR, qPCR and ELISA make the diagnosis of coinfection much simpler. PCR technology enhances the sensitivity of viral identification. However, PCR primers require the sequence information of the target virus, so PCR cannot identify novel viruses or unknown virus subspecies. The application of qPCR and ELISA technology makes up for the deficiency of PCR. By selecting genes or amino acid sites with high degeneracy, the versatility of detection is greatly improved and insufficient information regarding the unknown virus may be found. Novel coinfection identification methods are summarized as follows:
- (i)
- The application of multiplex reverse-transcription quantitative real-time PCR (MRT-qPCR) [150,151,152], an improved version of qRT-PCR, makes coinfection detection more convenient and rapid. Its disadvantage is that building a new system takes a lot of time.
- (ii)
- Application of digital droplet PCR (ddPCR) makes it possible to identify two highly similar viruses [153]. This method improves the accuracy and sensitivity of coinfection detection.
- (iii)
- The transmission electron microscopy detection method of a gold nanoparticle gene probe also has applications in coinfection detection [154]. This method makes detection more convenient, which is conducive to clinical detection.
- (iv)
- Fayyadh et al. used multicolor imaging with self-assembled quantum dot probes to image and successfully detect H1N1, H3N2, and H9N2 influenza viruses in coinfected cells [155]. This method provides a basis for in vitro detection of coinfection, which is more direct and easier to operate than traditional detection.
- (v)
- Srisomwat et al. developed a point-of-care testing (POCT) device for HIV/HCV DNA detection [156]. Enhanced electroluminescence was observed in the presence of the target DNA by increasing proton conductivity [156]. This method has high specificity and a low cross-reaction for coinfection detection.
Although the identification of co-infections has been improved, the ability to detect target pathogens remains limited. The application of a next-generation sequencing (NGS) platform has improved virus diagnosis and the discovery of new viruses. NGS does not require prior sequence information about the target genome and can detect most potential genomes in clinical samples [157,158,159].
4.2. Viral Separation and Purification
Viral purification is extremely difficult in viral coinfection. In bacterial coinfection, different bacteria could be rapidly purified from a mixed culture by colony purification. On the other hand, multiple viruses cannot be easily purified directly from clinical samples. The isolation methods of viral coinfection mainly include CPE [130], organic solvent treatment (enveloped virus) [160], hemadsorption (separation of hemagglutination virus) [161], endpoint dilution assay [162], antibody (Ab) neutralization [163], acid/alkali treatment (PH-sensitive virus) [164], and reverse genetic system rescue [130]. The advantages and disadvantages of these methods are summarized below:
- (i)
- The purification of viruses by CPE is a mainstream method for virus isolation in coinfection, but it requires the selection of suitable cell lines, where one virus can produce obvious CPE while the other virus does not produce obvious CPE. The disadvantage of this method is whether or not some traditional virus isolation cell lines are sensitive to another virus, and coinfection may affect the formation of CPE. At present, it is feasible to separate snakehead retrovirus (SnRV) from grouper nervous necrosis virus (GNNV) by SGF-1 [165]; FMDV from PPRV [130] or single serotype FMDV from multiple serotypes ofFMDV [142] by BHK21; IAV from respiratory viruses by suspended MDCK cells (MDCK-S) and adherent MDCK cells (MDCK-A) [166]; porcine epidemic diarrhea (PEDV) from porcine kobuvirus 1 (PKV) by Vero cells [157]; Hepatitis E virus (HEV) from porcine sapelovirus (PSV) by N1380 cells [167]; and porcine circovirus 2 (PCV2) from porcine parvovirus (PPV) by PK-15 [168].
- (ii)
- An endpoint dilution assay is used to isolate two viruses with a highly similar host range/orientation but different replication rates. However, the separation success rate is usually low. It needs subsequent molecular-level detection and multi-generation blind passages for verification. Beperet et al. successfully isolated two different subtypes of alphabaculoviruses from coinfection samples by an endpoint dilution assay [162]. Dormitorio et al. successfully detected avian influenza virus (AIV) from suspicious allantoine fluid samples using this method [169].
- (iii)
- The Ab neutralization method is suitable for different serotype viruses or two viruses with a distant genetic relationship. This method has a high success rate, but it needs to be verified by subsequent multi-generation blind passages. For the coinfection of multiple serotypes of the same virus, the serotype is generally determined first, and then the 2-dimensional microneutralization test (2D-MNT) corresponding to the serotype is carried out. Mahajan used 2D-MNT to isolate and purify multiple serotype viruses from coinfection samples of FMDV [142].
For coinfection, corresponding antibodies should be used, such as neutralizing PPRV in the coinfection of FMDV and PPRV [130], neutralizing NDV in coinfection of AIV and NDV [170], neutralizing CSFV from CSFV, and porcine astrovirus 5 (PAstV5) coinfection samples [171].
- (iv)
- The organic solvent treatment method has certain limitations. Whether an organic solvent can kill one virus without affecting another virus needs to be verified. The choice of organic solvent is crucial. At present, it is feasible to remove PPRV with an organic solvent in coinfection of FMDV and PPRV [130]. The use of 5% H2O2 can completely inactivate the infectious laryngotracheitis virus, while the infectivity of NDV, infectious bronchitis virus, and AIV is reduced without being fully inactivated [172].
- (v)
- Hemadsorption is suitable for virus isolation from non-hemagglutinating viruses. The integrity of this method for virus isolation is uncertain and the virus needs to be transferred to susceptible cell lines for amplification. At present, it is feasible to remove PPRV in coinfections of FMDV and PPRV [130]. Hemadsorption is useful for viruses such as IAV, parainfluenza virus, and mumps virus, which express their hemagglutinin proteins on the plasma membrane of infected cells [161].
- (vi)
- Acid/alkali treatment is suitable for the separation of one PH-sensitive virus and another non-PH-sensitive virus. However, due to the difference in PH sensitivity of the isolated virus and the misdetection of molecular detection methods, this method has some notable limitations. Acidic environments (PH < 6.6) can effectively inhibit AIV replication [173]. The optimum survival range of the plague virus is from pH 6 to pH 11, while that of NDV is from pH 2 to pH 11 [174]. Thus, we can isolate viruses from coinfection samples by acid/alkali treatment.
- (vii)
- Reverse genetic system rescues viruses. Some viruses have a mature reverse genetics system. We can isolate the complete genome fragments of the virus from the positive samples and then obtain complete or defective viruses. The disadvantage of this method is that constructing the system necessitates a considerable workload, and it is not suitable for the separation of two related viruses.
The successful isolation of viruses also depends on the cells used for virus purification. Sometimes, a single type of cell is not enough to isolate the virus [130,175,176]. Co-culture cells, i.e., a culture of multiple cell types together in a single layer, can solve the problem of isolating multiple viruses [161,177]. The mixture of MRC-5 and A549 cells can be used to detect cytomegalovirus (CMV), herpes simplex virus (HSV), and adenovirus in the same sample [177]. Mink lung and human adenocarcinoma cells (R-Mix) can be used for the rapid isolation of respiratory viruses (parainfluenza 1,2 and 3, influenza A and B, RSV, adenovirus, HSV, CMV and enterovirus) [178,179,180,181,182]. R-Mix cells also help in isolating highly pathogenic respiratory viruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV). Another method currently being used is R-Mi Too cell line (composed of MDCK and A549 cells), which does not support SARS-CoV infection [183] but is more sensitive than R-Mix cells in the detection of influenza B virus and adenovirus [184]. Both R-Mix and R-Mix Too cells promote the growth of different influenza virus strains [185,186]. In addition, a mixture of MRC-5 and CV1 cells contributes to multiple detections of HSV-1, HSV-2, and varicella-zoster virus (VZV) [187,188]. Finally, Vero/BHK-21 co-culture cells could simultaneously isolate PPRV and FMDV [130]. However, the cost of co-culture cells is usually much higher than that of a single cell culture.
5. Conclusions
Viral coinfection is common but complicated. Studies of coinfection and virus–virus interactions represent an emerging field in virology. Due to coinfections, one virus infection could impact the outcome of another virus. A faster viral coinfection detection and virus separation system should be established for further study. In the future, viral coinfection studies will improve diagnoses, the development of vaccines, and antiviral therapy.
To make it easier to study coinfections, we have outlined some of the literature on viral coinfections so that individuals can better select particular viruses of interest (Table 1).

Table 1.
Summary of viral coinfection *.
Author Contributions
C.W.: writing—original draft preparation. Y.D.: writing— review and editing. Y.Z.: review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financed by the National Key Research and Development Program of China (2021YFD1800200), National Natural Science Foundation of China (32000357, 32170539), Key Research and Development Program of Liaoning (2020JH2/10200035) and Liao Ning Revitalization Talents Program (XLYC2007114).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Salas-Benito, J.S.; De Nova-Ocampo, M. Viral Interference and Persistence in Mosquito-Borne Flaviviruses. J. Immunol. Res. 2015, 2015, 873404. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-J.; Lee, S.-D. Hepatitis B virus/hepatitis C virus coinfection: Epidemiology, clinical features, viral interactions and treatment. J. Gastroenterol. Hepatol. 2008, 23, 512–520. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Nakajima, N.; Sato, Y.; Takahashi, K.; Accola, M.; Chiba, S.; Fan, S.; Neumann, G.; Rehrauer, W.; Suzuki, T.; et al. SARS-CoV-2 Interference of Influenza Virus Replication in Syrian Hamsters. J. Infect. Dis. 2022, 225, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.D.; Sordillo, E.M.; Gitman, M.R.; Paniz Mondolfi, A.E. Coinfection in SARS-CoV-2 infected patients: Where are influenza virus and rhinovirus/enterovirus? J. Med. Virol. 2020, 92, 1699–1700. [Google Scholar] [CrossRef]
- Rojek, J.M.; Campbell, K.P.; Oldstone, M.B.; Kunz, S. Old World arenavirus infection interferes with the expression of functional alpha-dystroglycan in the host cell. Mol. Biol. Cell 2007, 18, 4493–4507. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.C.; Li, W.; Sui, J.; Marasco, W.; Choe, H.; Farzan, M. Influenza A virus neuraminidase limits viral superinfection. J. Virol. 2008, 82, 4834–4843. [Google Scholar] [CrossRef]
- Muñoz-González, S.; Perez-Simó, M.; Muñoz, M.; Bohorquez, J.A.; Rosell, R.; Summerfield, A.; Domingo, M.; Ruggli, N.; Ganges, L. Efficacy of a live attenuated vaccine in classical swine fever virus postnatally persistently infected pigs. Vet. Res. 2015, 46, 78. [Google Scholar] [CrossRef]
- Muñoz-González, S.; Pérez-Simó, M.; Colom-Cadena, A.; Cabezón, O.; Bohórquez, J.A.; Rosell, R.; Pérez, L.J.; Marco, I.; Lavín, S.; Domingo, M.; et al. Classical Swine Fever Virus vs. Classical Swine Fever Virus: The Superinfection Exclusion Phenomenon in Experimentally Infected Wild Boar. PLoS ONE 2016, 11, e0149469. [Google Scholar] [CrossRef]
- Goto, H.; Ihira, H.; Morishita, K.; Tsuchiya, M.; Ohta, K.; Yumine, N.; Tsurudome, M.; Nishio, M. Enhanced growth of influenza A virus by coinfection with human parainfluenza virus type 2. Med. Microbiol. Immunol. 2016, 205, 209–218. [Google Scholar] [CrossRef]
- Kent, R.J.; Crabtree, M.B.; Miller, B.R. Transmission of West Nile Virus by Culex quinquefasciatus Say Infected with Culex Flavivirus Izabal. PLoS Negl. Trop. Dis. 2010, 4, e671. [Google Scholar] [CrossRef]
- Bellecave, P.; Gouttenoire, J.; Gajer, M.; Brass, V.; Koutsoudakis, G.; Blum, H.E.; Bartenschlager, R.; Nassal, M.; Moradpour, D. Hepatitis B and C virus coinfection: A novel model system reveals the absence of direct viral interference. Hepatology 2009, 50, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Kanthong, N.; Khemnu, N.; Sriurairatana, S.; Pattanakitsakul, S.-N.; Malasit, P.; Flegel, T.W. Mosquito cells accommodate balanced, persistent co-infections with a densovirus and Dengue virus. Dev. Comp. Immunol. 2008, 32, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- López-Vázquez, C.; Alonso, M.C.; Dopazo, C.P.; Bandín, I. In vivo study of viral haemorrhagic septicaemia virus and infectious pancreatic necrosis virus coexistence in Senegalese sole (Solea senegalensis). J. Fish Dis. 2017, 40, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, P.; Chakraborty, S. Biology of viral satellites and their role in pathogenesis. Curr. Opin. Virol. 2018, 33, 96–105. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. The Subviral Agents. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 1211–1219. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Genus-Umbravirus. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 1191–1195. [Google Scholar] [CrossRef]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Haller, O. Jean Lindenmann: From viral interference to interferon and beyond (1924–2015). J. Interferon Cytokine Res. 2015, 35, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Dianzani, F. Viral interference and interferon. Ric. Clin. Lab. 1975, 5, 196–213. [Google Scholar] [CrossRef] [PubMed]
- Essaidi-Laziosi, M.; Geiser, J.; Huang, S.; Constant, S.; Kaiser, L.; Tapparel, C. Interferon-Dependent and Respiratory Virus-Specific Interference in Dual Infections of Airway Epithelia. Sci. Rep. 2020, 10, 10246. [Google Scholar] [CrossRef] [PubMed]
- Cribier, B.; Schmitt, C.; Rey, D.; Lang, J.M.; Kirn, A.; Stoll-Keller, F. Role of endogenous interferon in hepatitis C virus (HCV) infection and in coinfection by HIV and HCV. Res. Virol. 1996, 147, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.J.; Ijezie, E.C.; Balemba, O.B.; Miura, T.A. Attenuation of Influenza A Virus Disease Severity by Viral Coinfection in a Mouse Model. J. Virol. 2018, 92, e00881-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.S.; Olfat, F.; Phoon, M.C.; Hsu, J.P.; Howe, J.L.C.; Seet, J.E.; Chin, K.C.; Chow, V.T.K. In vivo and in vitro studies on the antiviral activities of viperin against influenza H1N1 virus infection. J. Gen. Virol. 2012, 93, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011, 10, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J. The antiviral activities of tetherin. Curr. Top. Microbiol. Immunol. 2013, 371, 67–104. [Google Scholar] [CrossRef] [PubMed]
- Lenschow, D.J. Antiviral Properties of ISG15. Viruses 2010, 2, 2154–2168. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, J.B.; Moran, J.V. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015, 11, e1005121. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Parise, M.C.; Ringeard, M.; Chelbi-Alix, M.K.; Hazan, U.; Nisole, S. Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology 2008, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Huang, J.; Zhang, C.; Huang, S.; Nunnari, G.; Wang, F.X.; Tong, X.; Gao, L.; Nikisher, K.; Zhang, H. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 2006, 80, 7645–7657. [Google Scholar] [CrossRef] [PubMed]
- Birdwell, L.D.; Zalinger, Z.B.; Li, Y.; Wright, P.W.; Elliott, R.; Rose, K.M.; Silverman, R.H.; Weiss, S.R. Activation of RNase L by Murine Coronavirus in Myeloid Cells Is Dependent on Basal Oas Gene Expression and Independent of Virus-Induced Interferon. J. Virol. 2016, 90, 3160–3172. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Nagata, K.; Kato, A.; Ishihama, A. Interferon-inducible mouse Mx1 protein that confers resistance to influenza virus is GTPase. J. Biol. Chem. 1991, 266, 21404–21408. [Google Scholar] [CrossRef] [PubMed]
- Streitenfeld, H.; Boyd, A.; Fazakerley, J.K.; Bridgen, A.; Elliott, R.M.; Weber, F. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J. Virol. 2003, 77, 5507–5511. [Google Scholar] [CrossRef]
- Lalle, E.; Sacchi, A.; Abbate, I.; Vitale, A.; Martini, F.; D’Offizi, G.; Antonucci, G.; Castilletti, C.; Poccia, F.; Capobianchi, M.R. Activation of interferon response genes and of plasmacytoid dendritic cells in HIV-1 positive subjects with GB virus C co-infection. Int. J. Immunopathol. Pharmacol. 2008, 21, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lv, J.; Liu, J.; Li, M.; Xie, J.; Lv, Q.; Deng, W.; Zhou, N.; Zhou, Y.; Song, J.; et al. Mucus production stimulated by IFN-AhR signaling triggers hypoxia of COVID-19. Cell Res. 2020, 30, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Leuven, J.T.V.; Gonzalez, A.J.; Ijezie, E.C.; Wixom, A.Q.; Clary, J.L.; Naranjo, M.N.; Ridenhour, B.J.; Miller, C.R.; Miura, T.A. Rhinovirus Reduces the Severity of Subsequent Respiratory Viral Infections by Interferon-Dependent and -Independent Mechanisms. mSphere 2021, 6, e00479-21. [Google Scholar] [CrossRef]
- Polacino, P.; Kaplan, G.; Palma, E.L. Homologous interference by a foot-and-mouth disease virus strain attenuated for cattle. Arch. Virol. 1985, 86, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.H.; Brown, D.T. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 1985, 54, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, A.; Koo, R.; Stollar, V. Evolution and properties of Aedes albopictus cell cultures persistently infected with sindbis virus. Virology 1977, 82, 69–83. [Google Scholar] [CrossRef]
- Karpf, A.R.; Lenches, E.; Strauss, E.G.; Strauss, J.H.; Brown, D.T. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 1997, 71, 7119–7123. [Google Scholar] [CrossRef]
- Marcus, P.I.; Carver, D.H. Intrinsic interference: A new type of viral interference. J. Virol. 1967, 1, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Zebovitz, E.; Brown, A. Interference among group A arboviruses. J. Virol. 1968, 2, 1283–1289. [Google Scholar] [CrossRef]
- Brinton, M.A. Host factors involved in West Nile virus replication. Ann. N. Y. Acad. Sci. 2001, 951, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Riis, B.; Rattan, S.I.; Clark, B.F.; Merrick, W.C. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990, 15, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wei, T.; Abbott, C.M.; Harrich, D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. MMBR 2013, 77, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kelen, K.; Beyaert, R.; Inzé, D.; De Veylder, L. Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 143–168. [Google Scholar] [CrossRef] [PubMed]
- De Nova-Ocampo, M.; Villegas-Sepúlveda, N.; del Angel, R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3’ untranslated region of dengue 4 virus RNA. Virology 2002, 295, 337–347. [Google Scholar] [CrossRef]
- Davis, W.G.; Blackwell, J.L.; Shi, P.Y.; Brinton, M.A. Interaction between the cellular protein eEF1A and the 3’-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007, 81, 10172–10187. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. Translation elongation factor-1 alpha interacts with the 3’ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997, 71, 6433–6444. [Google Scholar] [CrossRef]
- Yocupicio-Monroy, M.; Padmanabhan, R.; Medina, F.; del Angel, R.M. Mosquito La protein binds to the 3’ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 2007, 357, 29–40. [Google Scholar] [CrossRef] [PubMed]
- García-Montalvo, B.M.; Medina, F.; del Angel, R.M. La protein binds to NS5 and NS3 and to the 5’ and 3’ ends of Dengue 4 virus RNA. Virus Res. 2004, 102, 141–150. [Google Scholar] [CrossRef]
- Gomila, R.C.; Martin, G.W.; Gehrke, L. NF90 binds the dengue virus RNA 3’ terminus and is a positive regulator of dengue virus replication. PLoS ONE 2011, 6, e16687. [Google Scholar] [CrossRef]
- Kenney, J.L.; Solberg, O.D.; Langevin, S.A.; Brault, A.C. Characterization of a novel insect-specific flavivirus from Brazil: Potential for inhibition of infection of arthropod cells with medically important flaviviruses. J. Gen. Virol. 2014, 95, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.R.; Suomalainen, M.; Varadarajan, S.; Garoff, H.; Helenius, A. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology 1997, 231, 59–71. [Google Scholar] [CrossRef]
- New, C.; Lee, Z.-Y.; Tan, K.S.; Wong, A.H.; Wang, D.Y.; Tran, T. Tetraspanins: Host Factors in Viral Infections. Int. J. Mol. Sci. 2021, 22, 11609. [Google Scholar] [CrossRef]
- Kim, G.N.; Kang, C.Y. Utilization of homotypic and heterotypic proteins of vesicular stomatitis virus by defective interfering particle genomes for RNA replication and virion assembly: Implications for the mechanism of homologous viral interference. J. Virol. 2005, 79, 9588–9596. [Google Scholar] [CrossRef]
- Chotkowski, H.L.; Ciota, A.T.; Jia, Y.; Puig-Basagoiti, F.; Kramer, L.D.; Shi, P.Y.; Glaser, R.L. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology 2008, 377, 197–206. [Google Scholar] [CrossRef]
- Sánchez-Vargas, I.; Scott, J.C.; Poole-Smith, B.K.; Franz, A.W.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009, 5, e1000299. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Jupatanakul, N.; Ramirez, J.L.; Kang, S.; Romero-Vivas, C.M.; Mohammed, H.; Dimopoulos, G. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl. Trop. Dis. 2013, 7, e2295. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hong, H.; Yue, J.; Wu, Y.; Li, X.; Jiang, L.; Li, L.; Li, Q.; Gao, G.; Yang, X. Inhibitory effect of small interfering RNA on dengue virus replication in mosquito cells. Virol. J. 2010, 7, 270. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P. Flavivirus RNAi suppression: Decoding non-coding RNA. Curr. Opin. Virol. 2014, 7, 55–60. [Google Scholar] [CrossRef]
- Lemm, J.A.; Rice, C.M. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 1993, 67, 1916–1926. [Google Scholar] [CrossRef] [PubMed]
- Lemm, J.A.; Rümenapf, T.; Strauss, E.G.; Strauss, J.H.; Rice, C.M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994, 13, 2925–2934. [Google Scholar] [CrossRef]
- Shirako, Y.; Strauss, J.H. Regulation of Sindbis virus RNA replication: Uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994, 68, 1874–1885. [Google Scholar] [CrossRef]
- Nunes, F.M.; Aleixo, A.C.; Barchuk, A.R.; Bomtorin, A.D.; Grozinger, C.M.; Simões, Z.L. Non-Target Effects of Green Fluorescent Protein (GFP)-Derived Double-Stranded RNA (dsRNA-GFP) Used in Honey Bee RNA Interference (RNAi) Assays. Insects 2013, 4, 90–103. [Google Scholar] [CrossRef]
- Flenniken, M.L.; Andino, R. Non-Specific dsRNA-Mediated Antiviral Response in the Honey Bee. PLoS ONE 2013, 8, e77263. [Google Scholar] [CrossRef] [PubMed]
- Steck, F.T.; Rubin, H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 1966, 29, 642–653. [Google Scholar] [CrossRef]
- Le Guern, M.; Levy, J.A. Human immunodeficiency virus (HIV) type 1 can superinfect HIV-2-infected cells: Pseudotype virions produced with expanded cellular host range. Proc. Natl. Acad. Sci. USA 1992, 89, 363–367. [Google Scholar] [CrossRef]
- Michel, N.; Allespach, I.; Venzke, S.; Fackler, O.T.; Keppler, O.T. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. CB 2005, 15, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Hrecka, K.; Swigut, T.; Schindler, M.; Kirchhoff, F.; Skowronski, J. Nef proteins from diverse groups of primate lentiviruses downmodulate CXCR4 to inhibit migration to the chemokine stromal derived factor 1. J. Virol. 2005, 79, 10650–10659. [Google Scholar] [CrossRef] [PubMed]
- Geleziunas, R.; Bour, S.; Wainberg, M.A. Cell surface down-modulation of CD4 after infection by HIV-1. FASEB J. 1994, 8, 593–600. [Google Scholar] [CrossRef]
- Willey, R.L.; Maldarelli, F.; Martin, M.A.; Strebel, K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992, 66, 7193–7200. [Google Scholar] [CrossRef]
- Breiner, K.M.; Schaller, H.; Knolle, P.A. Endothelial cell-mediated uptake of a hepatitis B virus: A new concept of liver targeting of hepatotropic microorganisms. Hepatology 2001, 34, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Nethe, M.; Berkhout, B.; van der Kuyl, A.C. Retroviral superinfection resistance. Retrovirology 2005, 2, 52. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J.; Schnorr, J.J.; Brinckmann, U.; Dunster, L.M.; Baczko, K.; Liebert, U.G.; Schneider-Schaulies, S.; ter Meulen, V. Receptor usage and differential downregulation of CD46 by measles virus wild-type and vaccine strains. Proc. Natl. Acad. Sci. USA 1995, 92, 3943–3947. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Joyce, M.A.; Addison, W.R.; Fischer, K.P.; Tyrrell, D.L. Superinfection exclusion in duck hepatitis B virus infection is mediated by the large surface antigen. J. Virol. 2004, 78, 7925–7937. [Google Scholar] [CrossRef] [PubMed]
- Bratt, M.A.; Rubin, H. Specific interference among strains of Newcastle disease virus. II. Comparison of interference by active and inactive virus. Virology 1968, 35, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Benson, R.E.; Sanfridson, A.; Ottinger, J.S.; Doyle, C.; Cullen, B.R. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 1993, 177, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Rikkonen, M.; Peränen, J.; Kääriäinen, L. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 1992, 189, 462–473. [Google Scholar] [CrossRef]
- Ranki, M.; Ulmanen, I.; Kääriäinen, L. Semliki Forest virus-specific nonstructural protein is associated with ribosomes. FEBS Lett. 1979, 108, 299–302. [Google Scholar] [CrossRef]
- Simon, K.O.; Cardamone, J.J., Jr.; Whitaker-Dowling, P.A.; Youngner, J.S.; Widnell, C.C. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology 1990, 177, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Whitaker-Dowling, P.; Youngner, J.S.; Widnell, C.C.; Wilcox, D.K. Superinfection exclusion by vesicular stomatitis virus. Virology 1983, 131, 137–143. [Google Scholar] [CrossRef]
- Zou, G.; Zhang, B.; Lim, P.Y.; Yuan, Z.; Bernard, K.A.; Shi, P.Y. Exclusion of West Nile virus superinfection through RNA replication. J. Virol. 2009, 83, 11765–11776. [Google Scholar] [CrossRef] [PubMed]
- Geib, T.; Sauder, C.; Venturelli, S.; Hässler, C.; Staeheli, P.; Schwemmle, M. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 2003, 77, 4283–4290. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, V.; Hoffmann, S.; Herian, U.; Penin, F.; Bartenschlager, R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 2003, 77, 3007–3019. [Google Scholar] [CrossRef] [PubMed]
- Schaller, T.; Appel, N.; Koutsoudakis, G.; Kallis, S.; Lohmann, V.; Pietschmann, T.; Bartenschlager, R. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 2007, 81, 4591–4603. [Google Scholar] [CrossRef] [PubMed]
- Claus, C.; Tzeng, W.P.; Liebert, U.G.; Frey, T.K. Rubella virus-induced superinfection exclusion studied in cells with persisting replicons. J. Gen. Virol. 2007, 88, 2769–2773. [Google Scholar] [CrossRef] [PubMed]
- Whitaker-Dowling, P.; Youngner, J.S. Viral interference-dominance of mutant viruses over wild-type virus in mixed infections. Microbiol. Rev. 1987, 51, 179–191. [Google Scholar] [CrossRef]
- Beperet, I.; Irons, S.L.; Simón, O.; King, L.A.; Williams, T.; Possee, R.D.; López-Ferber, M.; Caballero, P. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J. Virol. 2014, 88, 3548–3556. [Google Scholar] [CrossRef]
- Frielle, D.W.; Kim, P.B.; Keene, J.D. Inhibitory effects of vesicular stomatitis virus on cellular and influenza viral RNA metabolism and protein synthesis. Virology 1989, 172, 274–284. [Google Scholar] [CrossRef]
- Norkin, L.C. Persistent infections of green monkey kidney cells initiated with temperature-sensitive mutants of simian virus 40. Virology 1980, 107, 375–388. [Google Scholar] [CrossRef]
- Norkin, L.C. Rhesus monkeys kidney cells persistently infected with Simian Virus 40: Production of defective interfering virus and acquisition of the transformed phenotype. Infect. Immun. 1976, 14, 783–792. [Google Scholar] [CrossRef]
- Ahmed, R.; Chakraborty, P.R.; Fields, B.N. Genetic variation during lytic reovirus infection: High-passage stocks of wild-type reovirus contain temperature-sensitive mutants. J. Virol. 1980, 34, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.M.; Wilkie, M.L. Persistent infection of Aedes albopictus C6/36 cells by Bunyamwera virus. Virology 1986, 150, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Kowal, K.J.; Stollar, V. Differential sensitivity of infectious and defective-interfering particles of Sindbis virus to ultraviolet irradiation. Virology 1980, 103, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Peleg, J.; Stollar, V. Homologous interference in Aedes aegypti cell cultures infected with Sindbis virus. Arch. Fur Die Gesamte Virusforsch. 1974, 45, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Shenk, T.E.; Koshelnyk, K.A.; Stollar, V. Temperature-sensitive virus from Aedes albopictus cells chronically infected with Sindbis virus. J. Virol. 1974, 13, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Ju, G.; Birrer, M.; Udem, S.; Bloom, B.R. Complementation analysis of measles virus mutants isolated from persistently infected lymphoblastoid cell lines. J. Virol. 1980, 33, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Frielle, D.W.; Huang, D.D.; Youngner, J.S. Persistent infection with influenza A virus: Evolution of virus mutants. Virology 1984, 138, 103–117. [Google Scholar] [CrossRef]
- Sivaram, A.; Barde, P.V.; Gokhale, M.D.; Singh, D.K.; Mourya, D.T. Evidence of co-infection of chikungunya and densonucleosis viruses in C6/36 cell lines and laboratory infected Aedes aegypti (L.) mosquitoes. Parasites Vectors 2010, 3, 95. [Google Scholar] [CrossRef]
- Wei, W.; Shao, D.; Huang, X.; Li, J.; Chen, H.; Zhang, Q.; Zhang, J. The pathogenicity of mosquito densovirus (C6/36DNV) and its interaction with dengue virus type II in Aedes albopictus. Am. J. Trop. Med. Hyg. 2006, 75, 1118–1126. [Google Scholar] [CrossRef]
- Burivong, P.; Pattanakitsakul, S.-N.; Thongrungkiat, S.; Malasit, P.; Flegel, T.W. Markedly reduced severity of Dengue virus infection in mosquito cell cultures persistently infected with Aedes albopictus densovirus (AalDNV). Virology 2004, 329, 261–269. [Google Scholar] [CrossRef]
- Dasgupta, R.; Selling, B.; Rueckert, R. Flock house virus: A simple model for studying persistent infection in cultured Drosophila cells. Arch. Virology. Suppl. 1994, 9, 121–132. [Google Scholar] [CrossRef]
- Rechavi, O.; Minevich, G.; Hobert, O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 2011, 147, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, R.; Free, H.M.; Zietlow, S.L.; Paskewitz, S.M.; Aksoy, S.; Shi, L.; Fuchs, J.; Hu, C.; Christensen, B.M. Replication of flock house virus in three genera of medically important insects. J. Med. Entomol. 2007, 44, 102–110. [Google Scholar] [CrossRef]
- Goic, B.; Vodovar, N.; Mondotte, J.A.; Monot, C.; Frangeul, L.; Blanc, H.; Gausson, V.; Vera-Otarola, J.; Cristofari, G.; Saleh, M.C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 2013, 14, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Whitaker-Dowling, P.; Youngner, J.S. Vaccinia rescue of VSV from interferon-induced resistance: Reversal of translation block and inhibition of protein kinase activity. Virology 1983, 131, 128–136. [Google Scholar] [CrossRef]
- Cheng, X.M.; Uchida, T.; Xia, Y.C.; Umarova, R.; Liu, C.J.; Chen, P.J.; Gaggar, A.; Suri, V.; Mucke, M.M.; Vermehren, J.; et al. Diminished hepatic IFN response following HCV clearance triggers HBV reactivation in coinfection. J. Clin. Investig. 2020, 130, 3205–3220. [Google Scholar] [CrossRef]
- Hao, X.Q.; Li, Y.C.; Chen, H.; Chen, B.; Liu, R.H.; Wu, Y.D.; Xiao, X.Y.; Zhou, P.; Li, S.J. Canine Circovirus Suppresses the Type I Interferon Response and Protein Expression but Promotes CPV-2 Replication. Int. J. Mol. Sci. 2022, 23, 6382. [Google Scholar] [CrossRef]
- He, B.; Paterson, R.G.; Stock, N.; Durbin, J.E.; Durbin, R.K.; Goodbourn, S.; Randall, R.E.; Lamb, R.A. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: The multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 2002, 303, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Wansley, E.K.; Grayson, J.M.; Parks, G.D. Apoptosis induction and interferon signaling but not IFN-β promoter induction by an SV5 P/V mutant are rescued by coinfection with wild-type SV5. Virology 2003, 316, 41–54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Du, X.; Zhou, D.; Zhou, J.; Xue, J.; Cheng, Z. Marek’s Disease Virus and Reticuloendotheliosis Virus Coinfection Enhances Viral Replication and Alters Cellular Protein Profiles. Front. Vet. Sci. 2022, 9, 854007. [Google Scholar] [CrossRef]
- Li, L.; Zhuang, P.P.; Cheng, Z.Q.; Yang, J.; Bi, J.M.; Wang, G.H. Avian leukosis virus subgroup J and reticuloendotheliosis virus coinfection induced TRIM62 regulation of the actin cytoskeleton. J. Vet. Sci. 2020, 21, e49. [Google Scholar] [CrossRef] [PubMed]
- McCall, L.I.; Siqueira-Neto, J.L.; McKerrow, J.H. Location, Location, Location: Five Facts about Tissue Tropism and Pathogenesis. PLoS Pathog. 2016, 12, e1005519. [Google Scholar] [CrossRef] [PubMed]
- Ure, A.E.; Forslund, O. Characterization of Human Papillomavirus Type 154 and Tissue Tropism of Gammapapillomaviruses. PLoS ONE 2014, 9, e89342. [Google Scholar] [CrossRef] [PubMed]
- Flegel, T.W. Update on viral accommodation, a model for host-viral interaction in shrimp and other arthropods. Dev. Comp. Immunol. 2007, 31, 217–231. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef]
- Johansson, M.W.; Söderhäll, K. The prophenoloxidase activating system and associated proteins in invertebrates. Prog. Mol. Subcell. Biol. 1996, 15, 46–66. [Google Scholar] [CrossRef]
- Namikoshi, A.; Wu, J.L.; Yamashita, T.; Nishizawa, T.; Nishioka, T.; Arimoto, M.; Muroga, K. Vaccination trials with Penaeus japonicus to induce resistance to white spot syndrome virus. Aquaculture 2004, 229, 25–35. [Google Scholar] [CrossRef]
- Venegas, C.A.; Nonaka, L.; Mushiake, K.; Nishizawa, T.; Murog, K. Quasi-immune response of Penaeus japonicus to penaeid rod-shaped DNA virus (PRDV). Dis. Aquat. Org. 2000, 42, 83–89. [Google Scholar] [CrossRef]
- Khanobdee, K.; Soowannayan, C.; Flegel, T.W.; Ubol, S.; Withyachumnarnkul, B. Evidence for apoptosis correlated with mortality in the giant black tiger shrimp Penaeus monodon infected with yellow head virus. Dis. Aquat. Org. 2002, 48, 79–90. [Google Scholar] [CrossRef][Green Version]
- Sahtout, A.H.; Hassan, M.D.; Shariff, M. DNA fragmentation, an indicator of apoptosis, in cultured black tiger shrimp Penaeus monodon infected with white spot syndrome virus (WSSV). Dis. Aquat. Org. 2001, 44, 155–159. [Google Scholar] [CrossRef]
- Wongprasert, K.; Khanobdee, K.; Glunukarn, S.S.; Meeratana, P.; Withyachumnarnkul, B. Time-course and levels of apoptosis in various tissues of black tiger shrimp Penaeus monodon infected with white-spot syndrome virus. Dis. Aquat. Org. 2003, 55, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Shell, E.J.; Fokam, E.B.; Mason, P.W.; Hanley, K.A.; Estes, D.M.; Weaver, S.C. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology 2007, 358, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Rudnick, A.; Chan, Y.C. Dengue Type 2 Virus in Naturally Infected Aedes albopictus Mosquitoes in Singapore. Science 1965, 149, 638–639. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Holmes, E.C.; Fokam, E.B.; Faye, O.; Diallo, M.; Sall, A.A.; Weaver, S.C. Evolutionary processes among sylvatic dengue type 2 viruses. J. Virol. 2007, 81, 9591–9595. [Google Scholar] [CrossRef] [PubMed]
- Linde, A.; Rotzén-Östlund, M.; Zweygberg-Wirgart, B.; Rubinova, S.; Brytting, M. Does viral interference affect spread of influenza? Eurosurveillance 2009, 14, 19354. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, T.; Robison, A.; Young, M.C.; Black, W.C.; Foy, B.D.; Ebel, G.D.; Ruckert, C. Sequential Infection of Aedes aegypti Mosquitoes with Chikungunya Virus and Zika Virus Enhances Early Zika Virus Transmission. Insects 2018, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Wernery, U.; Kaaden, O.R. Foot-and-mouth disease in camelids: A review. Vet. J. 2004, 168, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Barua, S.; Riyesh, T.; Chaubey, K.K.; Rawat, K.D.; Khandelwal, N.; Mishra, A.K.; Sharma, N.; Chandel, S.S.; Sharma, S.; et al. Complexities in Isolation and Purification of Multiple Viruses from Mixed Viral Infections: Viral Interference, Persistence and Exclusion. PLoS ONE 2016, 11, e0156110. [Google Scholar] [CrossRef]
- Goka, E.A.; Vallely, P.J.; Mutton, K.J.; Klapper, P.E. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol. Infect. 2015, 143, 37–47. [Google Scholar] [CrossRef]
- Goka, E.; Vallely, P.; Mutton, K.; Klapper, P. Influenza A viruses dual and multiple infections with other respiratory viruses and risk of hospitalisation and mortality. Influenza Other Respir Viruses 2013, 7, 1079–1087. [Google Scholar] [CrossRef]
- Tang, M.B.; Yu, C.P.; Chen, S.C.; Chen, C.H. Co-Infection of Adenovirus, Norovirus and Torque Teno Virus in Stools of Patients with Acute Gastroenteritis. Southeast Asian J. Trop. Med. Public Health 2014, 45, 1326–1336. [Google Scholar] [PubMed]
- Amin, J.; Law, M.G.; Bartlett, M.; Kaldor, J.M.; Dore, G.J. Causes of death after diagnosis of hepatitis B or hepatitis C infection: A large community-based linkage study. Lancet 2006, 368, 938–945. [Google Scholar] [CrossRef]
- Liang, L.; He, C.; Lei, M.; Li, S.; Hao, Y.; Zhu, H.; Duan, Q. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol. 2005, 24, 485–490. [Google Scholar] [CrossRef]
- Calixto, R.; Oliveira, G.; Lima, M.; Andrade, A.C.; Trindade, G.D.; De Oliveira, D.B.; Kroon, E.G. A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies. Viruses 2018, 10, 15. [Google Scholar] [CrossRef]
- Guan, Z.; Chen, C.; Li, Y.; Yan, D.; Zhang, X.; Jiang, D.; Yang, S.; Li, L. Impact of Coinfection With SARS-CoV-2 and Influenza on Disease Severity: A Systematic Review and Meta-Analysis. Front. Public Heal. 2021, 9, 773130. [Google Scholar] [CrossRef]
- Xiang, T.D.; Zheng, X. Interaction between hepatitis B virus and SARS-CoV-2 infections. World J. Gastroenterol. 2021, 27, 782–793. [Google Scholar] [CrossRef]
- Martinez-Roig, A.; Salvado, M.; Caballero-Rabasco, M.A.; Sanchez-Buenavida, A.; Lopez-Segura, N.; Bonet-Alcaina, M. Viral Coinfection in Childhood Respiratory Tract Infections. Arch. Bronconeumol. 2015, 51, 5–9. [Google Scholar] [CrossRef]
- Zeremski, M.; Martinez, A.D.; Talal, A.H. Editorial Commentary: Management of Hepatitis C Virus in HIV-Infected Patients in the Era of Direct-Acting Antivirals. Clin. Infect. Dis. 2014, 58, 880–882. [Google Scholar] [CrossRef]
- Mercado-Reyes, M.; Acosta-Reyes, J.; Navarro-Lechuga, E.; Corchuelo, S.; Rico, A.; Parra, E.; Tolosa, N.; Pardo, L.; González, M.; Martìn-Rodriguez-Hernández, J.; et al. Dengue, chikungunya and zika virus coinfection: Results of the national surveillance during the zika epidemic in Colombia. Epidemiol. Infect. 2019, 147, e77. [Google Scholar] [CrossRef]
- Mahajan, S.; Sharma, G.K.; Subramaniam, S.; Biswal, J.K.; Pattnaik, B. Selective isolation of foot-and-mouth disease virus from coinfected samples containing more than one serotype. Braz. J. Microbiol. 2021, 52, 2447–2454. [Google Scholar] [CrossRef]
- Scotta, M.C.; Chakr, V.C.; de Moura, A.; Becker, R.G.; de Souza, A.P.; Jones, M.H.; Pinto, L.A.; Sarria, E.E.; Pitrez, P.M.; Stein, R.T.; et al. Respiratory viral coinfection and disease severity in children: A systematic review and meta-analysis. J. Clin. Virol. 2016, 80, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Semple, M.G.; Cowell, A.; Dove, W.; Greensill, J.; McNamara, P.S.; Halfhide, C.; Shears, P.; Smyth, R.L.; Hart, C.A. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J. Infect. Dis. 2005, 191, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Faye, O.; Prot, M.; Casademont, I.; Fall, G.; Fernandez-Garcia, M.D.; Diagne, M.M.; Kipela, J.M.; Fall, I.S.; Holmes, E.C.; et al. Autochthonous Japanese Encephalitis with Yellow Fever Coinfection in Africa. New Engl. J. Med. 2017, 376, 1483–1485. [Google Scholar] [CrossRef] [PubMed]
- Salassa, B.; Daziano, E.; Bonino, F.; Lavarini, C.; Smedile, A.; Chiaberge, E.; Rosina, F.; Brunetto, M.R.; Pessione, E.; Spezia, C. Serological diagnosis of hepatitis B and delta virus (HBV/HDV) coinfection. J. Hepatol. 1991, 12, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Masyeni, S.; Santoso, M.S.; Widyaningsih, P.D.; Asmara, D.G.W.; Nainu, F.; Harapan, H.; Sasmono, R.T. Serological cross-reaction and coinfection of dengue and COVID-19 in Asia: Experience from Indonesia. Int. J. Infect. Dis. 2021, 102, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Costa-Hurtado, M.; Afonso, C.L.; Miller, P.J.; Shepherd, E.; DeJesus, E.; Smith, D.; Pantin-Jackwood, M.J. Effect of Infection with a Mesogenic Strain of Newcastle Disease Virus on Infection with Highly Pathogenic Avian Influenza Virus in Chickens. Avian. Dis. 2016, 60, 269–278. [Google Scholar] [CrossRef]
- Bara, J.J.; Muturi, E.J. Effect of mixed infections of Sindbis and La Crosse viruses on replication of each virus in vitro. Acta Trop. 2014, 130, 71–75. [Google Scholar] [CrossRef]
- Li, X.; Zhang, K.R.; Pei, Y.; Xue, J.; Ruan, S.F.; Zhang, G.Z. Development and Application of an MRT-qPCR Assay for Detecting Coinfection of Six Vertically Transmitted or Immunosuppressive Avian Viruses. Front. Microbiol. 2020, 11, 1581. [Google Scholar] [CrossRef]
- Xu, Z.X.; Peng, Y.; Yang, M.H.; Li, X.H.; Wang, J.; Zou, R.R.; Liang, J.H.; Fang, S.S.; Liu, Y.X.; Yang, Y. Simultaneous detection of Zika, chikungunya, dengue, yellow fever, West Nile, and Japanese encephalitis viruses by a two-tube multiplex real-time RT-PCR assay. J. Med. Virol. 2022, 94, 2528–2536. [Google Scholar] [CrossRef]
- Sanchez-Ponce, Y.; Varela-Fascinetto, G.; Romo-Vazquez, J.C.; Lopez-Martinez, B.; Sanchez-Huerta, J.L.; Parra-Ortega, I.; Fuentes-Panana, E.M.; Morales-Sanchez, A. Simultaneous Detection of Beta and Gamma Human Herpesviruses by Multiplex qPCR Reveals Simple Infection and Coinfection Episodes Increasing Risk for Graft Rejection in Solid Organ Transplantation. Viruses 2018, 10, 730. [Google Scholar] [CrossRef]
- Leibovitch, E.C.; Brunetto, G.S.; Caruso, B.; Fenton, K.; Ohayon, J.; Reich, D.S.; Jacobson, S. Coinfection of Human Herpesviruses 6A (HHV-6A) and HHV-6B as Demonstrated by Novel Digital Droplet PCR Assay. PLoS ONE 2014, 9, e92328. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Luo, X.; Ning, Q. Detection of HBV and HCV coinfection by TEM with Au nanoparticle gene probes. J. Huazhong Univ. Sci. Technol. 2007, 27, 532–534. [Google Scholar] [CrossRef] [PubMed]
- Fayyadh, T.K.; Ma, F.Y.; Qin, C.; Zhang, X.W.; Li, W.; Zhang, X.E.; Zhang, Z.P.; Cui, Z.Q. Simultaneous detection of multiple viruses in their co-infected cells using multicolour imaging with self-assembled quantum dot probes. Microchim. Acta 2017, 184, 2815–2824. [Google Scholar] [CrossRef]
- Srisomwat, C.; Yakoh, A.; Avihingsanon, A.; Chuaypen, N.; Tangkijvanich, P.; Vilaivan, T.; Chailapakul, O. An alternative label-free DNA sensor based on the alternating-current electroluminescent device for simultaneous detection of human immunodeficiency virus and hepatitis C co-infection. Biosens. Bioelectron. 2022, 196, 113719. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, M.-E.; Trujillo-Ortega, M.-E.; Alcaraz-Estrada, S.-L.; Lozano-Aguirre-Beltran, L.; Sandoval-Jaime, C.; Taboada-Ramirez, B.I.; Sarmiento-Silva, R.-E. Molecular Detection and Characterization of Porcine Epidemic Diarrhea Virus and Porcine Aichivirus C Coinfection in Mexico. Viruses 2021, 13, 738. [Google Scholar] [CrossRef]
- Bialasiewicz, S.; McVernon, J.; Nolan, T.; Lambert, S.B.; Zhao, G.; Wang, D.; Nissen, M.D.; Sloots, T.P. Detection of a divergent Parainfluenza 4 virus in an adult patient with influenza like illness using next-generation sequencing. BMC Infect. Dis. 2014, 14, 275. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Bester, R.; Burger, J.T.; Maree, H.J. Next-generation sequencing for virus detection: Covering all the bases. Virol. J. 2016, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- van Engelenburg, F.A.; Terpstra, F.G.; Schuitemaker, H.; Moorer, W.R. The virucidal spectrum of a high concentration alcohol mixture. J. Hosp. Infect. 2002, 51, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Leland Diane, S.; Ginocchio Christine, C. Role of Cell Culture for Virus Detection in the Age of Technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Beperet, I.; Simón, O.; López-Ferber, M.; Lent, J.v.; Williams, T.; Caballero, P.; Johnson, K.N. Mixtures of Insect-Pathogenic Viruses in a Single Virion: Towards the Development of Custom-Designed Insecticides. Appl. Environ. Microbiol. 2021, 87, e02180-20. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-F.; Su, C.-L.; Shu, P.-Y.; Yang, C.-F.; Liao, T.-L.; Cheng, C.-H.; Hu, H.-C.; Huang, J.-H. Concurrent Isolation of Chikungunya Virus and Dengue Virus from a Patient with Coinfection Resulting from a Trip to Singapore. J. Clin. Microbiol. 2010, 48, 4586–4589. [Google Scholar] [CrossRef]
- Davidson, I.; Nagar, S.; Haddas, R.; Ben-Shabat, M.; Golender, N.; Lapin, E.; Altory, A.; Simanov, L.; Ribshtein, I.; Panshin, A.; et al. Avian Influenza Virus H9N2 Survival at Different Temperatures and pHs. Avian Dis. 2010, 54, 725–728. [Google Scholar] [CrossRef]
- Lee, K.W.; Chi, S.C.; Cheng, T.M. Interference of the life cycle of fish nodavirus with fish retrovirus. J. Gen. Virol. 2002, 83, 2469–2474. [Google Scholar] [CrossRef]
- Harada, Y.; Takahashi, H.; Trusheim, H.; Roth, B.; Mizuta, K.; Hirata-Saito, A.; Ogane, T.; Odagiri, T.; Tashiro, M.; Yamamoto, N. Comparison of suspension MDCK cells, adherent MDCK cells, and LLC-MK2 cells for selective isolation of influenza viruses to be used as vaccine seeds. Influenza Other Respir. Viruses 2020, 14, 204–209. [Google Scholar] [CrossRef]
- Zhang, W.; Kataoka, M.; Doan, H.Y.; Wu, F.-T.; Takeda, N.; Muramatsu, M.; Li, T.-C. Isolation and Characterization of Hepatitis E Virus Subtype 4b Using a PLC/PRF/5 Cell-Derived Cell Line Resistant to Porcine Sapelovirus Infection. Jpn. J. Infect. Dis. 2021, 74, 573–575. [Google Scholar] [CrossRef]
- Kim, J.; Chae, C. A comparison of virus isolation, polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine circovirus 2 and porcine parvovirus in experimentally and naturally coinfected pigs. J. Vet. Diagn. Investig. 2004, 16, 45–50. [Google Scholar] [CrossRef]
- Dormitorio, T.V.; Giambrone, J.J. Limiting dilution studies to detect avian influenza viruses from questionable allantoic fluid samples. J. Dairy Sci. 2010, 93, 544. [Google Scholar]
- El Zowalatys, M.E.; Chander, Y.; Redig, P.T.; El Latif, H.K.A.; El Sayed, M.A.; Goyal, S.M. Selective isolation of Avian influenza virus (AIV) from cloacal samples containing AIV and Newcastle disease virus. J. Vet. Diagn. Investig. 2011, 23, 330–332. [Google Scholar] [CrossRef]
- Mi, S.J.; Guo, S.B.; Xing, C.N.; Xiao, C.T.; He, B.; Wu, B.; Xia, X.Z.; Tu, C.C.; Gong, W.J. Isolation and Characterization of Porcine Astrovirus 5 from a Classical Swine Fever Virus-Infected Specimen. J. Virol. 2021, 95, e01513-20. [Google Scholar] [CrossRef]
- Neighbor, N.K.; Newberry, L.A.; Bayyari, G.R.; Skeeles, J.K.; Beasley, J.N.; McNew, R.W. The effect of microaerosolized hydrogen peroxide on bacterial and viral poultry pathogens. Poult. Sci. 1994, 73, 1511–1516. [Google Scholar] [CrossRef]
- Brown, J.D.; Goekjian, G.; Poulson, R.; Valeika, S.; Stallknecht, D.E. Avian influenza virus in water: Infectivity is dependent on pH, salinity and temperature. Vet. Microbiol. 2009, 136, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Moses, H.E.; Brandly, C.A.; Jones, E.E. The pH Stability of Viruses of Newcastle Disease and Fowl Plague. Science 1947, 105, 477–479. [Google Scholar] [CrossRef]
- Hematian, A.; Sadeghifard, N.; Mohebi, R.; Taherikalani, M.; Nasrolahi, A.; Amraei, M.; Ghafourian, S. Traditional and Modern Cell Culture in Virus Diagnosis. Osong Public Health Res. Perspect. 2016, 7, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Waner, J.L. Mixed viral infections: Detection and management. Clin. Microbiol. Rev. 1994, 7, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Brumback, B.G.; Wade, C.D. Simultaneous culture for adenovirus, cytomegalovirus, and herpes simplex virus in same shell vial by using three-color fluorescence. J. Clin. Microbiol. 1994, 32, 2289–2290. [Google Scholar] [CrossRef]
- Weinberg, A.; Brewster, L.; Clark, J.; Simoes, E. Evaluation of R-Mix shell vials for the diagnosis of viral respiratory tract infections. J. Clin. Virol. 2004, 30, 100–105. [Google Scholar] [CrossRef]
- Lim, G.; Park, T.S.; Suh, J.T.; Lee, H.J. Comparison of R-mix virus culture and multiplex reverse transcriptase-PCR for the rapid detection of respiratory viruses. Korean J. Lab. Med. 2010, 30, 289–294. [Google Scholar] [CrossRef][Green Version]
- Barenfanger, J.; Drake, C.; Mueller, T.; Troutt, T.; O’Brien, J.; Guttman, K. R-Mix cells are faster, at least as sensitive and marginally more costly than conventional cell lines for the detection of respiratory viruses. J. Clin. Virol. 2001, 22, 101–110. [Google Scholar] [CrossRef]
- Dunn, J.J.; Woolstenhulme, R.D.; Langer, J.; Carroll, K.C. Sensitivity of respiratory virus culture when screening with R-mix fresh cells. J. Clin. Microbiol. 2004, 42, 79–82. [Google Scholar] [CrossRef]
- St George, K.; Patel, N.M.; Hartwig, R.A.; Scholl, D.R.; Jollick, J.A., Jr.; Kauffmann, L.M.; Evans, M.R.; Rinaldo, C.R., Jr. Rapid and sensitive detection of respiratory virus infections for directed antiviral treatment using R-Mix cultures. J. Clin. Virol. 2002, 24, 107–115. [Google Scholar] [CrossRef]
- Gillim-Ross, L.; Taylor, J.; Scholl, D.R.; Ridenour, J.; Masters, P.S.; Wentworth, D.E. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J. Clin. Microbiol. 2004, 42, 3196–3206. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Poudel, B. Tools to detect influenza virus. Yonsei Med. J. 2013, 54, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.Y.; Chan, P.K.; Peiris, M.; Tsang, D.N.; Que, T.L.; Shortridge, K.F.; Cheung, P.T.; To, W.K.; Ho, E.T.; Sung, R.; et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 1998, 351, 467–471. [Google Scholar] [CrossRef]
- Choi, W.S.; Noh, J.Y.; Baek, J.H.; Seo, Y.B.; Lee, J.; Song, J.Y.; Park, D.W.; Lee, J.S.; Cheong, H.J.; Kim, W.J. Suboptimal effectiveness of the 2011-2012 seasonal influenza vaccine in adult Korean populations. PLoS ONE 2015, 10, e0098716. [Google Scholar] [CrossRef]
- Schmutzhard, J.; Merete Riedel, H.; Zweygberg Wirgart, B.; Grillner, L. Detection of herpes simplex virus type 1, herpes simplex virus type 2 and varicella-zoster virus in skin lesions. Comparison of real-time PCR, nested PCR and virus isolation. J. Clin. Virol. 2004, 29, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Hite, S.; Duane, V.; Yan, H. CV-1 and MRC-5 mixed cells for simultaneous detection of herpes simplex viruses and varicella zoster virus in skin lesions. J. Clin. Virol. 2002, 24, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Woreta, T.A.; Chalasani, N. Fatty Liver Disease in Human Immunodeficiency Virus-Hepatitis B Virus Coinfection: A Cause for Concern COMMENT. Clin. Infect. Dis. 2021, 73, E3286–E3287. [Google Scholar] [CrossRef]
- Kim, Z.; Lee, J.H. Coinfection with severe acute respiratory syndrome coronavirus-2 and other respiratory viruses at a tertiary hospital in Korea. J Clin Lab Anal 2021, 35, e23868. [Google Scholar] [CrossRef]
- Zhong, P.P.; Zhang, H.L.; Chen, X.F.; Lv, F.F. Clinical characteristics of the lower respiratory tract infection caused by a single infection or coinfection of the human parainfluenza virus in children. J. Med. Virol. 2019, 91, 1625–1632. [Google Scholar] [CrossRef]
- Mavilia, M.G.; Wu, G.Y. HBV-HCV Coinfection: Viral Interactions, Management, and Viral Reactivation. J. Clin. Transl. Hepatol. 2018, 6, 296–305. [Google Scholar] [CrossRef]
- Almajhdi, F.N.; Ali, G. Report on Influenza A and B Viruses: Their Coinfection in a Saudi Leukemia Patient. Biomed Res. Int. 2013, 2013, 290609. [Google Scholar] [CrossRef]
- Yapali, S.; Bayrakci, B.; Gunsar, F.; Ersoz, G.; Karasu, Z.; Akarca, U.S. Delta virus coinfection does not increase, but hcv coinfection increase the hbsag loss, in chronic hbv infection. J. Hepatol. 2011, 54, S159–S160. [Google Scholar] [CrossRef]
- Kim, E.H.; Nguyen, T.Q.; Casel, M.A.B.; Rollon, R.; Kim, S.M.; Kim, Y.I.; Yu, K.M.; Jang, S.G.; Yang, J.; Poo, H.; et al. Coinfection with SARS-CoV-2 and Influenza A Virus Increases Disease Severity and Impairs Neutralizing Antibody and CD4(+) T Cell Responses. J. Virol. 2022, 96, e01873-21. [Google Scholar] [CrossRef]
- Pires, C.A.A.; Quaresma, J.A.S.; Aarao, T.L.D.; de Souza, J.R.; Macedo, G.M.M.; Neto, F.; Xavier, M.B. Expression of interleukin-1 beta and interleukin-6 in leprosy reactions in patients with human immunodeficiency virus coinfection. Acta Trop. 2017, 172, 213–216. [Google Scholar] [CrossRef]
- Kanthong, N.; Khemnu, N.; Pattanakitsakul, S.-N.; Malasit, P.; Flegel, T.W. Persistent, triple-virus co-infections in mosquito cells. BMC Microbiol. 2010, 10, 14. [Google Scholar] [CrossRef]
- Cook, J.K.; Huggins, M.B.; Orbell, S.J.; Mawditt, K.; Cavanagh, D. Infectious bronchitis virus vaccine interferes with the replication of avian pneumovirus vaccine in domestic fowl. Avian Pathol. J. WVPA 2001, 30, 233–242. [Google Scholar] [CrossRef]
- Gelb, J., Jr.; Ladman, B.S.; Licata, M.J.; Shapiro, M.H.; Campion, L.R. Evaluating viral interference between infectious bronchitis virus and Newcastle disease virus vaccine strains using quantitative reverse transcription-polymerase chain reaction. Avian Dis. 2007, 51, 924–934. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pantin-Jackwood, M.J.; Costa-Hurtado, M.; Miller, P.J.; Afonso, C.L.; Spackman, E.; Kapczynski, D.R.; Shepherd, E.; Smith, D.; Swayne, D.E. Experimental co-infections of domestic ducks with a virulent Newcastle disease virus and low or highly pathogenic avian influenza viruses. Vet. Microbiol. 2015, 177, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sloutskin, A.; Yee, M.B.; Kinchington, P.R.; Goldstein, R.S. Varicella-zoster virus and herpes simplex virus 1 can infect and replicate in the same neurons whether co- or superinfected. J. Virol. 2014, 88, 5079–5086. [Google Scholar] [CrossRef]
- White, P.A.; Li, Z.; Zhai, X.; Marinos, G.; Rawlinson, W.D. Mixed viral infection identified using heteroduplex mobility analysis (HMA). Virology 2000, 271, 382–389. [Google Scholar] [CrossRef]
- Diallo, I.S.; Taylor, J.; Gibson, J.; Hoad, J.; De Jong, A.; Hewitson, G.; Corney, B.G.; Rodwell, B.J. Diagnosis of a naturally occurring dual infection of layer chickens with fowlpox virus and gallid herpesvirus 1 (infectious laryngotracheitis virus). Avian Pathol. J. WVPA 2010, 39, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Otta, S.K.; Arulraj, R.; Ezhil Praveena, P.; Manivel, R.; Panigrahi, A.; Bhuvaneswari, T.; Ravichandran, P.; Jithendran, K.P.; Ponniah, A.G. Association of dual viral infection with mortality of Pacific white shrimp (Litopenaeus vannamei) in culture ponds in India. VirusDisease 2014, 25, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Vaziry, A.; Silim, A.; Bleau, C.; Frenette, D.; Lamontagne, L. Dual infections with low virulent chicken infectious anaemia virus (lvCIAV) and intermediate infectious bursal disease virus (iIBDV) in young chicks increase lvCIAV in thymus and bursa while decreasing lymphocyte disorders induced by iIBDV. Avian Pathol. J. WVPA 2013, 42, 88–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ge, X.-Y.; Wang, N.; Zhang, W.; Hu, B.; Li, B.; Zhang, Y.-Z.; Zhou, J.-H.; Luo, C.-M.; Yang, X.-L.; Wu, L.-J.; et al. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016, 31, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Zampiero, A.; Bianchini, S.; Mori, A.; Scala, A.; Tagliabue, C.; Sciarrabba, C.S.; Fossali, E.; Piralla, A.; Principi, N. Epidemiology and Clinical Characteristics of Respiratory Infections Due to Adenovirus in Children Living in Milan, Italy, during 2013 and 2014. PLoS ONE 2016, 11, e0152375. [Google Scholar] [CrossRef] [PubMed]
- González Álvarez, D.A.; López Cortés, L.F.; Cordero, E. Impact of HIV on the severity of influenza. Expert Rev. Respir. Med. 2016, 10, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Fan, C.; Liu, S.; Zhou, J.; Jin, Y.; Zheng, X.; Wang, Q.; Liu, J.; Yang, H.; Gu, J.; et al. Cellular proteomic analysis of porcine circovirus type 2 and classical swine fever virus coinfection in porcine kidney-15 cells using isobaric tags for relative and absolute quantitation-coupled LC-MS/MS. Electrophoresis 2017, 38, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Shinjoh, M.; Omoe, K.; Saito, N.; Matsuo, N.; Nerome, K. In vitro growth profiles of respiratory syncytial virus in the presence of influenza virus. Acta Virol. 2000, 44, 91–97. [Google Scholar] [PubMed]
- Hurt, A.C.; Nor’e, S.S.; McCaw, J.M.; Fryer, H.R.; Mosse, J.; McLean, A.R.; Barr, I.G. Assessing the viral fitness of oseltamivir-resistant influenza viruses in ferrets, using a competitive-mixtures model. J. Virol. 2010, 84, 9427–9438. [Google Scholar] [CrossRef] [PubMed]
- Dobrescu, I.; Levast, B.; Lai, K.; Delgado-Ortega, M.; Walker, S.; Banman, S.; Townsend, H.; Simon, G.; Zhou, Y.; Gerdts, V.; et al. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Vet. Microbiol. 2014, 169, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Rozeboom, L.E.; Kassira, E.N. Dual infections of mosquitoes with strains of West Nile virus. J. Med. Entomol. 1969, 6, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Pesko, K.; Mores, C.N. Effect of sequential exposure on infection and dissemination rates for West Nile and St. Louis encephalitis viruses in Culex quinquefasciatus. Vector-Borne Zoonotic Dis. 2009, 9, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, D.; Castro, A.; Haines, H. Demonstration of interference between dengue virus types in cultured mosquito cells using monoclonal antibody probes. J. Gen. Virol. 1982, 59, 273–282. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).