Tumor Suppressor p53 Inhibits Hepatitis B Virus Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation in Human Hepatocellular Carcinoma Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Culture and Transfection
2.3. HBV Cell Culture Systems
2.4. Quantitative Real-Time PCR of HBV DNA
2.5. Southern Blot Analysis of Intracellular HBV DNA
2.6. HBV e Antigen Enzyme-Linked Immunosorbent Assay
2.7. Immunofluorescence Analysis
2.8. Luciferase Reporter Assay
2.9. Western Blot Analysis
2.10. Immunoprecipitation
2.11. Statistical Analysis
3. Results
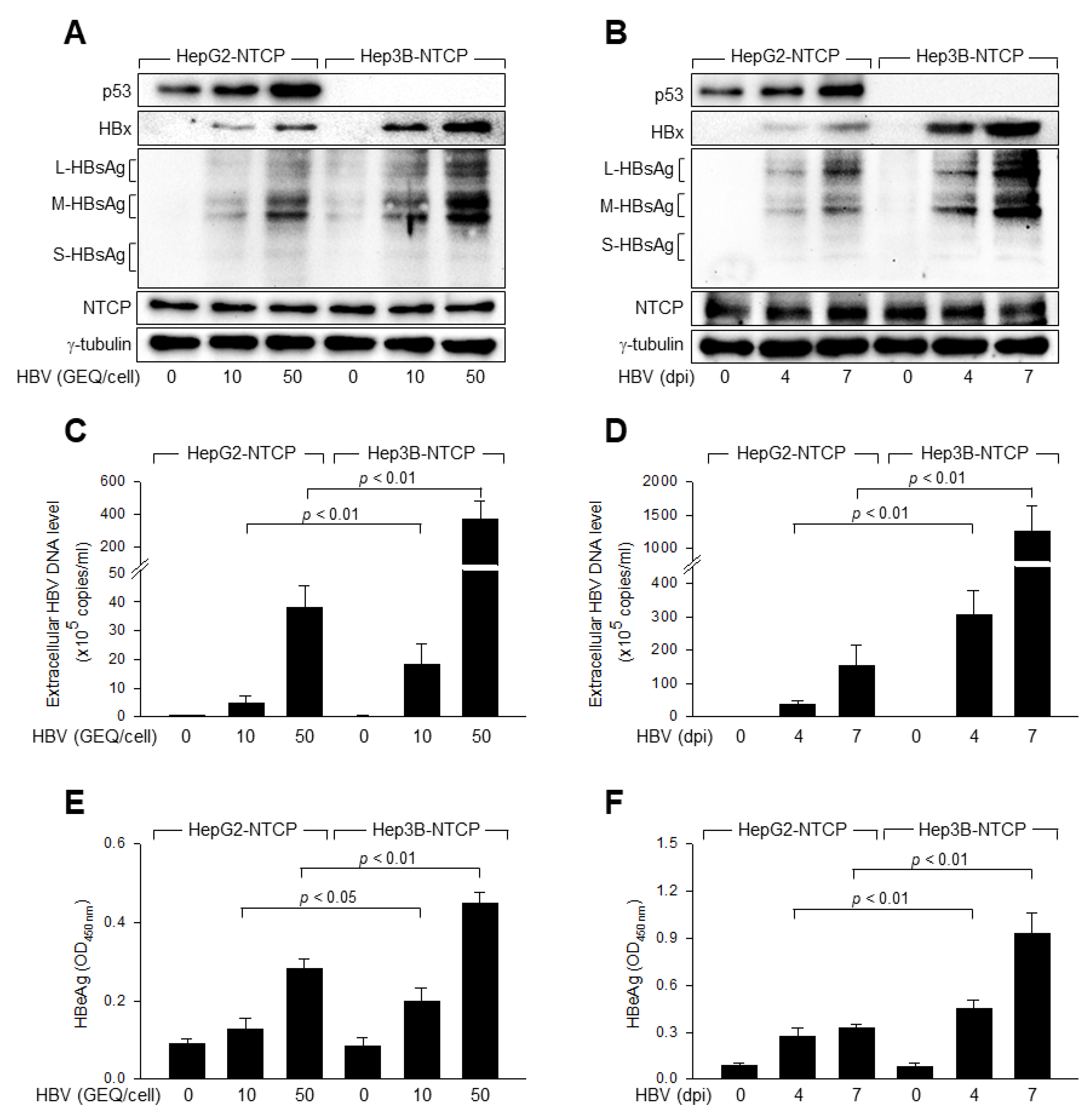
3.1. p53 Inhibits HBV Replication in Human Hepatoma Cells
3.2. p53 Transcriptional Activity Is Not Essential for the Inhibition of HBV Replication
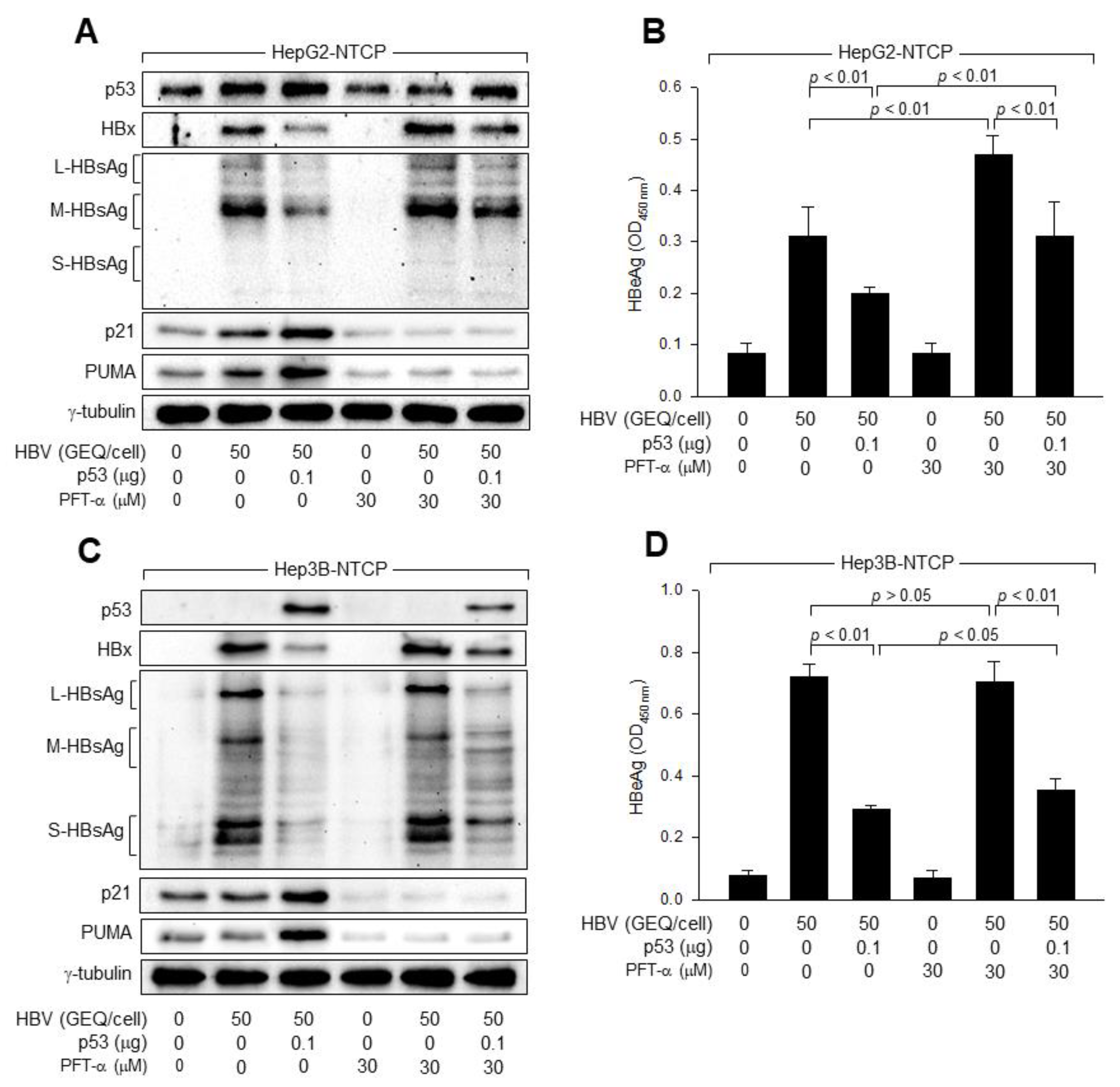
3.3. p53 Prevents HBx from Stimulating HBV Replication in Human Hepatoma Cells
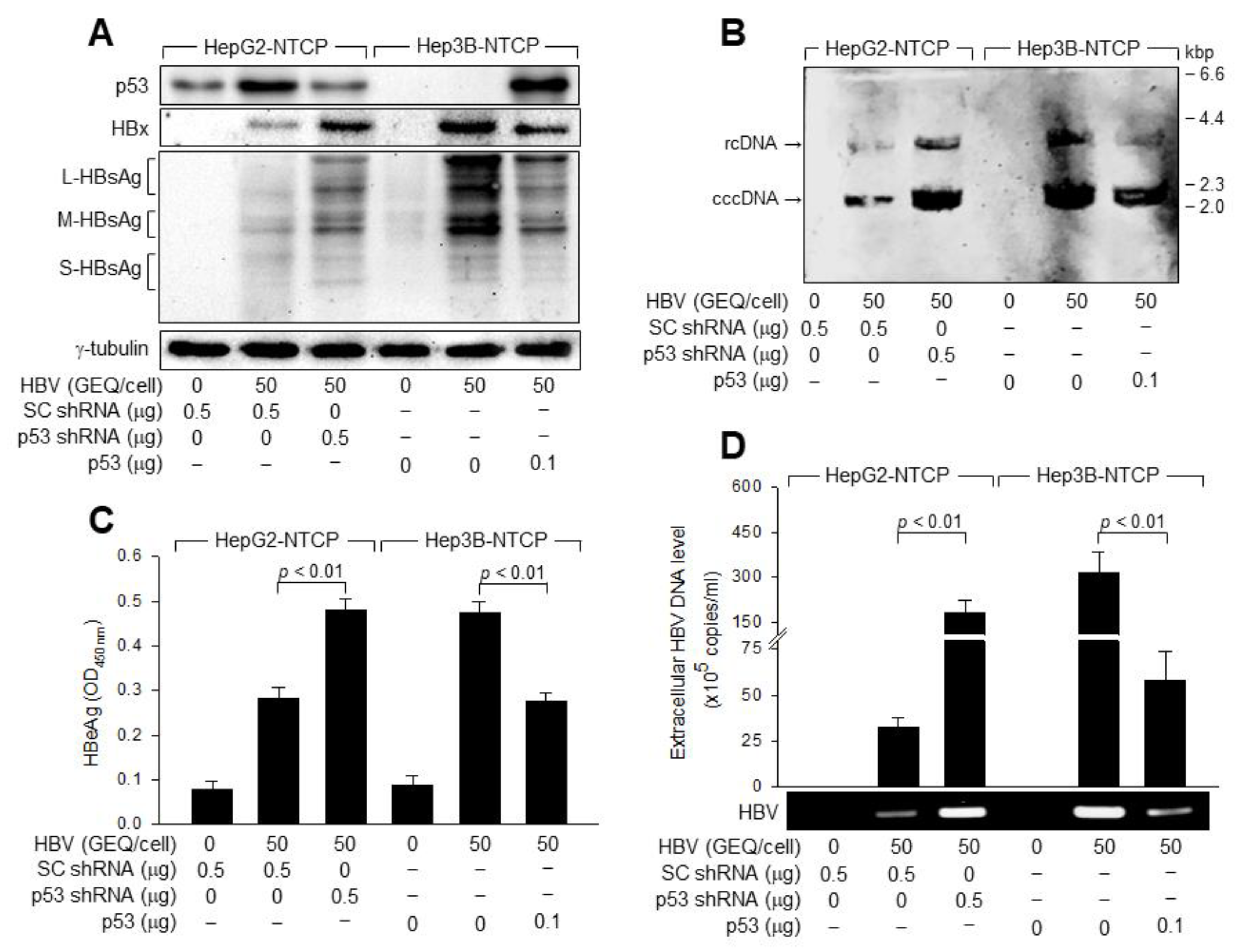
3.4. p53 Downregulates HBx to Inhibit HBV Replication in Human Hepatoma Cells
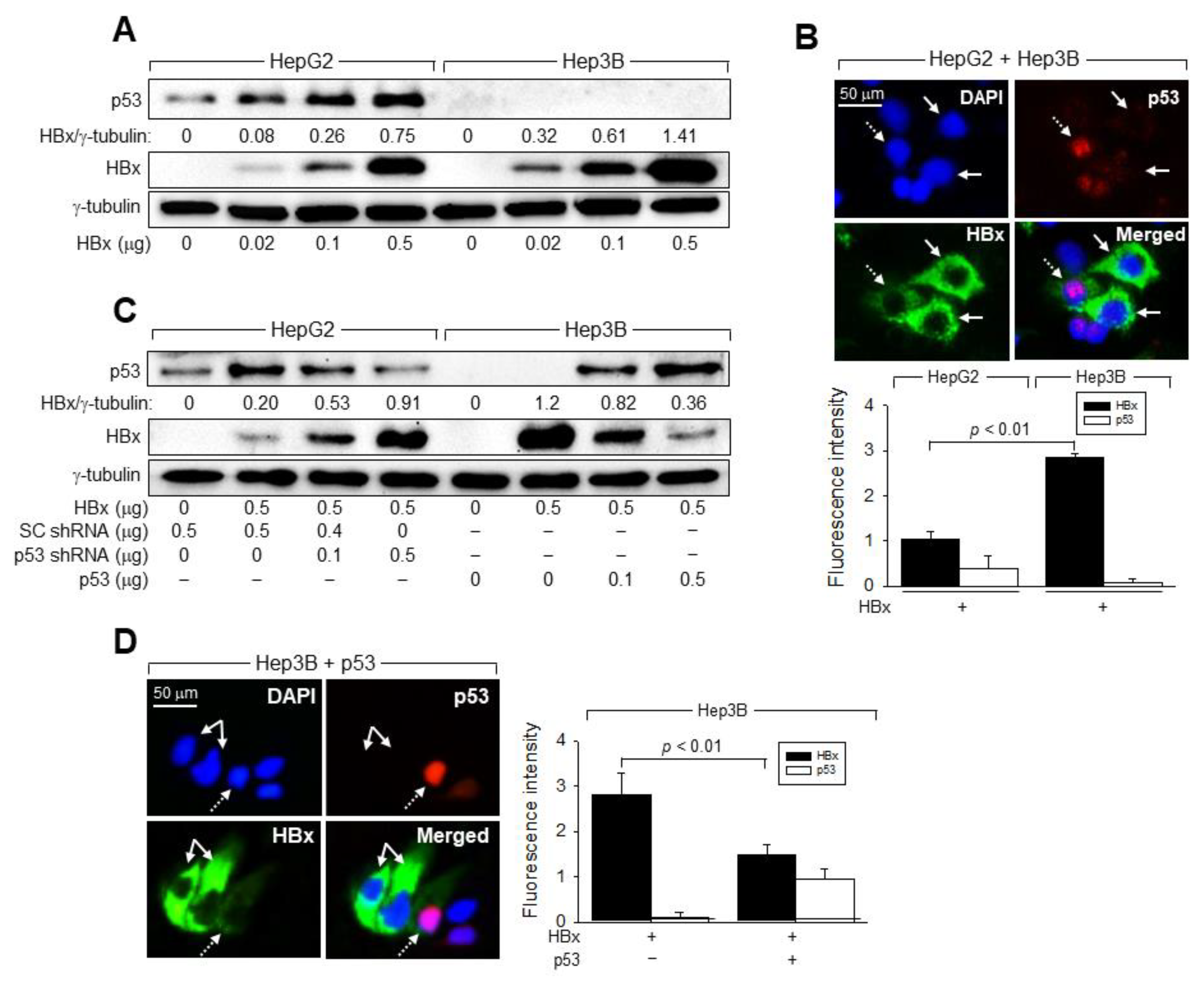
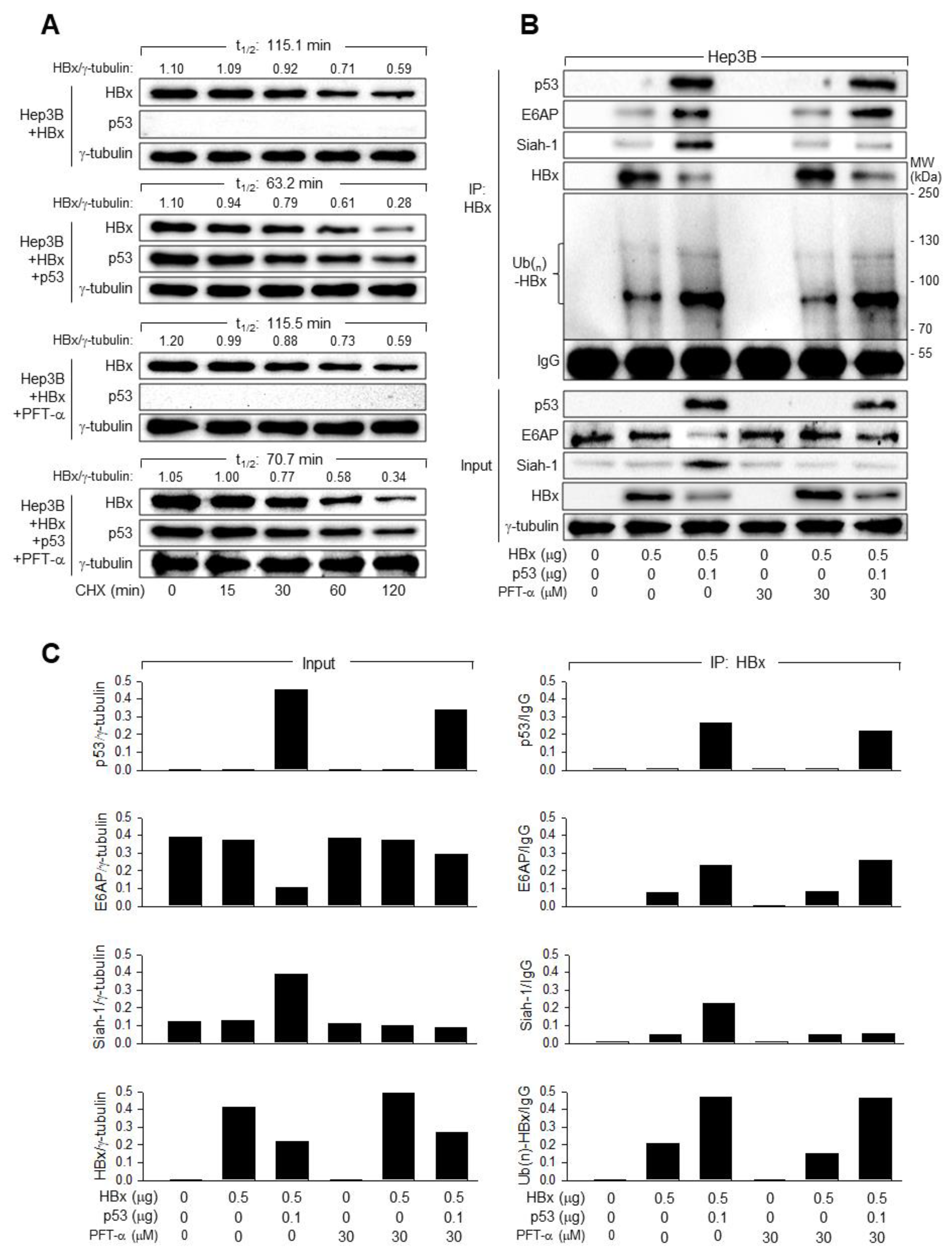
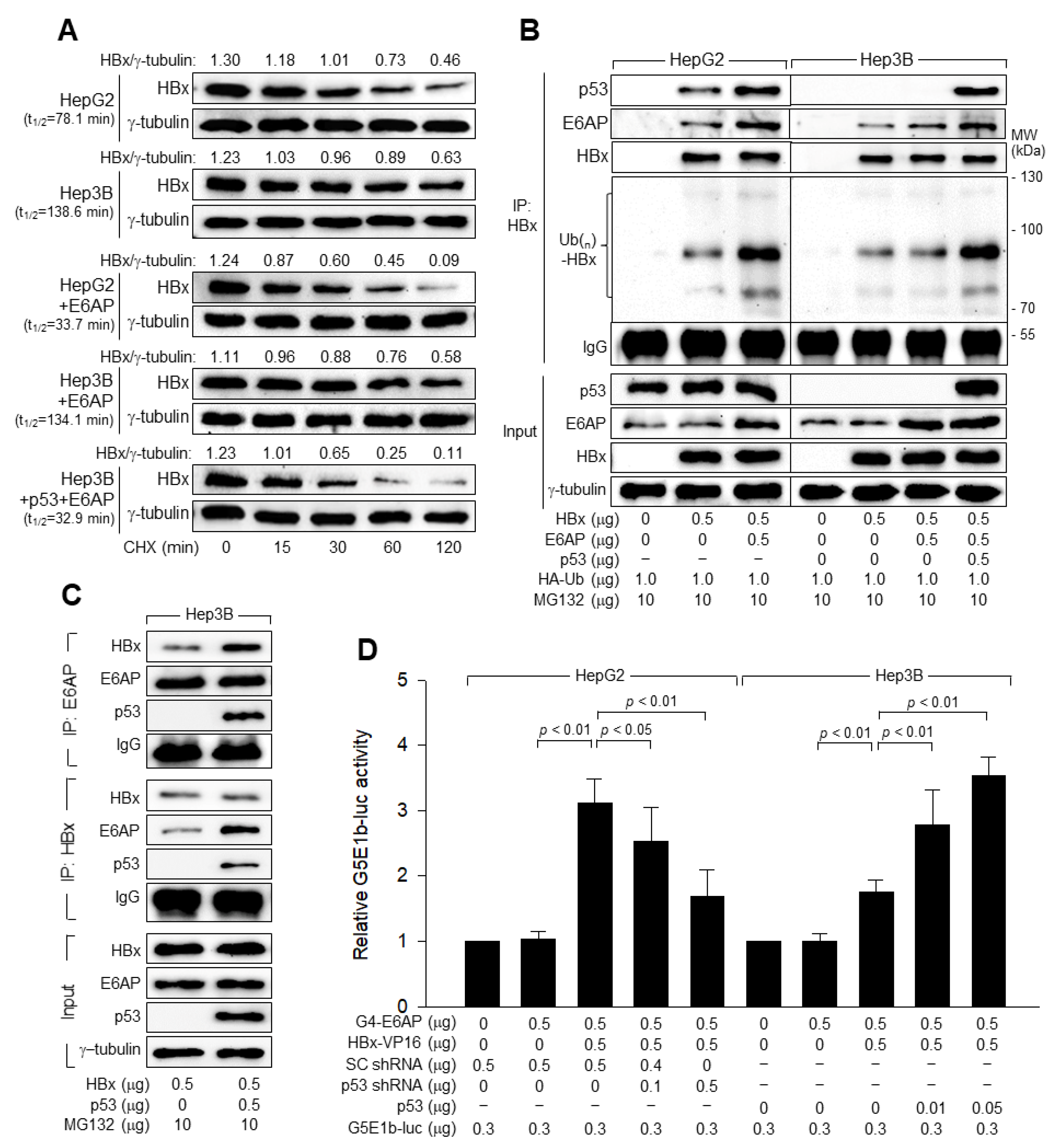
3.5. p53 Induces Ubiquitination and Proteasomal Degradation of HBx
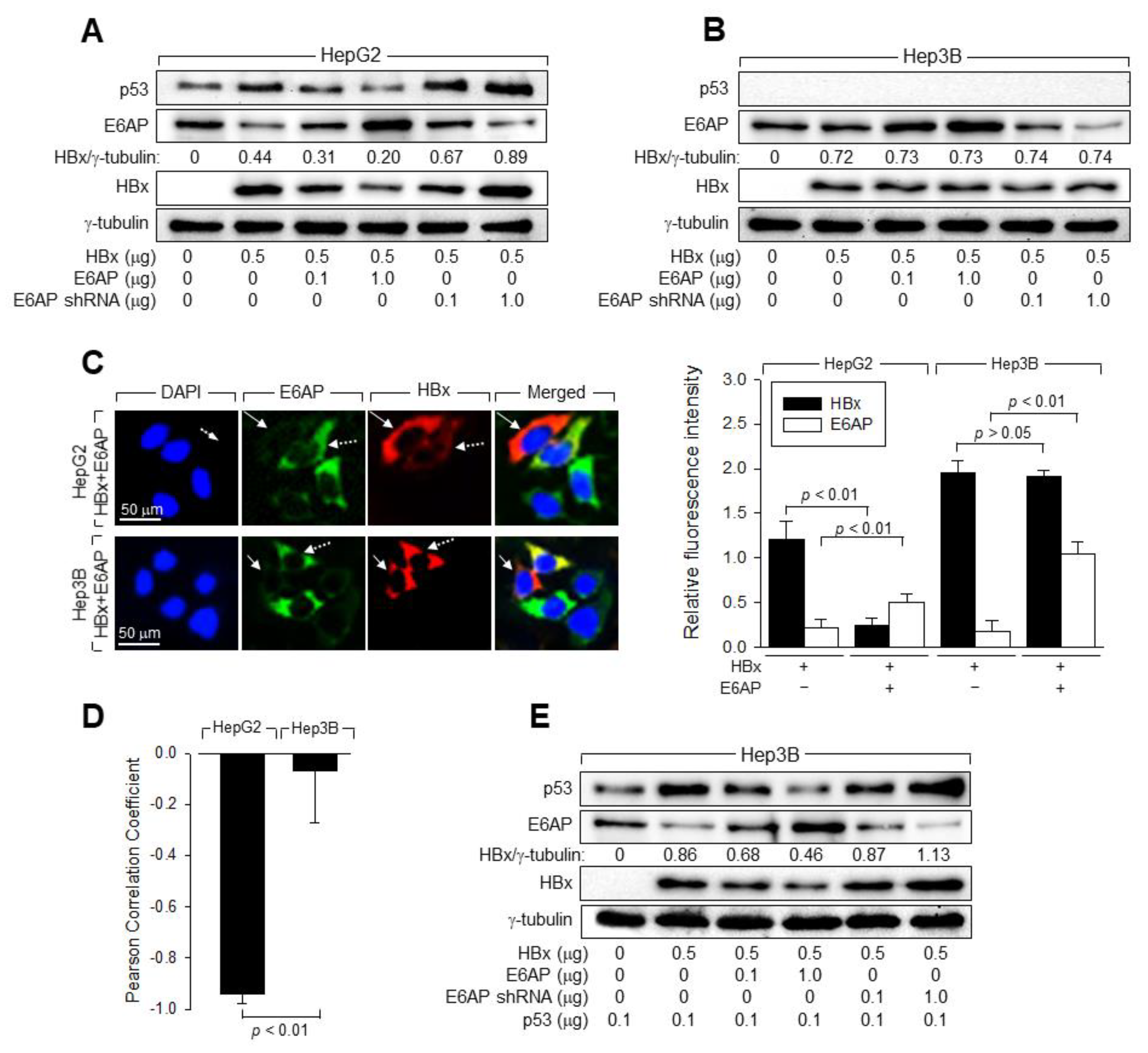
3.6. E6AP Is Responsible for the p53-Induced Ubiquitination of HBx
3.7. p53 and E6AP Help Each Other in Binding to HBx in Human Hepatoma Cells
3.8. p53 Inhibits HBV Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef] [PubMed]
- Lamontagne, R.J.; Bagga, S.; Bouchard, M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma. Res. 2016, 2, 163–186. [Google Scholar] [CrossRef] [PubMed]
- Locarnini, S.; Zoulim, F. Molecular genetics of HBV infection. Antivir. Ther. 2010, 15 (Suppl. 3), 3–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Koh, S.S.; Lee, C.G. Hepatitis B Virus X Protein and Hepatocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 940. [Google Scholar] [CrossRef]
- Tsuge, M.; Hiraga, N.; Akiyama, R.; Tanaka, S.; Matsushita, M.; Mitsui, F.; Abe, H.; Kitamura, S.; Hatakeyama, T.; Kimura, T.; et al. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J. Gen. Virol. 2010, 91, 1854–1864. [Google Scholar] [CrossRef]
- Xu, Z.; Yen, T.S.; Wu, L.; Madden, C.R.; Tan, W.; Slagle, B.L.; Ou, J.H. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 2002, 76, 2579–2584. [Google Scholar] [CrossRef]
- Keasler, V.V.; Hodgson, A.J.; Madden, C.R.; Slagle, B.L. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology 2009, 390, 122–129. [Google Scholar] [CrossRef]
- Keasler, V.V.; Hodgson, A.J.; Madden, C.R.; Slagle, B.L. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J. Virol. 2007, 81, 2656–2662. [Google Scholar] [CrossRef]
- Tang, H.; Delgermaa, L.; Huang, F.; Oishi, N.; Liu, L.; He, F.; Zhao, L.; Murakami, S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 2005, 79, 5548–5556. [Google Scholar] [CrossRef]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral. Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef]
- Bouchard, M.J.; Wang, L.H.; Schneider, R.J. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 2001, 294, 2376–2378. [Google Scholar] [CrossRef] [PubMed]
- Rawat, S.; Bouchard, M.J. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J. Virol. 2015, 89, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Luo, H. Interplay between the virus and the ubiquitin-proteasome system: Molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, Z.; Doo, E.; Coux, O.; Goldberg, A.L.; Liang, T.J. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 1999, 73, 7231–7240. [Google Scholar] [CrossRef] [PubMed]
- Schek, N.; Bartenschlager, R.; Kuhn, C.; Schaller, H. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene 1991, 6, 1735–1744. [Google Scholar]
- Zhao, J.; Wang, C.; Wang, J.; Yang, X.; Diao, N.; Li, Q.; Wang, W.; Xian, L.; Fang, Z.; Yu, L. E3 ubiquitin ligase Siah-1 facilitates poly-ubiquitylation and proteasomal degradation of the hepatitis B viral X protein. FEBS Lett. 2011, 585, 2943–2950. [Google Scholar] [CrossRef]
- Yeom, S.; Kim, S.S.; Jeong, H.; Jang, K.L. Hepatitis B virus X protein activates E3 ubiquitin ligase Siah-1 to control virus propagation via a negative feedback loop. J. Gen. Virol. 2017, 98, 1774–1784. [Google Scholar] [CrossRef]
- Ling, M.T.; Chiu, Y.T.; Lee, T.K.; Leung, S.C.; Fung, M.K.; Wang, X.; Wong, K.F.; Wong, Y.C. Id-1 induces proteasome-dependent degradation of the HBX protein. J. Mol. Biol. 2008, 382, 34–43. [Google Scholar] [CrossRef]
- Kido, T.; Ou, J.H.; Lau, Y.F. The X-linked tumor suppressor TSPX interacts and promotes degradation of the hepatitis B viral protein HBx via the proteasome pathway. PLoS One 2011, 6, e22979. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell. Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Sato, Y.; Tsurumi, T. Genome guardian p53 and viral infections. Rev. Med. Virol. 2013, 23, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, Y.H.; Huh, Y.S.; Moon, H.; Yun, Y. X-gene product antagonizes the p53-mediated inhibition of hepatitis B virus replication through regulation of the pregenomic/core promoter. J. Biol. Chem. 1995, 270, 31405–31412. [Google Scholar] [CrossRef] [PubMed]
- Truant, R.; Antunovic, J.; Greenblatt, J.; Prives, C.; Cromlish, J.A. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 1995, 69, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Forrester, K.; Yeh, H.; Feitelson, M.A.; Gu, J.R.; Harris, C.C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 1994, 91, 2230–2234. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell Biol. 1993, 13, 775–784. [Google Scholar]
- Kwun, H.J.; Jang, K.L. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004, 32, 2202–2213. [Google Scholar] [CrossRef]
- Cha, M.Y.; Ryu, D.K.; Jung, H.S.; Chang, H.E.; Ryu, W.S. Stimulation of hepatitis B virus genome replication by HBx is linked to both nuclear and cytoplasmic HBx expression. J. Gen. Virol. 2009, 90, 978–986. [Google Scholar] [CrossRef]
- Sadowski, I.; Ptashne, M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989, 17, 7539. [Google Scholar] [CrossRef]
- Lee, C.W.; Sorensen, T.S.; Shikama, N.; La Thangue, N.B. Functional interplay between p53 and E2F through co-activator p300. Oncogene 1998, 16, 2695–2710. [Google Scholar] [CrossRef][Green Version]
- Mendy, M.E.; Kaye, S.; van der Sande, M.; Rayco-Solon, P.; Waight, P.A.; Shipton, D.; Awi, D.; Snell, P.; Whittle, H.; McConkey, S.J. Application of real-time PCR to quantify hepatitis B virus DNA in chronic carriers in The Gambia. Virol. J. 2006, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Nie, H.; Yan, R.; Guo, J.T.; Block, T.M.; Guo, H. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol. Biol. 2013, 1030, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kawaguchi, K.; Honda, M.; Hashimoto, S.; Shirasaki, T.; Okada, H.; Orita, N.; Shimakami, T.; Yamashita, T.; Sakai, Y.; et al. Notch signaling facilitates hepatitis B virus covalently closed circular DNA transcription via cAMP response element-binding protein with E3 ubiquitin ligase-modulation. Sci. Rep. 2019, 9, 1621. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Yoon, H.; Jeong, Y.; Jang, K.L. Tumour suppressor p53 inhibits hepatitis C virus replication by inducing E6AP-mediated proteasomal degradation of the viral core protein. FEBS Lett. 2022, 596, 2525–2537. [Google Scholar] [CrossRef]
- Qiu, G.H.; Xie, X.; Xu, F.; Shi, X.; Wang, Y.; Deng, L. Distinctive pharmacological differences between liver cancer cell lines HepG2 and Hep3B. Cytotechnology 2015, 67, 1–12. [Google Scholar] [CrossRef]
- Cha, S.; Jang, K.L. Hepatitis B virus X protein stimulates cell growth by downregulating p16 levels via PA28gamma-mediated proteasomal degradation. J. Gen. Virol. 2020, 101, 963–971. [Google Scholar] [CrossRef]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef]
- Han, J.; Kim, H.; Jeong, H.; Yoon, H.; Jang, K.L. Proteasomal activator 28 gamma stabilizes hepatitis B virus X protein by competitively inhibiting the Siah-1-mediated proteasomal degradation. Biochem. Biophys. Res. Commun. 2021, 578, 97–103. [Google Scholar] [CrossRef]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 2012, 1, e00049. [Google Scholar] [CrossRef]
- Gerlich, W.H.; Heermann, K.H.; Lu, X. Functions of hepatitis B surface proteins. Arch. Virol. Suppl. 1992, 4, 129–132. [Google Scholar] [CrossRef]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef]
- Komarov, P.G.; Komarova, E.A.; Kondratov, R.V.; Christov-Tselkov, K.; Coon, J.S.; Chernov, M.V.; Gudkov, A.V. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 1999, 285, 1733–1737. [Google Scholar] [CrossRef] [PubMed]
- Sohn, D.; Graupner, V.; Neise, D.; Essmann, F.; Schulze-Osthoff, K.; Janicke, R.U. Pifithrin-alpha protects against DNA damage-induced apoptosis downstream of mitochondria independent of p53. Cell Death Differ. 2009, 16, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Cha, S.; Jang, K.L. HBx natural variants containing Ser-101 instead of Pro-101 evade ubiquitin-dependent proteasomal degradation by activating proteasomal activator 28 gamma expression. J. Gen. Virol. 2019, 100, 1554–1566. [Google Scholar] [CrossRef]
- Collot-Teixeira, S.; Bass, J.; Denis, F.; Ranger-Rogez, S. Human tumor suppressor p53 and DNA viruses. Rev. Med. Virol. 2004, 14, 301–319. [Google Scholar] [CrossRef]
- Takaoka, A.; Hayakawa, S.; Yanai, H.; Stoiber, D.; Negishi, H.; Kikuchi, H.; Sasaki, S.; Imai, K.; Shibue, T.; Honda, K.; et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003, 424, 516–523. [Google Scholar] [CrossRef]
- Munoz-Fontela, C.; Macip, S.; Martinez-Sobrido, L.; Brown, L.; Ashour, J.; Garcia-Sastre, A.; Lee, S.W.; Aaronson, S.A. Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 2008, 205, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Taura, M.; Eguma, A.; Suico, M.A.; Shuto, T.; Koga, T.; Komatsu, K.; Komune, T.; Sato, T.; Saya, H.; Li, J.D.; et al. p53 regulates Toll-like receptor 3 expression and function in human epithelial cell lines. Mol. Cell Biol. 2008, 28, 6557–6567. [Google Scholar] [CrossRef]
- Hummer, B.T.; Li, X.L.; Hassel, B.A. Role for p53 in gene induction by double-stranded RNA. J. Virol. 2001, 75, 7774–7777. [Google Scholar] [CrossRef]
- Rivas, C.; Aaronson, S.A.; Munoz-Fontela, C. Dual Role of p53 in Innate Antiviral Immunity. Viruses 2010, 2, 298–313. [Google Scholar] [CrossRef]
- Uchida, T.; Takahashi, K.; Tatsuno, K.; Dhingra, U.; Eliason, J.F. Inhibition of hepatitis-B-virus core promoter by p53: Implications for carcinogenesis in hepatocytes. Int. J. Cancer 1996, 67, 892–897. [Google Scholar] [CrossRef]
- Ori, A.; Zauberman, A.; Doitsh, G.; Paran, N.; Oren, M.; Shaul, Y. p53 binds and represses the HBV enhancer: An adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998, 17, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Carmona, S.; Ely, A.; Crowther, C.; Moolla, N.; Salazar, F.H.; Marion, P.L.; Ferry, N.; Weinberg, M.S.; Arbuthnot, P. Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Mol. Ther. 2006, 13, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Park, S.G.; Min, J.Y.; Chung, C.; Hsieh, A.; Jung, G. Tumor suppressor protein p53 induces degradation of the oncogenic protein HBx. Cancer Lett. 2009, 282, 229–237. [Google Scholar] [CrossRef]
- Kwak, J.; Shim, J.H.; Tiwari, I.; Jang, K.L. Hepatitis C virus core protein inhibits E6AP expression via DNA methylation to escape from ubiquitin-dependent proteasomal degradation. Cancer Lett. 2016, 380, 59–68. [Google Scholar] [CrossRef]
- Kim, J.H.; Sohn, S.Y.; Benedict Yen, T.S.; Ahn, B.Y. Ubiquitin-dependent and -independent proteasomal degradation of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 2008, 366, 1036–1042. [Google Scholar] [CrossRef]
- Li, S.; Hong, X.; Wei, Z.; Xie, M.; Li, W.; Liu, G.; Guo, H.; Yang, J.; Wei, W.; Zhang, S. Ubiquitination of the HPV Oncoprotein E6 Is Critical for E6/E6AP-Mediated p53 Degradation. Front. Microbiol. 2019, 10, 2483. [Google Scholar] [CrossRef]
- Talis, A.L.; Huibregtse, J.M.; Howley, P.M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 1998, 273, 6439–6445. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, H.-Y.; Han, J.; Yoon, H.; Jang, K.L. Tumor Suppressor p53 Inhibits Hepatitis B Virus Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation in Human Hepatocellular Carcinoma Cell Lines. Viruses 2022, 14, 2313. https://doi.org/10.3390/v14102313
Lim H-Y, Han J, Yoon H, Jang KL. Tumor Suppressor p53 Inhibits Hepatitis B Virus Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation in Human Hepatocellular Carcinoma Cell Lines. Viruses. 2022; 14(10):2313. https://doi.org/10.3390/v14102313
Chicago/Turabian StyleLim, Ha-Yeon, Jiwoo Han, Hyunyoung Yoon, and Kyung Lib Jang. 2022. "Tumor Suppressor p53 Inhibits Hepatitis B Virus Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation in Human Hepatocellular Carcinoma Cell Lines" Viruses 14, no. 10: 2313. https://doi.org/10.3390/v14102313
APA StyleLim, H.-Y., Han, J., Yoon, H., & Jang, K. L. (2022). Tumor Suppressor p53 Inhibits Hepatitis B Virus Replication by Downregulating HBx via E6AP-Mediated Proteasomal Degradation in Human Hepatocellular Carcinoma Cell Lines. Viruses, 14(10), 2313. https://doi.org/10.3390/v14102313



