Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April–December 2021 in Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Study Design
2.2. Virus Amplification and Sequencing
2.3. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
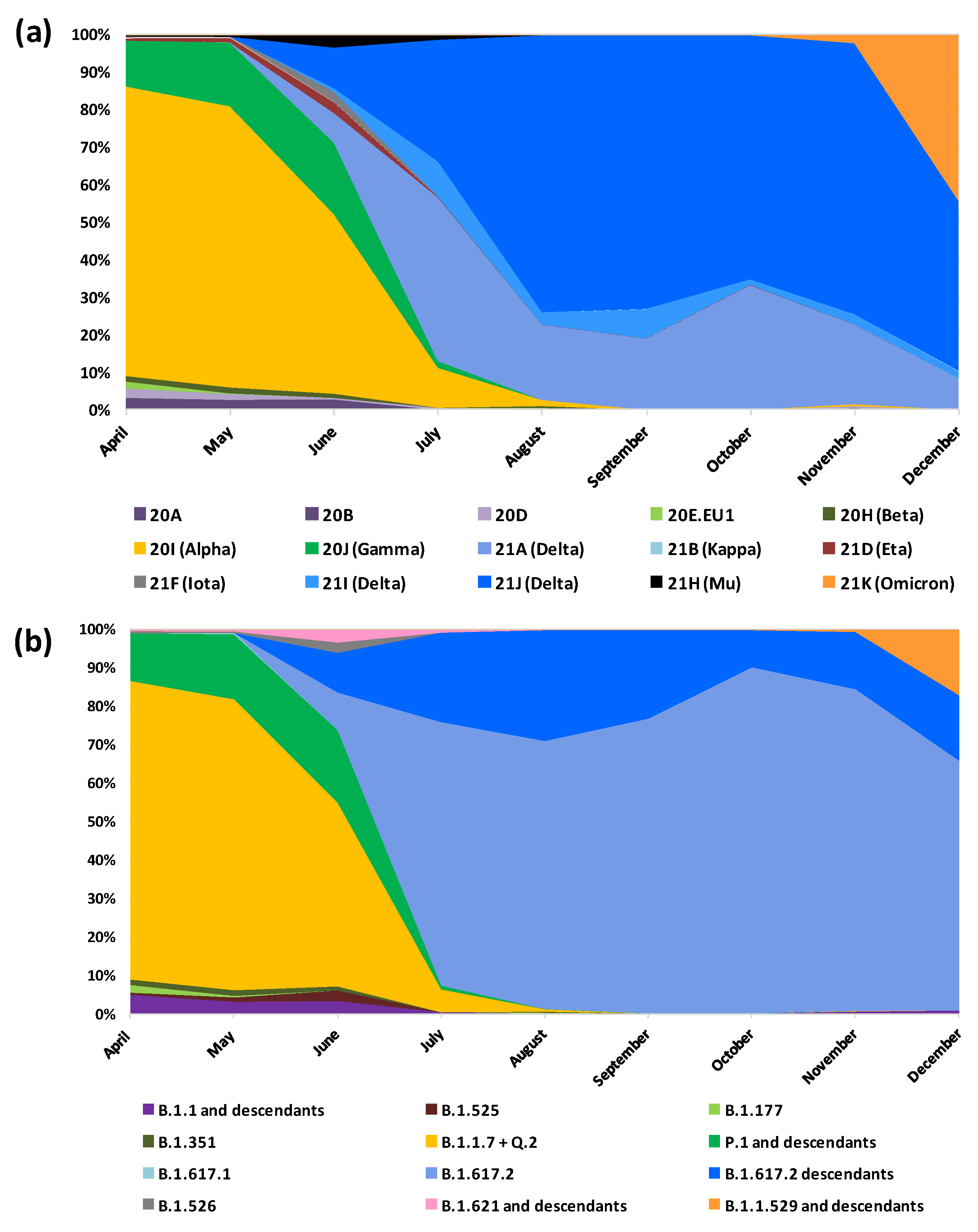
3.2. Lineages and Clades
3.3. Lineages and Clades over Time
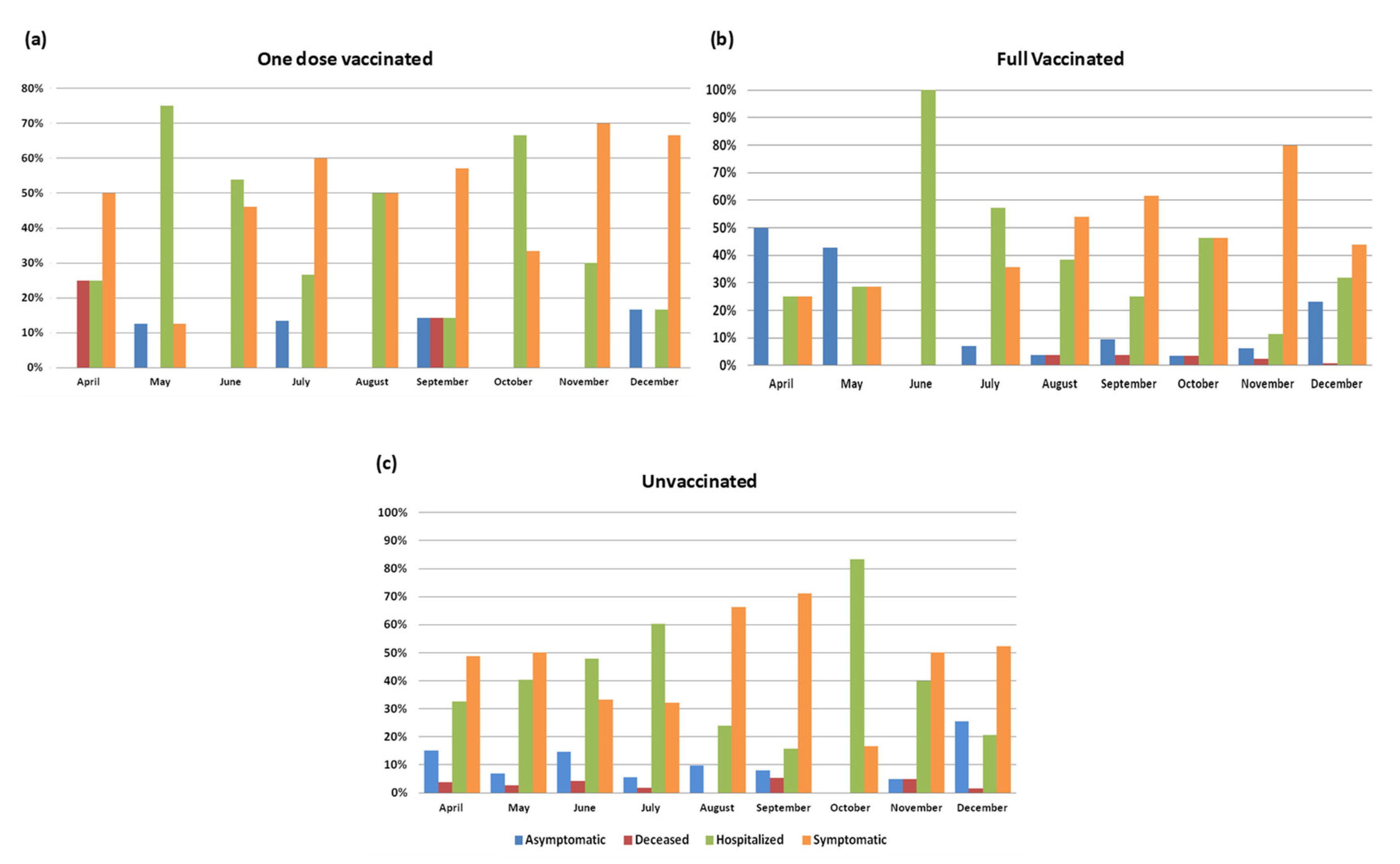
3.4. Vaccinated vs. Unvaccinated
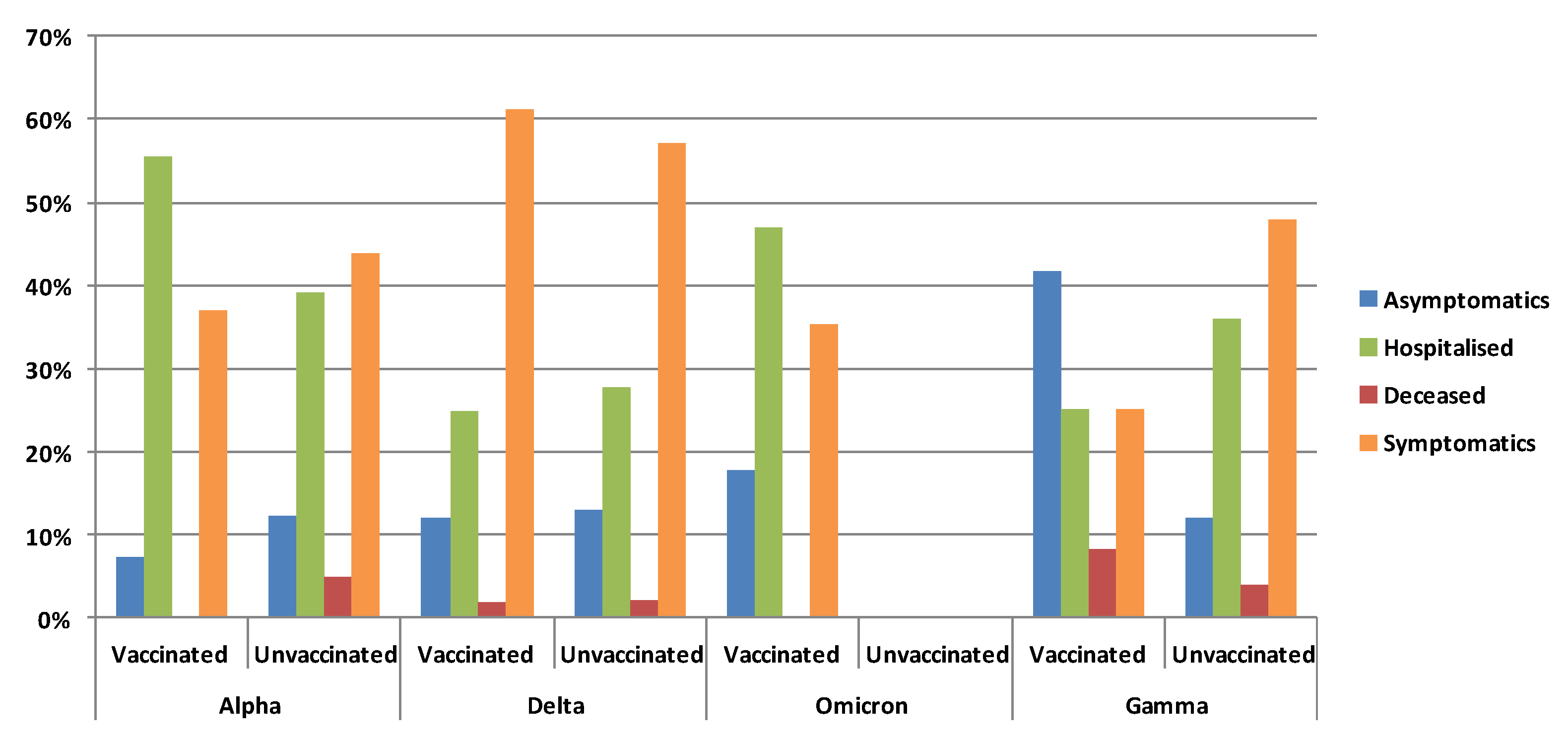
3.5. Clinical Status and Vaccination According to Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Velavan, T.P.; Meyer, C.G. The COVID-19 epidemic. Trop. Med. Int. Health TM IH 2020, 25, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Bal, A.; Destras, G.; Gaymard, A.; Stefic, K.; Marlet, J.; Eymieux, S.; Regue, H.; Semanas, Q.; d’Aubarede, C.; Billaud, G.; et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2021, 26, 2100008. [Google Scholar] [CrossRef] [PubMed]
- Gaymard, A.; Bosetti, P.; Feri, A.; Destras, G.; Enouf, V.; Andronico, A.; Burrel, S.; Behillil, S.; Sauvage, C.; Bal, A.; et al. Early assessment of diffusion and possible expansion of SARS-CoV-2 Lineage 20I/501Y.V1 (B.1.1.7, variant of concern 202012/01) in France, January to March 2021. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2021, 26, 2100133. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Ravi, V.; Devi, P.; Maurya, R.; Parveen, S.; Mishra, P.; Yadav, A.; Swaminathan, A.; Saifi, S.; Khare, K.; et al. Mutational dynamics across VOCs in International travellers and Community transmission underscores importance of Spike-ACE2 interaction. Microbiol. Res. 2022, 262, 127099. [Google Scholar] [CrossRef]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372, n579. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet. Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Caucci, S.; Corvaro, B.; Tiano, S.M.L.; Valenza, A.; Longo, R.; Marinelli, K.; Ferreri, M.L.; Spiridigliozzi, P.; Salvoni, G.; Bagnarelli, P.; et al. Weak Cross-Lineage Neutralization by Anti SARS-CoV-2 Spike Antibodies after Natural Infection or Vaccination Is Rescued by Repeated Immunological Stimulation. Vaccines 2021, 9, 1124. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Brustolini, O.J.; Cardoso, C.C.; Gerber, A.L.; Guimarães, A.P.C.; Leitão, I.C.; Mariani, D.; Ota, V.A.; et al. Intra-host evolution during SARS-CoV-2 prolonged infection. Virus Evol. 2021, 7, veab078. [Google Scholar] [CrossRef]
- Naveca, F.G.; Nascimento, V.; Souza, V.; Corado, A.L.; Nascimento, F.; Silva, G.; Mejía, M.C.; Brandão, M.J.; Costa, Á.; Duarte, D.; et al. Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil. Microbiol. Spectr. 2022, 10, e0236621. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; St Denis, K.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.; et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383. [Google Scholar] [CrossRef]
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Cardoso, C.C.; Brustolini, O.J.; Gerber, A.L.; Guimarães, A.P.C.; Mariani, D.; da Costa, R.M.; Ferreira, O.C., Jr.; et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021, 95, e00119-21. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Maggi, F. Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines. Rev. Med. Virol. 2021, 31, e2231. [Google Scholar] [CrossRef] [PubMed]
- Riemersma, K.K.; Haddock, L.A.; Wilson, N.A.; Minor, N.; Eickhoff, J.; Grogan, B.E.; Kita-Yarbro, A.; Halfmann, P.J.; Segaloff, H.E.; Kocharian, A.; et al. Shedding of Infectious SARS-CoV-2 Despite Vaccination. medRxiv 2022. 2021.2007.2031.21261387. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Bergna, A.; Menzo, S.; Zehender, G.; Caucci, S.; Ghisetti, V.; Rizzo, F.; Maggi, F.; Cerutti, F.; Giurato, G.; et al. Circulating SARS-CoV-2 variants in Italy, October 2020-March 2021. Virol. J. 2021, 18, 168. [Google Scholar] [CrossRef]
- EpiCentro. Bollettino n.3 25.6.2021. 2021. Available online: https://www.epicentro.iss.it/coronavirus/aggiornamenti (accessed on 3 November 2022).
- Han, A.X.; Kozanli, E.; Koopsen, J.; Vennema, H.; Hajji, K.; Kroneman, A.; van Walle, I.; Klinkenberg, D.; Wallinga, J.; Russell, C.A.; et al. Regional importation and asymmetric within-country spread of SARS-CoV-2 variants of concern in the Netherlands. eLife 2022, 11, e78770. [Google Scholar] [CrossRef] [PubMed]
- Wegrzynska, K.; Komiazyk, M.; Walory, J.; Kozinska, A.; Wasko, I.; Baraniak, A. Differentiation of SARS-CoV-2 Variants Using RT-qPCRs by Targeting Recurrent Mutation Sites: A Diagnostic Laboratory Experience from Multi-Center Regional Study, August 2020-December 2021, Poland. Int. J. Mol. Sci. 2022, 23, 9416. [Google Scholar] [CrossRef] [PubMed]
- Yakovleva, A.; Kovalenko, G.; Redlinger, M.; Liulchuk, M.G.; Bortz, E.; Zadorozhna, V.I.; Scherbinska, A.M.; Wertheim, J.O.; Goodfellow, I.; Meredith, L.; et al. Tracking SARS-COV-2 variants using Nanopore sequencing in Ukraine in 2021. Sci. Rep. 2022, 12, 15749. [Google Scholar] [CrossRef]
- Funk, T.; Pharris, A.; Spiteri, G.; Bundle, N.; Melidou, A.; Carr, M.; Gonzalez, G.; Garcia-Leon, A.; Crispie, F.; O’Connor, L.; et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: Data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2021, 26, 2100348. [Google Scholar] [CrossRef]
- Sievers, C.; Zacher, B.; Ullrich, A.; Huska, M.; Fuchs, S.; Buda, S.; Haas, W.; Diercke, M.; An der Heiden, M.; Kröger, S. SARS-CoV-2 Omicron variants BA.1 and BA.2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Euro Surveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2022, 27, 2200396. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.E.; Addetia, A.; Dang, H.V.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Sprouse, K.R.; Walls, A.C.; Mazzitelli, I.G.; Logue, J.K.; et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science 2022, 377, 890–894. [Google Scholar] [CrossRef]
- Lynch, S.M.; Guo, G.; Gibson, D.S.; Bjourson, A.J.; Rai, T.S. Role of Senescence and Aging in SARS-CoV-2 Infection and COVID-19 Disease. Cells 2021, 10, 3367. [Google Scholar] [CrossRef]
- Romero Starke, K.; Reissig, D.; Petereit-Haack, G.; Schmauder, S.; Nienhaus, A.; Seidler, A. The isolated effect of age on the risk of COVID-19 severe outcomes: A systematic review with meta-analysis. BMJ Glob. Health 2021, 6, e006434. [Google Scholar] [CrossRef]
- Houhamdi, L.; Gautret, P.; Hoang, V.T.; Fournier, P.E.; Colson, P.; Raoult, D. Characteristics of the first 1119 SARS-CoV-2 Omicron variant cases, in Marseille, France, November-December 2021. J. Med. Virol. 2022, 94, 2290–2295. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Bergna, A.; Toppo, S.; Morganti, M.; Menzo, S.; Ghisetti, V.; Bruzzone, B.; Codeluppi, M.; Fiore, V.; Rullo, E.V.; et al. Phylogeography and genomic epidemiology of SARS-CoV-2 in Italy and Europe with newly characterized Italian genomes between February-June 2020. Sci. Rep. 2022, 12, 5736. [Google Scholar] [CrossRef]
- Colson, P.; Fournier, P.E.; Chaudet, H.; Delerce, J.; Giraud-Gatineau, A.; Houhamdi, L.; Andrieu, C.; Brechard, L.; Bedotto, M.; Prudent, E.; et al. Analysis of SARS-CoV-2 Variants From 24,181 Patients Exemplifies the Role of Globalization and Zoonosis in Pandemics. Front. Microbiol. 2021, 12, 786233. [Google Scholar] [CrossRef] [PubMed]


| April (n = 357) | May (n = 267) | June (n = 184) | July (n = 304) | August (n = 443) | September (n = 409) | October (n = 323) | November (n = 764) | December (n = 1349) | Total (n = 4400) | |
|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | 162 | 57 | 25 | 118 | 248 | 219 | 211 | 514 | 981 | 2535 |
| NGS WG | 134 | 149 | 81 | 59 | 109 | 77 | 32 | 76 | 164 | 881 |
| NGS Spike | 42 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 46 |
| Sanger Spike | 19 | 60 | 78 | 127 | 86 | 113 | 80 | 174 | 201 | 938 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, A.; Bergna, A.; Della Ventura, C.; Menzo, S.; Bruzzone, B.; Sagradi, F.; Ceccherini-Silberstein, F.; Weisz, A.; Clementi, N.; Brindicci, G.; et al. Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April–December 2021 in Italy. Viruses 2022, 14, 2508. https://doi.org/10.3390/v14112508
Lai A, Bergna A, Della Ventura C, Menzo S, Bruzzone B, Sagradi F, Ceccherini-Silberstein F, Weisz A, Clementi N, Brindicci G, et al. Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April–December 2021 in Italy. Viruses. 2022; 14(11):2508. https://doi.org/10.3390/v14112508
Chicago/Turabian StyleLai, Alessia, Annalisa Bergna, Carla Della Ventura, Stefano Menzo, Bianca Bruzzone, Fabio Sagradi, Francesca Ceccherini-Silberstein, Alessandro Weisz, Nicola Clementi, Gaetano Brindicci, and et al. 2022. "Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April–December 2021 in Italy" Viruses 14, no. 11: 2508. https://doi.org/10.3390/v14112508
APA StyleLai, A., Bergna, A., Della Ventura, C., Menzo, S., Bruzzone, B., Sagradi, F., Ceccherini-Silberstein, F., Weisz, A., Clementi, N., Brindicci, G., Vicenti, I., Sasset, L., Caucci, S., Corvaro, B., Ippoliti, S., Acciarri, C., De Pace, V., Lanfranchi, L., Bellocchi, M. C., ... SARS-CoV- ITALIAN RESEARCH ENTERPRISE–(SCIRE) Collaborative Group. (2022). Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April–December 2021 in Italy. Viruses, 14(11), 2508. https://doi.org/10.3390/v14112508














