Evolution of Animal South American RVA Told by the NSP4 Gene E12 Genotype
Abstract
1. Introduction
2. Materials and Methods
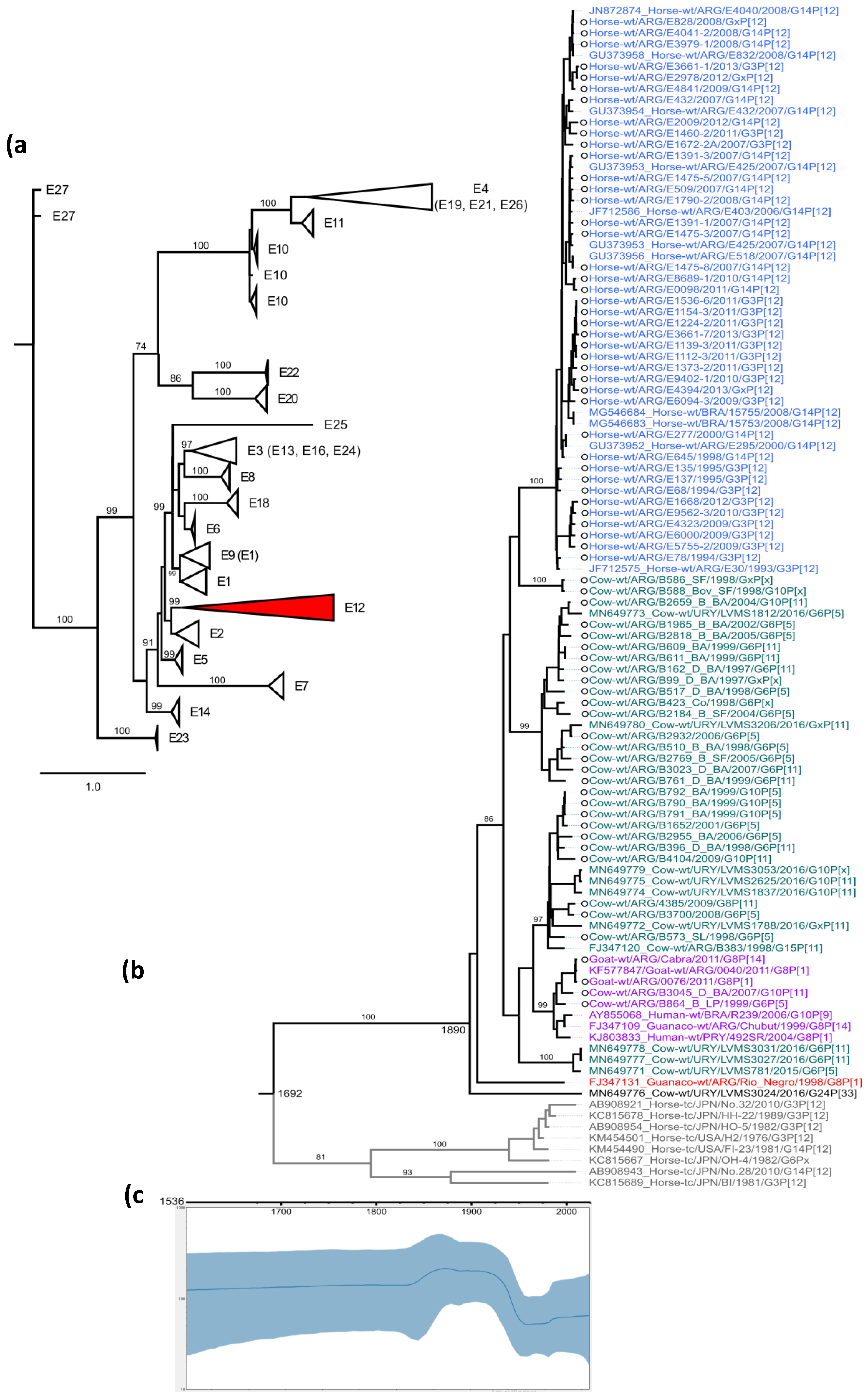
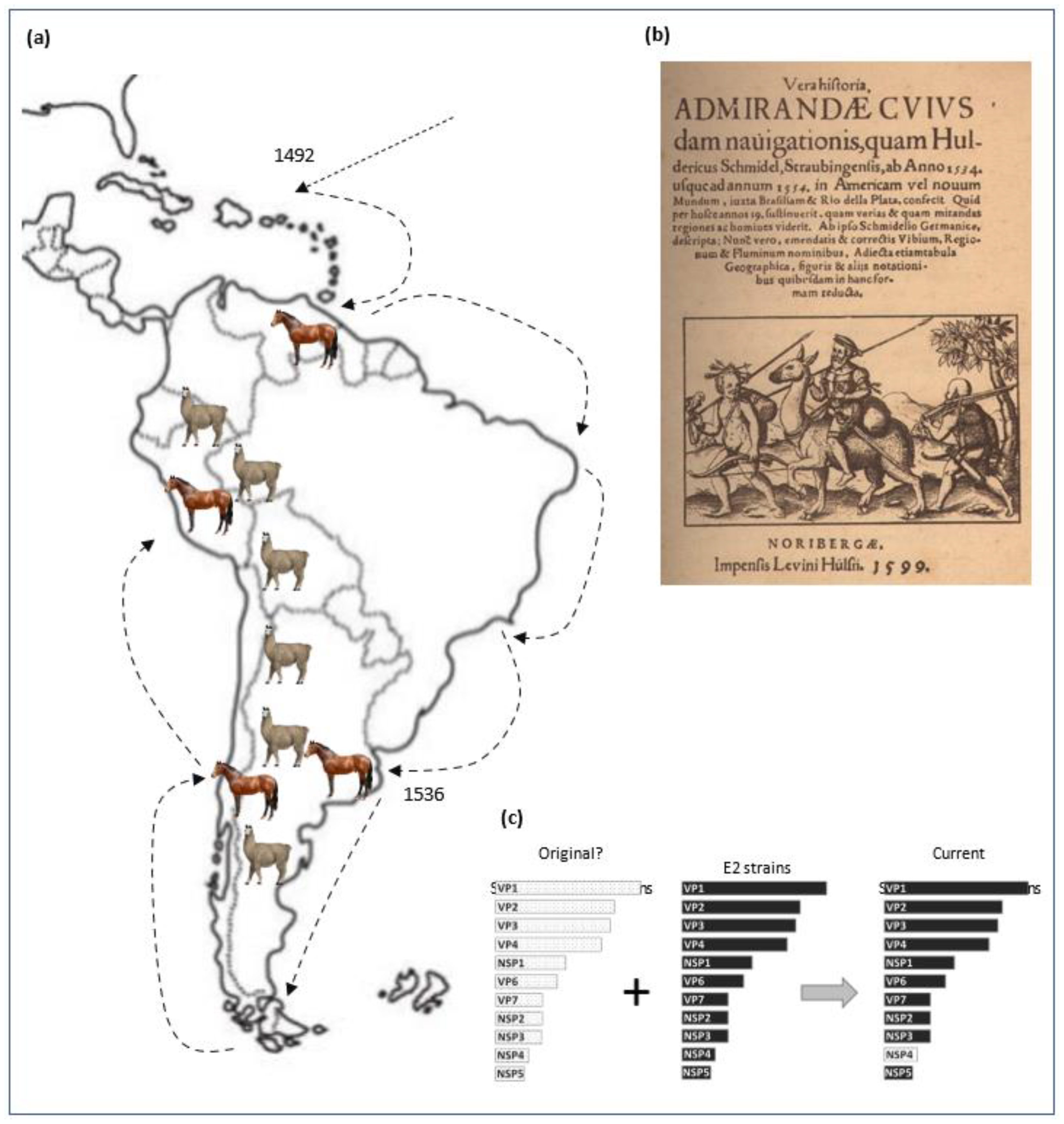
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Hyser, J.M.; Estes, M.K.; Prasad, B.V.V. Rotavirus Non-Structural Proteins: Structure and Function. Curr. Opin. Virol. 2012, 2, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J. Rotavirus Classification Working Group: RCWG. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg (accessed on 16 October 2022).
- Matthijnssens, J.; Potgieter, C.A.; Ciarlet, M.; Parreño, V.; Martella, V.; Bányai, K.; Garaicoechea, L.; Palombo, E.A.; Novo, L.; Zeller, M.; et al. Are Human P[14] Rotavirus Strains the Result of Interspecies Transmissions from Sheep or Other Ungulates That Belong to the Mammalian Order Artiodactyla ? J. Virol. 2009, 83, 2917–2929. [Google Scholar] [CrossRef] [PubMed]
- Castells, M.; Colina, R.; Miño, S.; Castells, F.; Castells, D.; Victoria, M.; Riet-Correa, F.; Caffarena, R.D.; Casaux, M.L.; Schild, C.; et al. Phylogenetic Analyses of Rotavirus a from Cattle in Uruguay Reveal the Circulation of Common and Uncommon Genotypes and Suggest Interspecies Transmission. Pathogens 2020, 9, 570. [Google Scholar] [CrossRef]
- Garaicoechea, L.; Miño, S.; Ciarlet, M.; Fernández, F.; Barrandeguy, M.; Parreño, V. Molecular Characterization of Equine Rotaviruses Circulating in Argentinean Foals during a 17-Year Surveillance Period (1992–2008). Vet. Microbiol. 2011, 148, 150–160. [Google Scholar] [CrossRef]
- Louge Uriarte, E.L.; Badaracco, A.; Matthijnssens, J.; Zeller, M.; Heylen, E.; Manazza, J.; Miño, S.; Van Ranst, M.; Odeón, A.; Parreño, V. The First Caprine Rotavirus Detected in Argentina Displays Genomic Features Resembling Virus Strains Infecting Members of the Bovidae and Camelidae. Vet. Microbiol. 2014, 171, 189–197. [Google Scholar] [CrossRef]
- Martinez, M.; Gia, T.; Eugenia, M.; Russomando, G.; Parreno, V.; Delwart, E.; Parra, G.I. Genomic Characterization of a Rotavirus G8P[1] Detected in a Child with Diarrhea Reveal Direct Animal-to-Human Transmission. Infect. Genet. Evol. 2014, 27, 402–407. [Google Scholar] [CrossRef]
- Volotão, E.M.; Soares, C.C.; Maranhão, A.G.; Rocha, L.N.; Hoshino, Y.; Santos, N. Rotavirus Surveillance in the City of Rio de Janeiro-Brazil during 2000-2004: Detection of Unusual Strains with G8P[4] or G10P[9] Specificities. J. Med. Virol. 2006, 78, 263–272. [Google Scholar] [CrossRef]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in Segmented RNA Viruses: Mechanisms and Outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef]
- Miño, S.; Barrandeguy, M.; Parreño, V.; Parra, G.I. Genetic Linkage of Capsid Protein-Encoding RNA Segments in Group A Equine Rotaviruses. J. Gen. Virol. 2016, 97, 912–921. [Google Scholar] [CrossRef][Green Version]
- Sinclair, J.R. Importance of a One Health Approach in Advancing Global Health Security and the Sustainable Development Goals. Rev. Sci. Tech. 2019, 38, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Improv. Ultrafast Bootstrap Approx. Diep 2017, 35, 518–522. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Badaracco, A.; Garaicoeachea, L.; Mathijnssens, J.; Parreño, V. Epidemiology of Rotavirus Infection in Cattle, Small Ruminants and Horses. In Rotavirus Infections: Epidemiology, Clinical Characteristics and Treatment Options; Zeni, C., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2014; pp. 79–116. ISBN 978-1-63321-442-2. [Google Scholar]
- Miño, S. Síndrome Diarreico En Potrillos, Diagnóstico y Caracterización de Los Agentes Virales Involucrados, Universidad de Buenos Aires. 2017. Available online: http://repositoriouba.sisbi.uba.ar/gsdl/collect/avaposgra/index/assoc/HWA_1923.dir/1923.PDF (accessed on 16 October 2022).
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T.A. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A Simple and Robust Statistical Test for Detecting the Presence of Recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Strimmer, K.; Von Haeseler, A. Likelihood-Mapping: A Simple Method to Visualize Phylogenetic Content of a Sequence Alignment. Proc. Natl. Acad. Sci. USA 1997, 94, 6815–6819. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Carvalho, L.M.; Pybus, O.G. Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Yuan, L.; Honma, S.; Ishida, S.I.; Yan, X.Y.; Kapikian, A.Z.; Hoshino, Y. Species-Specific but Not Genotype-Specific Primary and Secondary Isotype-Specific NSP4 Antibody Responses in Gnotobiotic Calves and Piglets Infected with Homologous Host Bovine (NSP4[A]) or Porcine (NSP4[B]) Rotavirus. Virology 2004, 330, 92–104. [Google Scholar] [CrossRef][Green Version]
- Moya, A.; Elena, S.F.; Bracho, A.; Miralles, R.; Barrio, E. The Evolution of RNA Viruses: A Population Genetics View. Proc. Natl. Acad. Sci. USA 2000, 97, 6967–6973. [Google Scholar] [CrossRef]
- Volz, E.M.; Koelle, K.; Bedford, T. Viral Phylodynamics. PLoS Comput. Biol. 2013, 9, e1002947. [Google Scholar] [CrossRef]
- Carrazzoni, J. El Bovino Criollo Argentino: Ayer Y Hoy. Acad. Nac. Agron. Vet. Buenos Aires 1998, 52, 1–19. Available online: https://www.produccion-animal.com.ar/informacion_tecnica/raza_criolla/15-el_bovino_criollo.pdf (accessed on 16 October 2022).
- Beteta Ortiz, M. Llegada Del Vacuno Español a Suramérica. In Anales de la Real Academia de Ciencias Veterinarias; Real Academia de Ciencias Veterinarias, 2000; pp. 99–116. Available online: https://www.racve.es/publicaciones/llegada-del-vacuno-espanol-a-suramerica/ (accessed on 16 October 2022).
- Gonzalez, D.D.; Dus Santos, M.J. Bovine Colostral Cells—The Often Forgotten Component of Colostrum. J. Am. Vet. Med. Assoc. 2017, 250, 998–1005. [Google Scholar] [CrossRef]
- Schmídel, U.; Mitre, B.; Lafone Quevedo, S.A. Viaje Al Río de La Plata: (1534–1554). Available online: http://www.cervantesvirtual.com/obra-visor/viaje-al-rio-de-la-plata-1534-1554/html/ff3a9778-82b1-11df-acc7-002185ce6064_82.html (accessed on 16 October 2022).
- Molina Martínez, M. Identidad e Imagen de Andalucía en la Edad Moderna; Inicio: Almería, España, 2016. [Google Scholar]
- Martínez, R.D.; Fernández, E.N.; Género, E.R.; Rumiano, F.J.L. El Ganado Bovino Criollo En Argentina. Arch. Zootec. 2000, 49, 353–361. [Google Scholar]
- Pellini, C. Origen del Caballo en Argentina Como Llega el Caballo a Argentina? Available online: https://historiaybiografias.com/caballos2/ (accessed on 16 October 2022).
- Barquez, R.M.; Mónica Díaz, M.; Ojeda, R.A. Mamíferos de Argentina Sistemática y Distribución Editores 2006 SAREM Sociedad Argentina Para El Estudio de Los Mamíferos. Asoc. Argentina para el Estud. los Mamíferos (SAREM). 2006. Available online: http://www.scielo.org.ar/scielo.php?script=sci_arttext&pid=S0327-93832007000100018 (accessed on 16 October 2022).
| Host | Country | |||
|---|---|---|---|---|
| Argentina | Brazil | Paraguay | Uruguay | |
| Humans | - | 1 a | 1 b | - |
| Cows | 30 * | - | - | 10 c |
| Horses | 49 * | 2 d | - | - |
| Guanacos | 2 e | - | - | - |
| Goats | 3 *,f | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miño, S.O.; Badaracco, A.; Louge Uriarte, E.; Ciarlet, M.; Parreño, V. Evolution of Animal South American RVA Told by the NSP4 Gene E12 Genotype. Viruses 2022, 14, 2506. https://doi.org/10.3390/v14112506
Miño SO, Badaracco A, Louge Uriarte E, Ciarlet M, Parreño V. Evolution of Animal South American RVA Told by the NSP4 Gene E12 Genotype. Viruses. 2022; 14(11):2506. https://doi.org/10.3390/v14112506
Chicago/Turabian StyleMiño, Samuel Orlando, Alejandra Badaracco, Enrique Louge Uriarte, Max Ciarlet, and Viviana Parreño. 2022. "Evolution of Animal South American RVA Told by the NSP4 Gene E12 Genotype" Viruses 14, no. 11: 2506. https://doi.org/10.3390/v14112506
APA StyleMiño, S. O., Badaracco, A., Louge Uriarte, E., Ciarlet, M., & Parreño, V. (2022). Evolution of Animal South American RVA Told by the NSP4 Gene E12 Genotype. Viruses, 14(11), 2506. https://doi.org/10.3390/v14112506






