Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Cells
2.3. Viral Isolation
2.4. Sample Selection
2.5. RNA Extraction and PCR Amplification for NGS
2.6. Library Preparation and Sequence Runs for NGS
2.7. NGS Data Analysis
2.8. Subclade Classification by Amino Acid Substitutions in HA
2.9. Evaluation of Genetic Match with Vaccine Strains
2.10. Subclade Classification and Reassortment Analysis Using Phylogenetic Trees
2.11. Evolutionary Analysis and Estimation of Nucleotide Substitutions Rates
3. Results
3.1. Characterization of Influenza Virus Isolates
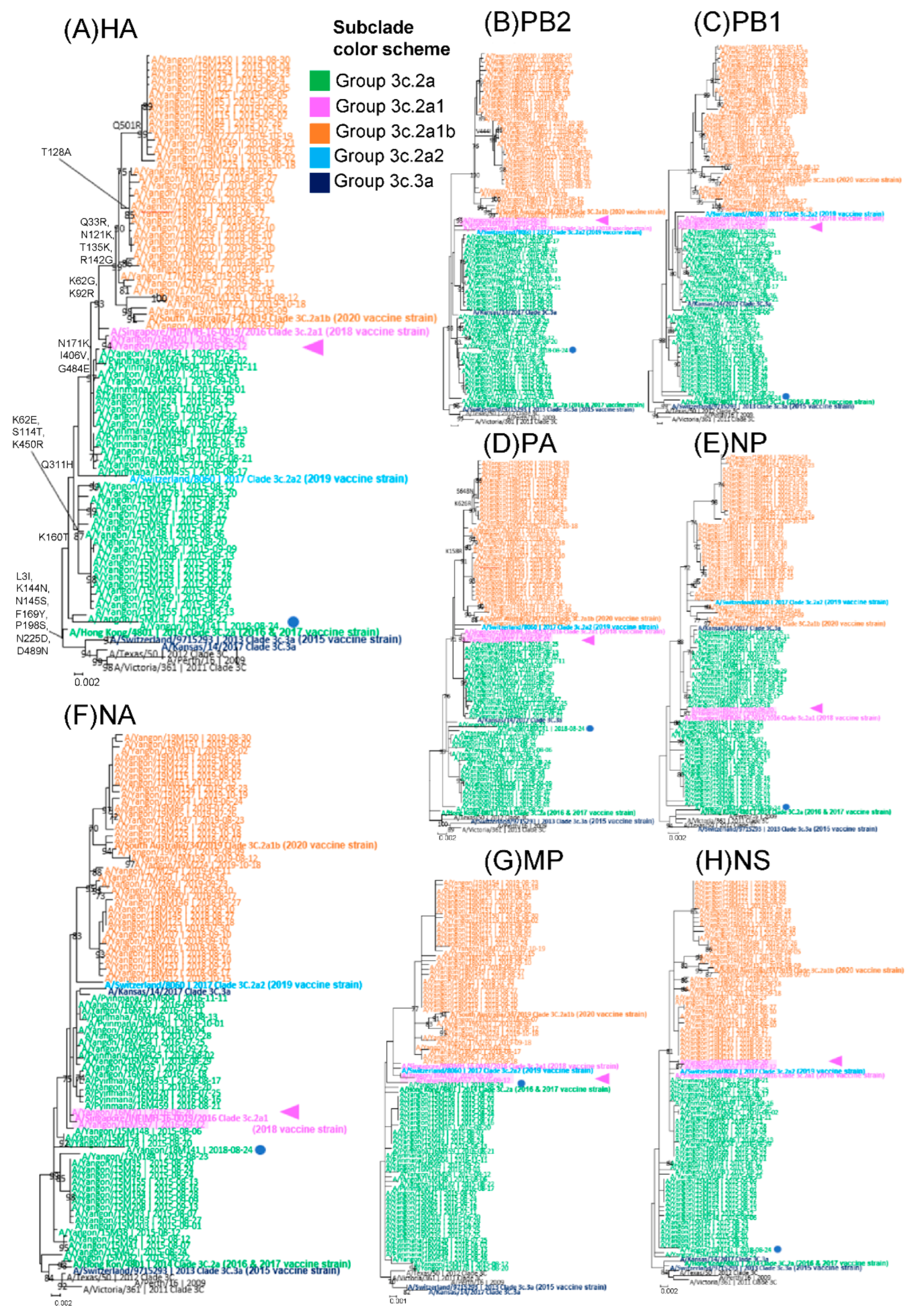
3.2. Subclade Analysis by Amino Acid Substitutions and Phylogenetic Tree Analysis of HA
3.3. Analysis of Comparison of Amino Acid Substitutions between Circulating Strains and A/Texas/50/2012 Vaccine Strain in HA
3.4. Comparison of Amino Acid Substitutions between Circulating Strains and Vaccine Strains in HA and Five Other Segments
3.5. Analysis of Whole-Genome Sequence Data and Identification of Reassortments
3.6. Nucleotide Substitution Rates Using the Bayesian MCMC Method
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Key Facts about Influenza (Flu). 2022. Available online: https://www.cdc.gov/flu/about/keyfacts.htm (accessed on 10 February 2022).
- World Health Organization. Influenza (Seasonal) Fact Sheet. 2018. Available online: www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 10 February 2022).
- Thompson, W.W.; Weintraub, E.; Dhankhar, P.; Cheng, P.Y.; Brammer, L.; Meltzer, M.I.; Bresee, J.S.; Shay, D.K. Estimates of US influenza-associated deaths made using four different methods. Influenza Other Respir. Viruses 2009, 3, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Nair, H.; Brooks, W.A.; Katz, M.; Roca, A.; Berkley, J.A.; Madhi, S.A.; Simmerman, J.M.; Gordon, A.; Sato, M.; Howie, S.; et al. Global burden of respiratory infections due to seasonal influenza in young children: A systematic review and meta-analysis. Lancet 2011, 378, 1917–1930. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005, 79, 2814–2822. [Google Scholar] [CrossRef]
- Olsen, B.; Munster, V.J.; Wallensten, A.; Waldenström, J.; Osterhaus, A.D.; Fouchier, R.A. Global patterns of influenza a virus in wild birds. Science 2006, 312, 384–388. [Google Scholar] [CrossRef]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef]
- Htwe, K.T.Z.; Dapat, C.; Shobugawa, Y.; Odagiri, T.; Hibino, A.; Kondo, H.; Yagami, R.; Saito, T.; Takemae, N.; Tamura, T.; et al. Phylogeographic analysis of human influenza A and B viruses in Myanmar, 2010–2015. PLoS ONE 2019, 14, e0210550. [Google Scholar] [CrossRef]
- Kyaw Win, S.M.; Saito, R.; Win, N.C.; Lasham, D.J.; Kyaw, Y.; Lin, N.; Thein, K.N.; Chon, I.; Odagiri, T.; Thein, W.; et al. Epidemic of influenza A(H1N1)pdm09 analyzed by full genome sequences and the first case of oseltamivir-resistant strain in Myanmar 2017. PLoS ONE 2020, 15, e0229601. [Google Scholar] [CrossRef]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef]
- Chen, R.; Holmes, E.C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008, 66, 655–663. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Holmes, E.C.; Joseph, U.; Fourment, M.; Su, Y.C.; Halpin, R.; Lee, R.T.; Deng, Y.M.; Gunalan, V.; Lin, X.; et al. The contrasting phylodynamics of human influenza B viruses. Elife 2015, 4, e05055. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, N.M.; Galvani, A.P.; Bush, R.M. Ecological and immunological determinants of influenza evolution. Nature 2003, 422, 428–433. [Google Scholar] [CrossRef]
- Rambaut, A.; Pybus, O.G.; Nelson, M.I.; Viboud, C.; Taubenberger, J.K.; Holmes, E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature 2008, 453, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Muscatello, D.J.; Newall, A.T.; Dwyer, D.E.; Macintyre, C.R. Mortality attributable to seasonal and pandemic influenza, Australia, 2003 to 2009, using a novel time series smoothing approach. PLoS ONE 2014, 8, e64734. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, L. The global impact of influenza on morbidity and mortality. Vaccine 1999, 17 (Suppl. 1), S3–S10. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Summary of the 2016–2017 Influenza Season. Available online: https://www.cdc.gov/flu/about/season/flu-season-2016-2017.htm (accessed on 10 June 2022).
- Centers for Disease Control and Prevention. Summary of the 2017–2018 Influenza Season. Available online: https://www.cdc.gov/flu/about/season/flu-season-2017-2018.htm (accessed on 10 June 2022).
- Goldstein, E.J.; Harvey, W.T.; Wilkie, G.S.; Shepherd, S.J.; MacLean, A.R.; Murcia, P.R.; Gunson, R.N. Integrating patient and whole-genome sequencing data to provide insights into the epidemiology of seasonal influenza A(H3N2) viruses. Microb. Genom. 2018, 4, e000137. [Google Scholar] [CrossRef]
- Baillie, G.J.; Galiano, M.; Agapow, P.M.; Myers, R.; Chiam, R.; Gall, A.; Palser, A.L.; Watson, S.J.; Hedge, J.; Underwood, A.; et al. Evolutionary dynamics of local pandemic H1N1/2009 influenza virus lineages revealed by whole-genome analysis. J. Virol. 2012, 86, 11–18. [Google Scholar] [CrossRef]
- Watson, S.J.; Langat, P.; Reid, S.M.; Lam, T.T.; Cotten, M.; Kelly, M.; Van Reeth, K.; Qiu, Y.; Simon, G.; Bonin, E.; et al. Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013. J. Virol. 2015, 89, 9920–9931. [Google Scholar] [CrossRef]
- Langat, P.; Raghwani, J.; Dudas, G.; Bowden, T.A.; Edwards, S.; Gall, A.; Bedford, T.; Rambaut, A.; Daniels, R.S.; Russell, C.A.; et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog. 2017, 13, e1006749. [Google Scholar] [CrossRef]
- Okoli, G.N.; Racovitan, F.; Abdulwahid, T.; Righolt, C.H.; Mahmud, S.M. Variable seasonal influenza vaccine effectiveness across geographical regions, age groups and levels of vaccine antigenic similarity with circulating virus strains: A systematic review and meta-analysis of the evidence from test-negative design studies after the 2009/10 influenza pandemic. Vaccine 2021, 39, 1225–1240. [Google Scholar]
- Ohmit, S.E.; Thompson, M.G.; Petrie, J.G.; Thaker, S.N.; Jackson, M.L.; Belongia, E.A.; Zimmerman, R.K.; Gaglani, M.; Lamerato, L.; Spencer, S.M.; et al. Influenza vaccine effectiveness in the 2011-2012 season: Protection against each circulating virus and the effect of prior vaccination on estimates. Clin. Infect. Dis. 2014, 58, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Ferdinands, J.M.; Gaglani, M.; Martin, E.T.; Monto, A.S.; Middleton, D.; Silveira, F.; Talbot, H.K.; Zimmerman, R.; Patel, M. Waning Vaccine Effectiveness Against Influenza-Associated Hospitalizations Among Adults, 2015-2016 to 2018–2019, United States Hospitalized Adult Influenza Vaccine Effectiveness Network. Clin. Infect. Dis. 2021, 73, 726–729. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Parkhouse, K.; Gumina, M.E.; Kim, K.; Diaz Perez, S.; Wilson, P.C.; Treanor, J.J.; Sant, A.J.; Cobey, S.; Hensley, S.E. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc. Natl. Acad. Sci. USA 2017, 114, 12578–12583. [Google Scholar] [CrossRef] [PubMed]
- Suderman, M.; Moniwa, M.; Alkie, T.N.; Ojkic, D.; Broes, A.; Pople, N.; Berhane, Y. Comparative Susceptibility of Madin-Darby Canine Kidney (MDCK) Derived Cell Lines for Isolation of Swine Origin Influenza A Viruses from Different Clinical Specimens. Viruses 2021, 13, 2346. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Barr, I.G.; Mosse, J.A.; Laurie, K.L. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J. Clin. Microbiol. 2008, 46, 2189–2194. [Google Scholar] [CrossRef]
- Chon, I.; Saito, R.; Hibino, A.; Yagami, R.; Dapat, C.; Odagiri, T.; Kondo, H.; Sato, I.; Kimura, S.; Kawashima, T.; et al. Effectiveness of the quadrivalent inactivated influenza vaccine in Japan during the 2015-2016 season: A test-negative case-control study comparing the results by real time PCR, virus isolation. Vaccine X 2019, 1, 100011. [Google Scholar] [CrossRef]
- Watson, S.J.; Welkers, M.R.; Depledge, D.P.; Coulter, E.; Breuer, J.M.; de Jong, M.D.; Kellam, P. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120205. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Westgeest, K.B.; Russell, C.A.; Lin, X.; Spronken, M.I.; Bestebroer, T.M.; Bahl, J.; van Beek, R.; Skepner, E.; Halpin, R.A.; de Jong, J.C.; et al. Genomewide analysis of reassortment and evolution of human influenza A(H3N2) viruses circulating between 1968 and 2011. J. Virol. 2014, 88, 2844–2857. [Google Scholar] [CrossRef]
- Rao, S.; Martin, J.; Ahearn, M.A.; Osborne, C.; Moss, A.; Dempsey, A.; Dominguez, S.R.; Weinberg, A.; Messacar, K.B. Neurologic Manifestations of Influenza A(H3N2) Infection in Children During the 2016–2017 Season. J. Pediatric Infect. Dis. Soc. 2020, 9, 71–74. [Google Scholar] [CrossRef]
- Nabakooza, G.; Pastusiak, A.; Kateete, D.P.; Lutwama, J.J.; Kitayimbwa, J.M.; Frost, S.D.W. Whole-genome analysis to determine the rate and patterns of intra-subtype reassortment among influenza type-A viruses in Africa. Virus Evol. 2022, 8, veac005. [Google Scholar] [CrossRef]
- Ampofo, W.K.; Azziz-Baumgartner, E.; Bashir, U.; Cox, N.J.; Fasce, R.; Giovanni, M.; Grohmann, G.; Huang, S.; Katz, J.; Mironenko, A.; et al. Strengthening the influenza vaccine virus selection and development process: Report of the 3rd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection held at WHO headquarters, Geneva, Switzerland, 1–3 April 2014. Vaccine 2015, 33, 4368–4382. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Chadha, M.; Al Mamun, A.; Rahman, M.; Sturm-Ramirez, K.; Chittaganpitch, M.; Pattamadilok, S.; Olsen, S.J.; Sampurno, O.D.; Setiawaty, V.; et al. Influenza seasonality and vaccination timing in tropical and subtropical areas of southern and south-eastern Asia. Bull. World Health Organ. 2014, 92, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Nyang’au, E.M.; Bulimo, W.D.; Mobegi, V.; Opanda, S.; Magiri, E. Genetic analysis of HA1 domain of influenza A/H3N2 viruses isolated in Kenya during the 2007-2013 seasons reveal significant divergence from WHO-recommended vaccine strains. Int. J. Infect. Dis. 2020, 95, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Boonnak, K.; Mansanguan, C.; Schuerch, D.; Boonyuen, U.; Lerdsamran, H.; Jiamsomboon, K.; Sae Wang, F.; Huntrup, A.; Prasertsopon, J.; Kosoltanapiwat, N.; et al. Molecular Characterization of Seasonal Influenza A and B from Hospitalized Patients in Thailand in 2018–2019. Viruses 2021, 13, 977. [Google Scholar] [CrossRef]
- Jagadesh, A.; Krishnan, A.; Nair, S.; Sivadas, S.; Arunkumar, G. Genetic characterization of hemagglutinin (HA) gene of influenza A viruses circulating in Southwest India during 2017 season. Virus Genes 2019, 55, 458–464. [Google Scholar] [CrossRef]
- Hampson, A.; Barr, I.; Cox, N.; Donis, R.O.; Siddhivinayak, H.; Jernigan, D.; Katz, J.; McCauley, J.; Motta, F.; Odagiri, T.; et al. Improving the selection and development of influenza vaccine viruses-Report of a WHO informal consultation on improving influenza vaccine virus selection, Hong Kong SAR, China, 18-20 November 2015. Vaccine 2017, 35, 1104–1109. [Google Scholar] [CrossRef]
- Potter, B.I.; Kondor, R.; Hadfield, J.; Huddleston, J.; Barnes, J.; Rowe, T.; Guo, L.; Xu, X.; Neher, R.A.; Bedford, T.; et al. Evolution and rapid spread of a reassortant A(H3N2) virus that predominated the 2017–2018 influenza season. Virus Evol. 2019, 5, vez046. [Google Scholar] [CrossRef]
- Kobasa, D.; Takada, A.; Shinya, K.; Hatta, M.; Halfmann, P.; Theriault, S.; Suzuki, H.; Nishimura, H.; Mitamura, K.; Sugaya, N.; et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 2004, 431, 703–707. [Google Scholar] [CrossRef]
- Melidou, A.; Gioula, G.; Exindari, M.; Ioannou, E.; Gkolfinopoulou, K.; Georgakopoulou, T.; Tsiodras, S.; Papa, A. Influenza A(H3N2) genetic variants in vaccinated patients in northern Greece. J. Clin. Virol. 2017, 94, 29–32. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, S.; Rao, H. Whole-genome sequences of two influenza A (H3N2) virus strains isolated from Qinghai, China. Biosafety Health 2021, 3, 81–86. [Google Scholar] [CrossRef]
- Takashita, E.; Daniels, R.S.; Fujisaki, S.; Gregory, V.; Gubareva, L.V.; Huang, W.; Hurt, A.C.; Lackenby, A.; Nguyen, H.T.; Pereyaslov, D.; et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2017–2018. Antivir. Res. 2020, 175, 104718. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi, A.; Shalhoub, S.; Rosenfeld, P.; Li, A.; Higgins, R.R.; Stogios, P.J.; Savchenko, A.; Bastien, N.; Li, Y.; Rotstein, C.; et al. Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob. Agents Chemother. 2014, 58, 7188–7197. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Pichon, M.; Valette, M.; Burfin, G.; Richard, M.; Lina, B.; Josset, L. Whole Genome Sequencing of A(H3N2) Influenza Viruses Reveals Variants Associated with Severity during the 2016–2017 Season. Viruses 2019, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Fodor, E. The RNA polymerase of influenza a virus: Mechanisms of viral transcription and replication. Acta Virol. 2013, 57, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.E.; Le Sage, V.; Padovani, G.H.; Calderon, M.; Wright, E.S.; Lakdawala, S.S. Parallel evolution between genomic segments of seasonal human influenza viruses reveals RNA-RNA relationships. Elife 2021, 10, e66525. [Google Scholar] [CrossRef]
- Hiromoto, Y.; Saito, T.; Lindstrom, S.E.; Li, Y.; Nerome, R.; Sugita, S.; Shinjoh, M.; Nerome, K. Phylogenetic analysis of the three polymerase genes (PB1, PB2 and PA) of influenza B virus. J. Gen. Virol. 2000, 81 Pt 4, 929–937. [Google Scholar] [CrossRef]
| Representative Strain | Clade | Amino Acid Mutations in Each Nucleotide Position | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2015 | 19 | 21 | 69 | 130 | 140 | 144 | 154 | 158 | 160 | 175 | 176 | 185 | 241 | 280 | 327 | 342 | 395 | 466 | 495 | 505 | |
| A/Switzerland/9715293/2013 a | 3C.3a | L | G | D | S | S | A | S | G | N | S | K | P | N | K | Q | R | G | K | G | D |
| 2015 Myanmar viruses | 3C.2a | I | R | N | T | R | T | A | R | S | Y | T | S | D | R | H | K | R | R | R | N |
| 2016 | 49 | 112 | 137 | 158 | 160 | 174 | 176 | 187 | 210 | 258 | 422 | 500 | |||||||||
| A/Hong Kong/4801/2014 b | 3C.2a | R | S | N | R | S | N | K | N | P | I | I | G | ||||||||
| 2016 Myanmar viruses | 3C.2a and 3C.2a1 | Q | N | K | G | G | H | T | K | L | M | V | E | ||||||||
| 2017 | 78 | 94 | 108 | 112 | 137 | 151 | 158 | 176 | 187 | 210 | 287 | 305 | 327 | 372 | 422 | 500 | |||||
| A/Hong Kong/4801/2014 b | 3C.2a | E | G | K | S | N | T | R | K | N | P | D | P | H | Q | I | G | ||||
| 2017 Myanmar viruses | 3C.2a1b | G | D | R | N | K | K | G | T | K | L | N | S | Q | L | V | E | ||||
| 2018 | 64 | 78 | 108 | 140 | 144 | 151 | 160 | 174 | 176 | 187 | 213 | 223 | 235 | 277 | 325 | 327 | 363 | 422 | 495 | 546 | |
| A/Singapore/INFIMH-16-0019/2016 c | 3C.2a1 | I | E | K | S | T | T | S | N | T | K | Q | K | S | R | V | H | V | V | E | A |
| 2018 Myanmar viruses | 3C.2a and 3C.2a1b | R | G | R | R | A | K | K | H | R | N | R | R | F | Q | I | Q | M | I | G | V |
| 2019 | 16 | 49 | 78 | 99 | 108 | 110 | 112 | 137 | 144 | 147 | 151 | 153 | 154 | 158 | 187 | 205 | 209 | 277 | 327 | 363 | |
| A/Switzerland/8060/2017 d | 3C.2a2 | A | R | E | K | K | Y | S | N | T | K | T | S | A | K | N | K | F | Q | H | V |
| 2019 Myanmar viruses | 3C.2a1b | T | Q | G | E | R | N | N | K | A | T | K | F | S | G | K | R | S | R | Q | M |
| 422 | 500 | 517 | 538 | 545 | 546 | ||||||||||||||||
| I | G | Q | I | V | A | ||||||||||||||||
| V | E | R | M | I | V | ||||||||||||||||
| Segment | Mean Rate of Substitution (Substitution/Site/Year) | |
|---|---|---|
| Mean | 95% Highest Posterior Density | |
| PB2 | 1.95 × 10−3 | 1.42 × 10−3 to 2.50 × 10−3 |
| PB1 | 1.64 × 10−3 | 1.11 × 10−3 to 2.18 × 10−3 |
| PA | 2.20 × 10−3 | 1.57 × 10−3 to 2.79 × 10−3 |
| HA | 3.37 × 10−3 | 2.52 × 10−3 to 4.21 × 10−3 |
| NP | 1.84 × 10−3 | 1.31 × 10−3 to 2.44 × 10−3 |
| NA | 2.89 × 10−3 | 2.13 × 10−3 to 3.71 × 10−3 |
| MP | 1.80 × 10−3 | 1.06 × 10−3 to 2.67 × 10−3 |
| NS | 4.19 × 10−5 | 3.25 × 10−5 to 5.16 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phyu, W.W.; Saito, R.; Kyaw, Y.; Lin, N.; Win, S.M.K.; Win, N.C.; Ja, L.D.; Htwe, K.T.Z.; Aung, T.Z.; Tin, H.H.; et al. Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019. Viruses 2022, 14, 2414. https://doi.org/10.3390/v14112414
Phyu WW, Saito R, Kyaw Y, Lin N, Win SMK, Win NC, Ja LD, Htwe KTZ, Aung TZ, Tin HH, et al. Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019. Viruses. 2022; 14(11):2414. https://doi.org/10.3390/v14112414
Chicago/Turabian StylePhyu, Wint Wint, Reiko Saito, Yadanar Kyaw, Nay Lin, Su Mon Kyaw Win, Nay Chi Win, Lasham Di Ja, Khin Thu Zar Htwe, Thin Zar Aung, Htay Htay Tin, and et al. 2022. "Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019" Viruses 14, no. 11: 2414. https://doi.org/10.3390/v14112414
APA StylePhyu, W. W., Saito, R., Kyaw, Y., Lin, N., Win, S. M. K., Win, N. C., Ja, L. D., Htwe, K. T. Z., Aung, T. Z., Tin, H. H., Pe, E. H., Chon, I., Wagatsuma, K., & Watanabe, H. (2022). Evolutionary Dynamics of Whole-Genome Influenza A/H3N2 Viruses Isolated in Myanmar from 2015 to 2019. Viruses, 14(11), 2414. https://doi.org/10.3390/v14112414







