Analysis of Exosomal MicroRNA Dynamics in Response to Rhinovirus Challenge in a Longitudinal Case-Control Study of Asthma
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Subjects and Sample Collection
2.2. Exosomal miRNA Extraction, Library Preparation, and Sequencing
2.3. miRNA-seq Annotation and Differential Expression (DE) Analysis
2.4. Time Series Clustering of DE miRNAs
2.5. miRNA–Traits Correlation Analysis
2.6. MiRNA Gene Target Prediction and Gene Network Construction
2.7. Statistical Analysis
3. Results
3.1. On Average the RV-Induced Cohort Showed No Significant Pulmonary Function Decline after RV Infection
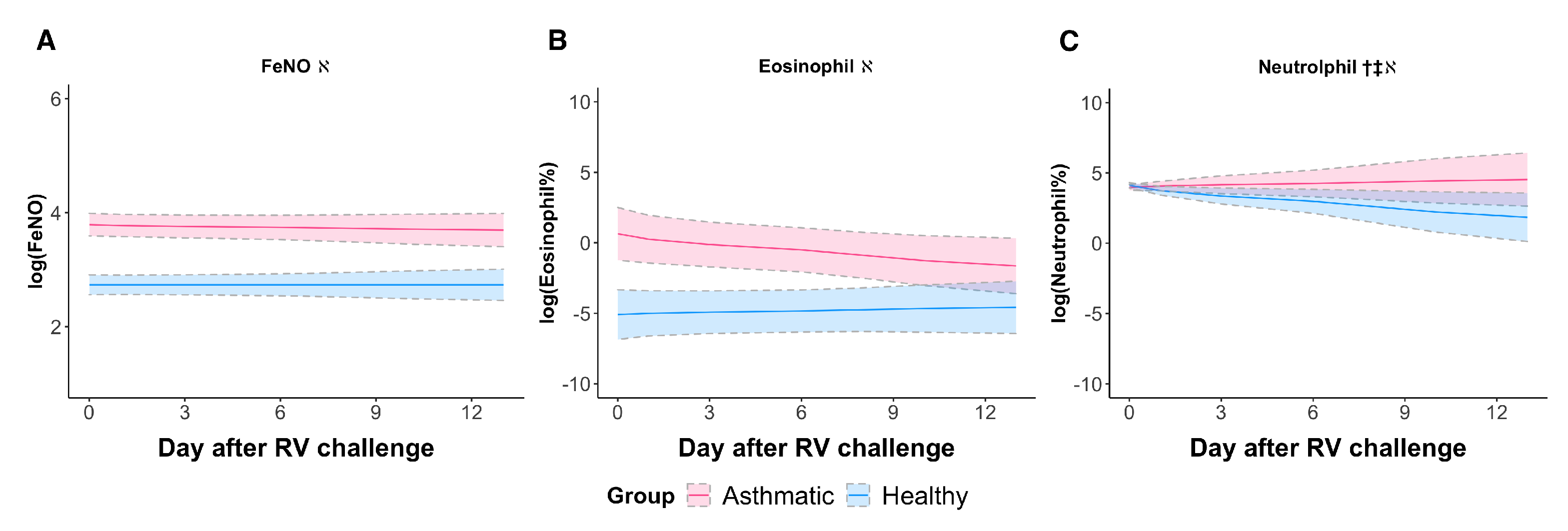
3.2. RV Elicited a Neutrophilic Response in Asthmatic Subjects When Sampled Longitudinally
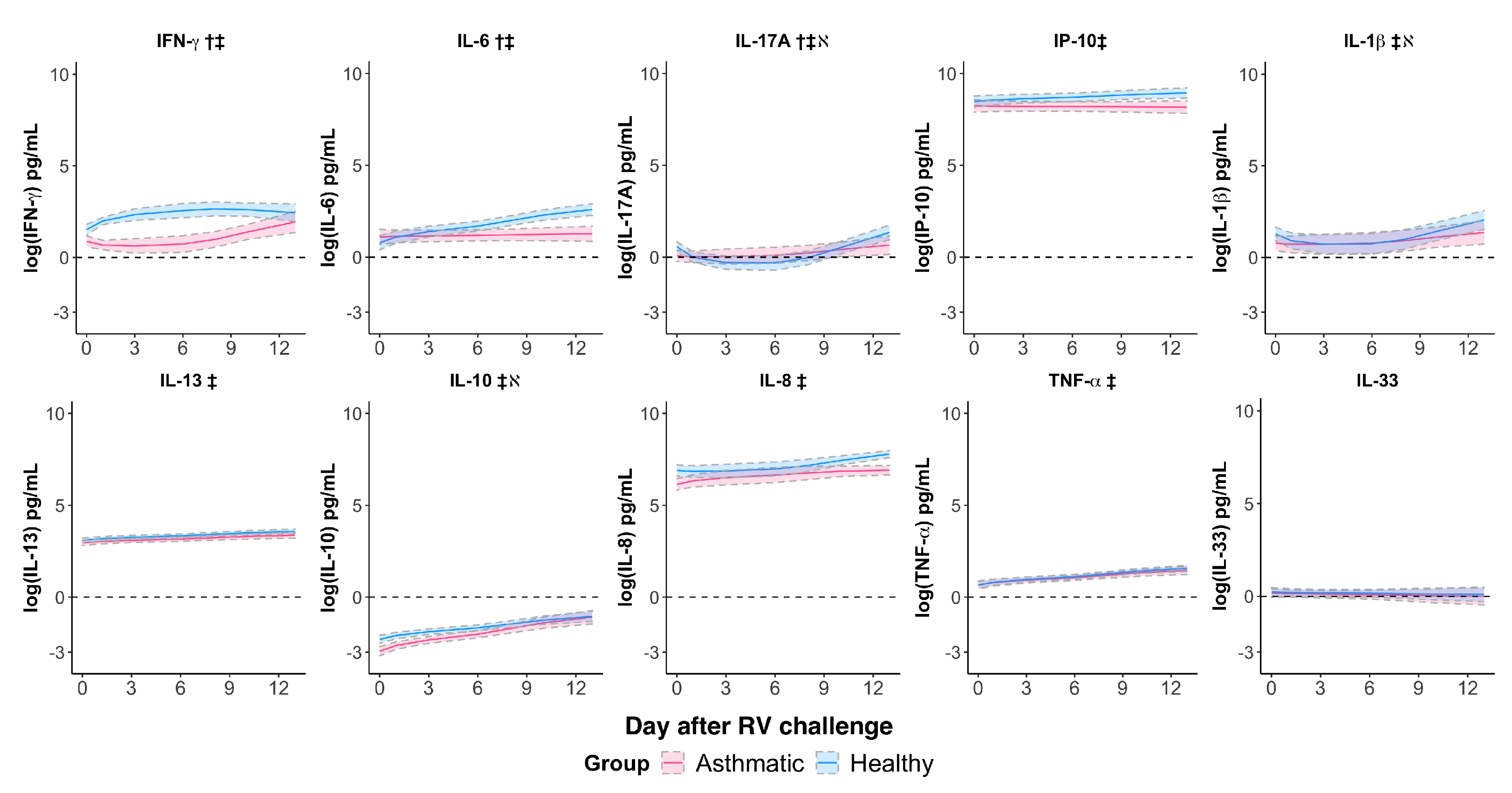
3.3. Rhinovirus Suppressed IFN- and IL-6 Responses and Impaired the IL-17A Response in Asthmatics
3.4. Identification of Differentially Expressed Systemic Exosomal miRNAs (ExoMiRNAs)
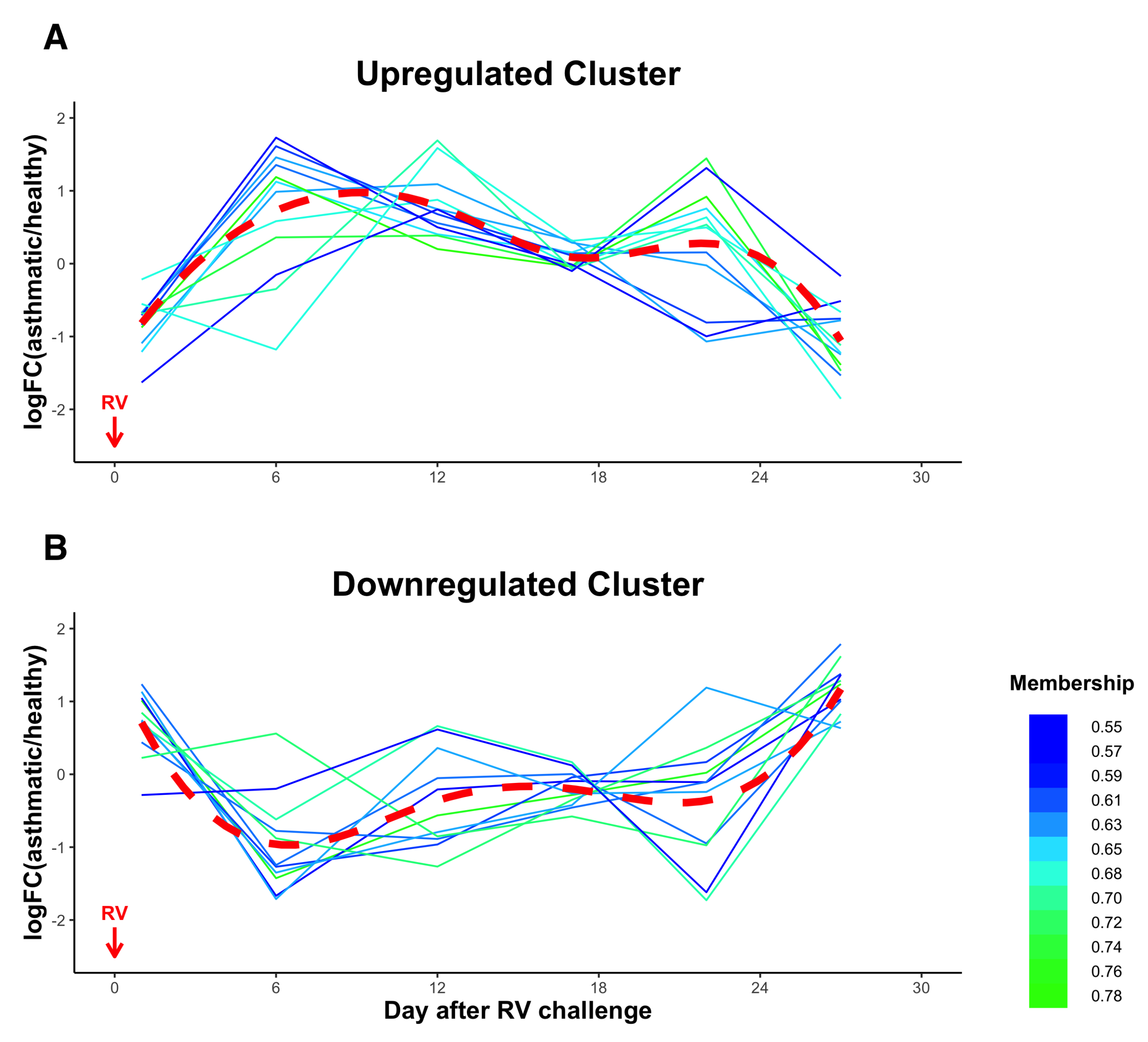
3.5. Temporal Clustering Analysis of DE miRNAs Revealed Two Clusters of miRNAs with Different Dynamics and Correlations with Clinical Traits
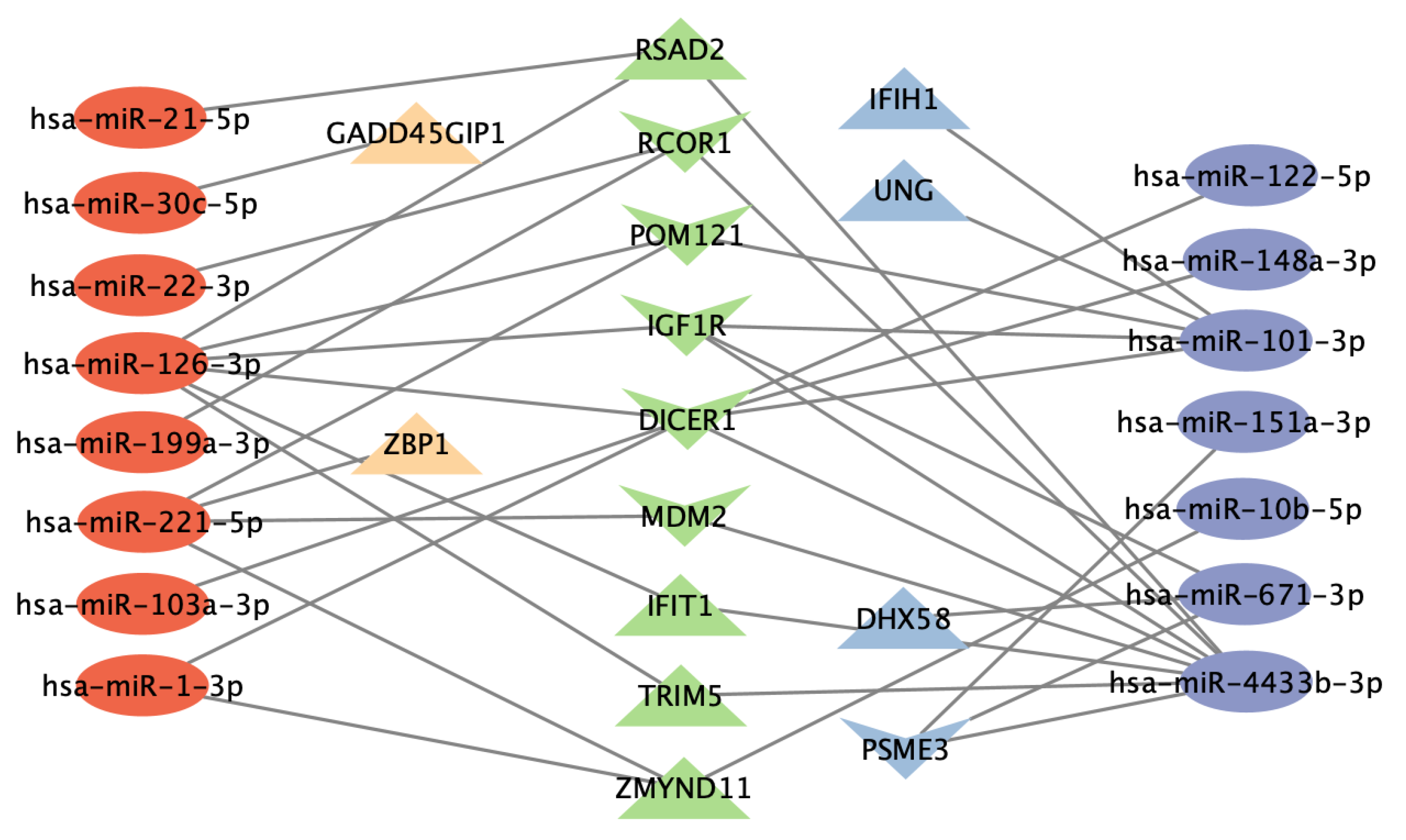
3.6. Functional Analysis of Gene Targets of DE miRNAs and Network Analysis Revealed Enriched Anti-Viral Defense Mechanisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| RV | rhinovirus |
| RSV | respiratory syncytial virus |
| FeNO | fractional exhaled nitric oxide |
| IFN- | interferon-gamma |
| IL | interleukin |
| IP-10 | IFN--inducible protein-10 |
| DE | differentially expressed |
| TNF- | tumor necrosis factor alpha |
| FVC | forced vital capacity |
| FEV | forced expiratory volume in the first second |
| PEF | peak expiratory flow |
| ExoMiRNAs | exosomal microRNAs |
| exoRNAs | exosomal RNAs |
| Th1 | T-helper cell type 1 |
| Th2 | T-helper cell type 2 |
| PACE | principal components analysis through conditional expectation |
| DEGs | differential expressed genes |
References
- Schatz, M.; Rosenwasser, L. The allergic asthma phenotype. J. Allergy Clin. Immunol. Pract. 2014, 2, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Frey, U.; Brodbeck, T.; Majumdar, A.; Robin Taylor, D.; Ian Town, G.; Silverman, M.; Suki, B. Risk of severe asthma episodes predicted from fluctuation analysis of airway function. Nature 2005, 438, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Gern, J.E.; Busse, W.W. Association of rhinovirus infections with asthma. Clin. Microbiol. Rev. 1999, 12, 9–18. [Google Scholar] [CrossRef]
- Contoli, M.; Message, S.D.; Laza-Stanca, V.; Edwards, M.R.; Wark, P.A.B.; Bartlett, N.W.; Kebadze, T.; Mallia, P.; Stanciu, L.A.; Parker, H.L.; et al. Role of deficient type III interferon-gamma production in asthma exacerbations. Nat. Med. 2006, 12, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.D.; Buchta, K.A.; Swenson, C.A.; Gern, J.E.; Busse, W.W. Rhinovirus-induced interferon-γ and Airway Responsiveness in Asthma. Am. J. Respir. Crit. Care Med. 2003, 168, 1091–1094. [Google Scholar] [CrossRef]
- Kim, S.R. Viral infection and airway epithelial immunity in asthma. Int. J. Mol. Sci. 2022, 23, 9914. [Google Scholar] [CrossRef]
- Schmit, T.; Ghosh, S.; Mathur, R.K.; Barnhardt, T.; Ambigapathy, G.; Wu, M.; Combs, C.; Khan, M.N. IL-6 deficiency exacerbates allergic asthma and abrogates the protective effect of allergic inflammation against Streptococcus pneumoniae Pathog. J. Immunol. Res. 2020, 205, 469–479. [Google Scholar]
- Lammers, A.; Brinkman, P.; Te Nijenhuis, L.H.; de Vries, R.; Dagelet, Y.W.F.; Duijvelaar, E.; Xu, B.; Abdel-Aziz, M.I.; Vijverberg, S.J.; Neerincx, A.H.; et al. Increased day-to-day fluctuations in exhaled breath profiles after a rhinovirus challenge in asthma. Allergy 2021, 76, 2488–2499. [Google Scholar] [CrossRef]
- Calhoun, W.J.; Dick, E.C.; Schwartz, L.B.; Busse, W.W. A common cold virus, rhinovirus 16, potentiates airway inflammation after segmental antigen bronchoprovocation in allergic subjects. J. Clin. Investig. 1994, 94, 2200–2208. [Google Scholar] [CrossRef]
- Fraenkel, D.J.; Bardin, P.G.; Sanderson, G.; Lampe, F.; Johnston, S.L.; Holgate, S.T. Lower airways inflammation during rhinovirus colds in normal and in asthmatic subjects. Am. J. Respir. Crit. Care Med. 1995, 151, 879–886. [Google Scholar]
- Pizzichini, M.M.M.; Pizzichini, E.; Efthimiadis, A.; Chauhan, A.J.; Johnston, S.L.; Hussack, P.; Mahony, J.; Dolovich, J.; Hargreave, F.E. Asthma and natural colds. Am. J. Respir. Crit. Care Med. 1998, 158, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Sastre, B.; Rodrigo-Muñoz, J.M.; del Pozo, V. Exosomes: A new approach to asthma pathology. Clin. Chim. 2019, 495, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef]
- Sastre, B.; Cañas, J.A.; Rodrigo-Muñoz, J.M.; del Pozo, V. Novel modulators of asthma and allergy: Exosomes and microRNAs. Front. Immunol. 2017, 8, 826. [Google Scholar] [CrossRef]
- Feng, S.; Zeng, D.; Zheng, J.; Zhao, D. MicroRNAs: Mediators and therapeutic targets to airway hyper reactivity after respiratory syncytial virus infection. Front. Microbiol. 2018, 9, 2177. [Google Scholar] [CrossRef]
- Mortaz, E.; Alipoor, S.D.; Varahram, M.; Jamaati, H.; Garssen, J.; Mumby, S.E.; Adcock, I.M. Exosomes in severe asthma: Update in their roles and potential in therapy. Biomed. Res. Int. 2018, 2018, 2862187. [Google Scholar] [CrossRef]
- Sinha, A.; Lutter, R.; Xu, B.; Dekker, T.; Dierdorp, B.; Sterk, P.J.; Frey, U.; Eckert, E.D. Loss of adaptive capacity in asthmatic patients revealed by biomarker fluctuation dynamics after rhinovirus challenge. eLife 2019, 8, e47969. [Google Scholar] [CrossRef]
- Sinha, A.; Lutter, R.; Dekker, T.; Dierdorp, B.; Sterk, P.J.; Frey, U.; Delgado-Eckert, E. Can Measurements of Inflammatory Biomarkers be Used to Spot Respiratory Viral Infections? Viruses 2020, 12, 1175. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gu, C. Smoothing spline ANOVA models: R package gss. J. Stat. Softw. 2014, 58, 1–25. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Yao, F.; Müller, H.G.; Wang, J.L. Functional data analysis for sparse longitudinal data. J. Am. Stat. Assoc. 2005, 100, 577–590. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, X.; Petersen, A.; Müller, H.G. Quantifying infinite-dimensional data: Functional data analysis in action. Stat. Biosci. 2017, 9, 582–604. [Google Scholar] [CrossRef]
- Wang, J.L.; Chiou, J.M.; Müller, H.G. Functional Data Analysis. Annu. Rev. Stat. Appl. 2016, 3, 257–295. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Ravi, A.; Chang, M.; van de Pol, M.; Yang, S.; Aliprantis, A.; Thornton, B.; Carayannopoulos, L.N.; Bautmans, A.; Robberechts, M.; De Lepeleire, I.; et al. Rhinovirus-16 induced temporal interferon responses in nasal epithelium links with viral clearance and symptoms. Clin. Exp. Allergy 2019, 49, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 10, gkac194. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Campbell, S.; Amir, M.; Mosure, S.A.; Bassette, M.A.; Eliason, A.; Sundrud, M.S.; Kamenecka, T.M.; Solt, L.A. Genetic and pharmacological inhibition of the nuclear receptor RORα regulates TH17 driven inflammatory disorders. Nat. Commun. 2021, 12, 76. [Google Scholar] [CrossRef]
- Scanlon, S.T.; McKenzie, A.N. Type 2 innate lymphoid cells: New players in asthma and allergy. Curr. Opin. Allergy Clin. Immunol. 2012, 24, 707–712. [Google Scholar] [CrossRef]
- Tajiri, T.; Matsumoto, H.; Niimi, A.; Ito, I.; Oguma, T.; Nakaji, H.; Inoue, H.; Iwata, T.; Nagasaki, T.; Kanemitsu, Y.; et al. Association of eosinophilic inflammation with FKBP51 expression in sputum cells in asthma. PLoS ONE 2013, 8, e65284. [Google Scholar]
- MacKay, C.R.; Wang, J.P.; Kurt-Jones, E.A. Dicer’s role as an antiviral: Still an enigma. Curr. Opin. Allergy Clin. Immunol. 2014, 26, 49–55. [Google Scholar] [CrossRef]
- Vázquez-Mera, S.; Martelo-Vidal, L.; Miguéns-Suárez, P.; Saavedra-Nieves, P.; Arias, P.; González-Fernández, C.; Mosteiro-Añón, M.; Corbacho-Abelaira, M.D.; Blanco-Aparicio, M.; Méndez-Brea, P.; et al. Serum exosome inflamma-miRs are surrogate biomarkers for asthma phenotype and severity. Allergy 2022. [Google Scholar] [CrossRef] [PubMed]
- Atashbasteh, M.; Mortaz, E.; Mahdaviani, S.A.; Jamaati, H.; Allameh, A. Expression levels of plasma exosomal miR-124, miR-125b, miR-133b, miR-130a and miR-125b-1-3p in severe asthma patients and normal individuals with emphasis on inflammatory factors. Allergy, Asthma Clin. Immunol. 2021, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Gon, Y.; Shimizu, T.; Mizumura, K.; Maruoka, S.; Hikichi, M. Molecular techniques for respiratory diseases: MicroRNA and extracellular vesicles. Respirology 2020, 25, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yin, Z.; Fan, J.; Zhang, S.; Yang, W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct. Target. Ther. 2019, 4, 47. [Google Scholar] [CrossRef]
- Leon-Icaza, S.A.; Zeng, M.; Rosas-Taraco, A.G. microRNAs in viral acute respiratory infections: Immune regulation, biomarkers, therapy, and vaccines. ExRNA 2019, 1, 1–7. [Google Scholar] [CrossRef]
- Spurrell, J.C.L.; Wiehler, S.; Zaheer, R.S.; Sanders, S.P.; Proud, D. Human airway epithelial cells produce IP-10 (CXCL10) in vitro and in vivo upon rhinovirus infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2005, 289, L85–L95. [Google Scholar] [CrossRef]
- Wark, P.A.; Bucchieri, F.; Johnston, S.L.; Gibson, P.G.; Hamilton, L.; Mimica, J.; Zummo, G.; Holgate, S.T.; Attia, J.; Thakkinstian, A.; et al. IFN-γ–induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J. Allergy Clin. Immunol. 2007, 120, 586–593. [Google Scholar] [CrossRef]
- Tau, G.; Rothman, P. Biologic functions of the IFN-γ receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef]
- Hansel, T.T.; Tunstall, T.; Trujillo-Torralbo, M.B.; Shamji, B.; del Rosario, A.; Dhariwal, J.; Kirk, P.D.W.; Stumpf, M.P.H.; Koopmann, J.; Telcian, A.; et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: Increased interferons (IFN-γ and IFN-λ) and type 2 inflammation (IL-5 and IL-13). EBioMedicine 2017, 19, 128–138. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Fitch, F.W. Anti-proliferative effect of IFN-gamma in immune regulation. I. IFN-gamma inhibits the proliferation of Th2 but not Th1 murine helper T lymphocyte clones. J. Immunol. 1988, 140, 4245–4252. [Google Scholar]
- Oriss, T.B.; McCarthy, S.A.; Morel, B.F.; Campana, M.A.; Morel, P.A. Crossregulation between T helper cell (Th)1 and Th2: Inhibition of Th2 proliferation by IFN-gamma involves interference with IL-1. J. Immunol. 1997, 158, 3666–3672. [Google Scholar] [PubMed]
- Jartti, T.; Paul-Anttila, M.; Lehtinen, P.; Parikka, V.; Vuorinen, T.; Simell, O.; Ruuskanen, O. Systemic T-helper and T-regulatory cell type cytokine responses in rhinovirus vs. respiratory syncytial virus induced early wheezing: An observational study. Respir. Res. 2009, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Message, S.D.; Laza-Stanca, V.; Mallia, P.; Parker, H.L.; Zhu, J.; Kebadze, T.; Contoli, M.; Sanderson, G.; Kon, O.M.; Papi, A.; et al. Rhinovirus induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc. Natl. Acad. Sci. USA 2008, 105, 13562–13567. [Google Scholar] [CrossRef] [PubMed]
- Muehling, L.M.; Heymann, P.W.; Wright, P.W.; Eccles, J.D.; Agrawal, R.; Carper, H.T.; Murphy, D.D.; Workman, L.J.; Word, C.R.; Ratcliffe, S.J.; et al. Human TH1 and TH2 cells targeting rhinovirus and allergen coordinately promote allergic asthma. J. Allergy Clin. Immunol. 2020, 146, 555–570. [Google Scholar] [CrossRef]
- Contoli, M.; Ito, K.; Padovani, A.; Poletti, D.; Marku, B.; Edwards, M.R.; Stanciu, L.A.; Gnesini, G.; Pastore, A.; Spanevello, A.; et al. Th2 cytokines impair innate immune responses to rhinovirus in respiratory epithelial cells. Allergy 2015, 70, 910–920. [Google Scholar] [CrossRef]
- Moriwaki, A.; Matsumoto, K.; Matsunaga, Y.; Fukuyama, S.; Matsumoto, T.; Kan-o, K.; Noda, N.; Asai, Y.; Nakanishi, Y.; Inoue, H. IL-13 suppresses double-stranded RNA-induced IFN-λ production in lung cells. Biochem. Biophys. Res. Commun. 2011, 404, 922–927. [Google Scholar] [CrossRef]
- Wiehler, S.; Proud, D. Interleukin-17A modulates human airway epithelial responses to human rhinovirus infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2007, 293, 505–515. [Google Scholar] [CrossRef]
- Grünberg, K.; Smits, H.H.; Timmers, M.C.; De Klerk, E.P.; Dolhain, R.J.; Dick, E.C.; Hiemstra, P.S.; Sterk, P.J. Experimental rhinovirus 16 infection effects on cell differentials and soluble markers in sputum in asthmatic subjects. Am. J. Respir. Crit. Care Med. 1997, 156, 609–616. [Google Scholar] [CrossRef]
- Fleming, H.E.; Little, F.F.; Schnurr, D.; Avila, P.C.; Wong, H.; Liu, J.; Yagi, S.; Boushey, H.A. Rhinovirus-16 colds in healthy and in asthmatic subjects. Am. J. Respir. Crit. Care. Med. 1999, 160, 100–108. [Google Scholar] [CrossRef]
- Pyle, C.J.; Uwadiae, F.I.; Swieboda, D.P.; Harker, J.A. Early IL-6 signalling promotes IL-27 dependent maturation of regulatory T cells in the lungs and resolution of viral immunopathology. PLoS Pathog. 2017, 13, e1006640. [Google Scholar] [CrossRef]
- Ravanetti, L.; Dijkhuis, A.; Dekker, T.; Pineros, Y.S.S.; Ravi, A.; Dierdorp, B.S.; Erjefält, J.S.; Mori, M.; Pavlidis, S.; Adcock, I.M.; et al. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J. Allergy Clin. Immunol. 2019, 143, 1355–1370.e16. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Makrinioti, H.; Rana, B.M.J.; Shamji, B.W.H.; Trujillo-Torralbo, M.B.; Footitt, J.; Jerico, d.R.; Telcian, A.G.; Nikonova, A.; Zhu, J.; et al. IL-33–dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am. J. Respir. Crit. Care Med. 2014, 190, 1373–1382. [Google Scholar] [CrossRef]
- Lewis, T.C.; Metitiri, E.E.; Mentz, G.B.; Ren, X.; Goldsmith, A.M.; Eder, B.N.; Wicklund, K.E.; Walsh, M.P.; Comstock, A.T.; Ricci, J.M.; et al. Impact of community respiratory viral infections in urban children with asthma. Ann. Allergy Asthma Immunol. 2019, 122, 175–183.e2. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, E.C.; Liang, K.K.; Lawrence, M.D.; Forsyth, K.D.; Dixon, D.L. Neutrophil infiltration and activation in bronchiolitic airways are independent of viral etiology. Pediatr. Pulmonol. 2017, 52, 238–246. [Google Scholar] [CrossRef] [PubMed]
- John, V.; Fahya, B.; Kimb, K.W.; Liub, J.; Homer, A.; Bousheya, B. Prominent neutrophilic inflammation in sputum from subjects with asthma exacerbation. J. Allergy Clin. Immunol. 1995, 95, 843–852. [Google Scholar]
- Henkels, K.M.; Frondorf, K.; Gonzalez-Mejia, M.E.; Doseff, A.L.; Gomez-Cambronero, J. IL-8-induced neutrophil chemotaxis is mediated by Janus kinase 3 (JAK3). FEBS Lett. 2011, 585, 159–166. [Google Scholar] [CrossRef]
- Zaheer, R.S.; Koetzler, R.; Holden, N.S.; Wiehler, S.; Proud, D. Selective transcriptional down-regulation of human rhinovirus-induced production of CXCL10 from airway epithelial cells via the MEK1 pathway. J. Immunol. Res. 2009, 182, 4854–4864. [Google Scholar] [CrossRef]
- Rupani, H.; Martinez-Nunez, R.T.; Dennison, P.; Lau, L.C.K.; Jayasekera, N.; Havelock, T.; Francisco-Garcia, A.S.; Grainge, C.; Howarth, P.H.; Sanchez-Elsner, T. Toll-like receptor 7 is reduced in severe asthma and linked to an altered microRNA profile. Am. J. Respir. Crit. Care Med. 2016, 194, 26–37. [Google Scholar] [CrossRef]
- Beeh, K.M.; Kanniess, F.; Wagner, F.; Schilder, C.; Naudts, I.; Hammann-Haenni, A.; Willers, J.; Stocker, H.; Mueller, P.; Bachmann, M.F.; et al. The novel TLR-9 agonist QbG10 shows clinical efficacy in persistent allergic asthma. J. Allergy Clin. Immunol. 2013, 131, 866–874. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Voronova, N.V.; Savost’Anov, K.V.; Turakulov, R.I. Loss-of-function mutations E6 27X and I923V of IFIH1 are associated with lower poly(I:C)–induced interferon-β production in peripheral blood mononuclear cells of type 1 diabetes patients. Hum. Immunol 2010, 71, 1128–1134. [Google Scholar] [CrossRef]
- Proud, D.; Turner, R.B.; Winther, B.; Wiehler, S.; Tiesman, J.P.; Reichling, T.D.; Juhlin, K.D.; Fulmer, A.W.; Ho, B.Y.; Walanski, A.A.; et al. Gene expression profiles during in vivo human rhinovirus infection. Am. J. Respir. Crit. Care Med. 2008, 178, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Alhamdan, F.; Marsh, L.M.; Pedersen, F.; Alhamwe, B.A.; Thölken, C.; Pfefferle, P.I.; Bahmer, T.; Greulich, T.; Potaczek, D.P.; Garn, H. Differential regulation of interferon signaling pathways in CD4+ T cells of the low type-2 obesity-associated asthma phenotype. Int. J. Mol. Sci. 2021, 22, 10144. [Google Scholar] [CrossRef] [PubMed]


| Demographic Features | Healthy (n = 12) | Asthmatic (n = 12) | p-Value |
|---|---|---|---|
| Age (years), mean ± sd | 21 ± 1.5 | 22.2 ± 2.2 | 0.181 |
| Female gender, n (%) | 7 (58.3%) | 8 (66.7%) | 1 |
| Ethnicity (Caucasian), n | 11 | 9 | 0.590 |
| BMI, mean ± sd | 22.2 ± 1.6 | 22.8 ± 3.1 | 0.576 |
| Smoking (pack years), n | 1 (0.17 PY) | NA | NA |
| Height (cm), mean ± sd | 177.7 ± 8.6 | 172.5 ± 13.0 | 0.260 |
| Weight (kg), mean ± sd | 70.4 ± 10.1 | 67.8 ± 12.4 | 0.575 |
| Baseline FEV1% predicted, mean ± sd | 105.7 ± 11.6 | 101.0 ± 10.0 | 0.507 |
| Baseline FVC% predicted, mean ± sd | 104.2 ± 10.5 | 104.2 ± 10.2 | 0.931 |
| Baseline PEF% predicted, mean ± sd | 108.4 ± 14.0 | 104.7 ± 12.2 | 0.624 |
| Time × Group † | Time ‡ | Group ℵ | |||||
|---|---|---|---|---|---|---|---|
| (SE) | p-Value | (SE) | p-Value | (SE) | p-Value | ||
| Pulmonary function | FVC% | () | 0.3269 | −0.0111 () | 0.0146 | 0.0202 (0.05698) | 0.723 |
| FEV% | () | 0.9403 | () | 0.126 | (0.04953) | 0.995 | |
| PEF% | −0.0114 () | 0.1683 | () | 0.599 | −0.0294 (0.09050) | 0.745 | |
| Inflammatory biomarkers | FeNO | −0.0159 (0.0303) | 0.606 | (0.0214) | 0.999 | 1.072 (0.143) | <0.0001 |
| Eosinophil% | −0.465 (0.255) | 0.0706 | 0.0853 (0.172) | 0.621 | 6.193 (1.454) | 0.0003 | |
| Neutrophil% | 0.471 (0.220) | 0.0435 | −0.379 (0.148) | 0.0182 | −0.619 (0.282) | 0.0399 | |
| Cytokine | IFN- § | 0.139 (0.0455) | 0.0057 | −0.0643 (0.0298) | 0.0424 | 0.311 (0.485) | 0.527 |
| IP-10 | −0.0902 (0.0482) | 0.0744 | 0.0784 (0.0310) | 0.0191 | −0.142 (0.266) | 0.598 | |
| IL-13 | −0.0122 (0.0199) | 0.548 | 0.0825 (0.01289) | <0.0001 | −0.126 (0.110) | 0.265 | |
| IL-17A § | −0.112 (0.0355) | 0.0047 | 0.140 (0.0226) | <0.0001 | −1.239 (0.358) | 0.0022 | |
| IL-10 | 0.101 (0.0518) | 0.0644 | 0.209 (0.0324) | <0.0001 | −0.748 (0.205) | 0.0014 | |
| IL-1 § | −0.0700 (0.0348) | 0.0562 | 0.102 (0.0220) | 0.0001 | −0.966 (0.362) | 0.0141 | |
| IL-6 | −0.272 (0.0602) | 0.0002 | 0.301 (0.0386) | <0.0001 | 0.584 (0.328) | 0.0884 | |
| IL-8 § | −0.0544 (0.0283) | 0.0680 | 0.0409 (0.0188) | 0.0401 | −1.118 (0.322) | 0.0806 | |
| TNF- | −0.0196 (0.0339) | 0.569 | 0.151 (0.0212) | <0.0001 | 0.0270 (0.171) | 0.876 | |
| IL-33 | −0.00681 (0.0622) | 0.913 | −0.0219 (0.0412) | 0.597 | −0.0510 (0.214) | 0.814 | |
| Upregulated Cluster | Downregulated Cluster |
|---|---|
| hsa-let-7f-5p | hsa-miR-122-5p |
| hsa-miR-26a-5p | hsa-miR-148a-3p |
| hsa-let-7a-5p | hsa-miR-101-3p |
| hsa-miR-92a-3p | hsa-miR-151a-3p |
| hsa-miR-21-5p | hsa-miR-375-3p |
| hsa-miR-126-3p | hsa-miR-10b-5p |
| hsa-miR-30c-5p | hsa-miR-423-5p |
| hsa-miR-22-3p | hsa-miR-671-3p |
| hsa-miR-199a-3p | hsa-miR-127-3p |
| hsa-miR-221-5p | hsa-miR-4433b-3p |
| hsa-miR-103a-3p | hsa-miR-99a-5p |
| hsa-miR-1-3p |
| GO Term | Fisher Exact p-Value | |
|---|---|---|
| Upregulated Cluster | defense response to virus | |
| negative regulation of viral genome replication | ||
| response to virus | ||
| type I interferon signaling pathway | ||
| epidermal growth factor receptor signaling pathway | ||
| positive regulation of phosphatidylinositol 3-kinase signaling | ||
| protein phosphorylation | ||
| RNA phosphodiester bond hydrolysis, endonucleolytic | ||
| response to glucose | ||
| glycosaminoglycan metabolic process | ||
| heparan sulfate proteoglycan biosynthetic process | ||
| macromolecular complex assembly | ||
| viral process | ||
| protein polyubiquitination | ||
| cellular response to insulin stimulus | ||
| Downregulated Cluster | negative regulation of viral genome replication | |
| response to virus | ||
| type I interferon signaling pathway | ||
| viral process | ||
| protein polyubiquitination | ||
| defense response | ||
| positive regulation of interferon-beta production | ||
| phospholipid transport | ||
| positive regulation of protein export from nucleus | ||
| brain development | ||
| proteasome-mediated ubiquitin-dependent protein catabolic process | ||
| cellular response to virus | ||
| negative regulation of apoptotic process | ||
| negative regulation of viral entry into host cell |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Sinha, A.; Lutter, R.; Yang, J.; Ascoli, C.; Sterk, P.J.; Nemsick, N.K.; Perkins, D.L.; Finn, P.W. Analysis of Exosomal MicroRNA Dynamics in Response to Rhinovirus Challenge in a Longitudinal Case-Control Study of Asthma. Viruses 2022, 14, 2444. https://doi.org/10.3390/v14112444
Wang W, Sinha A, Lutter R, Yang J, Ascoli C, Sterk PJ, Nemsick NK, Perkins DL, Finn PW. Analysis of Exosomal MicroRNA Dynamics in Response to Rhinovirus Challenge in a Longitudinal Case-Control Study of Asthma. Viruses. 2022; 14(11):2444. https://doi.org/10.3390/v14112444
Chicago/Turabian StyleWang, Wangfei, Anirban Sinha, René Lutter, Jie Yang, Christian Ascoli, Peter J. Sterk, Nicole K. Nemsick, David L. Perkins, and Patricia W. Finn. 2022. "Analysis of Exosomal MicroRNA Dynamics in Response to Rhinovirus Challenge in a Longitudinal Case-Control Study of Asthma" Viruses 14, no. 11: 2444. https://doi.org/10.3390/v14112444
APA StyleWang, W., Sinha, A., Lutter, R., Yang, J., Ascoli, C., Sterk, P. J., Nemsick, N. K., Perkins, D. L., & Finn, P. W. (2022). Analysis of Exosomal MicroRNA Dynamics in Response to Rhinovirus Challenge in a Longitudinal Case-Control Study of Asthma. Viruses, 14(11), 2444. https://doi.org/10.3390/v14112444








