Protein Degradation by Gammaherpesvirus RTAs: More Than Just Viral Transactivators
Abstract
1. Introduction
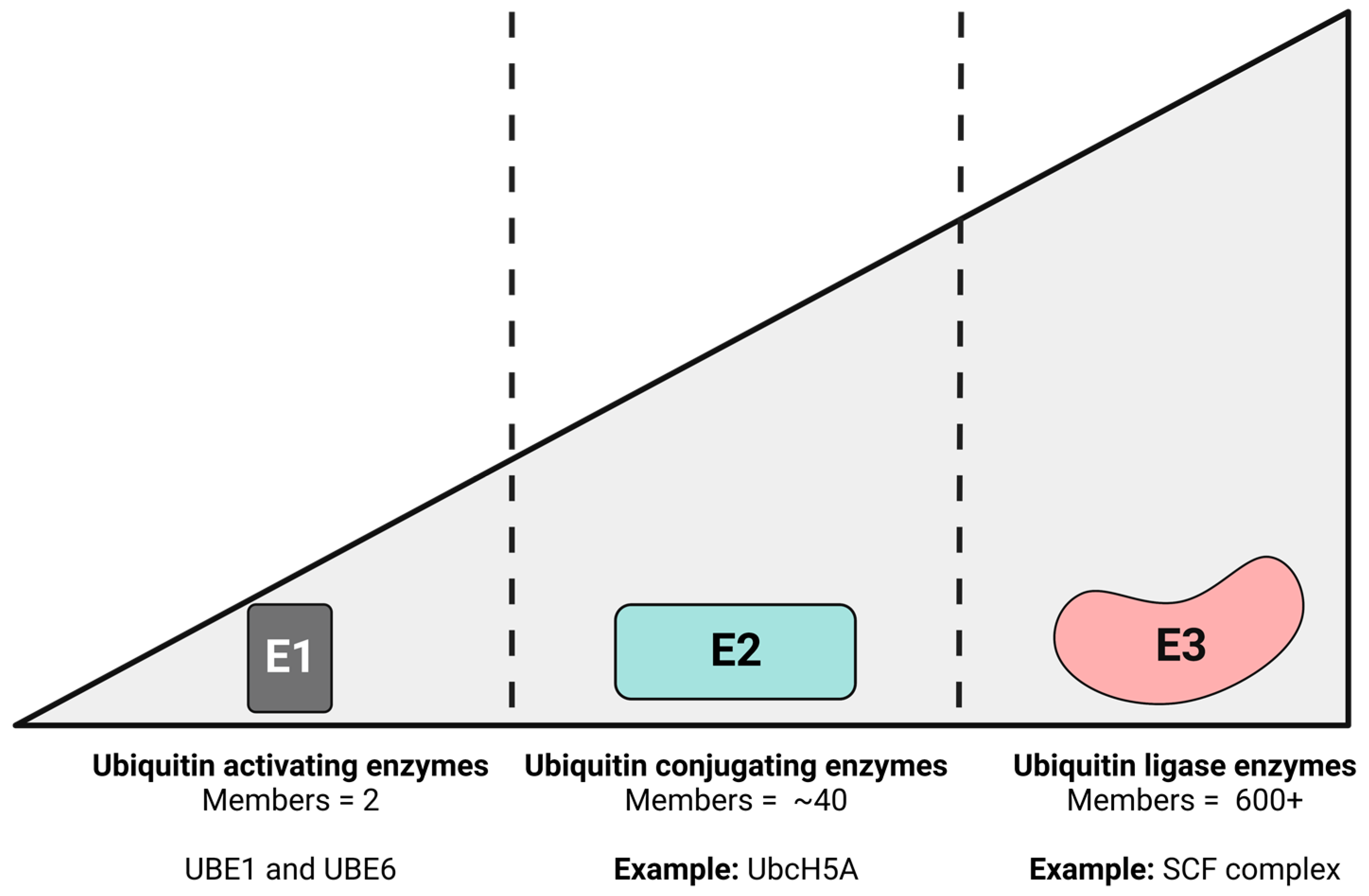
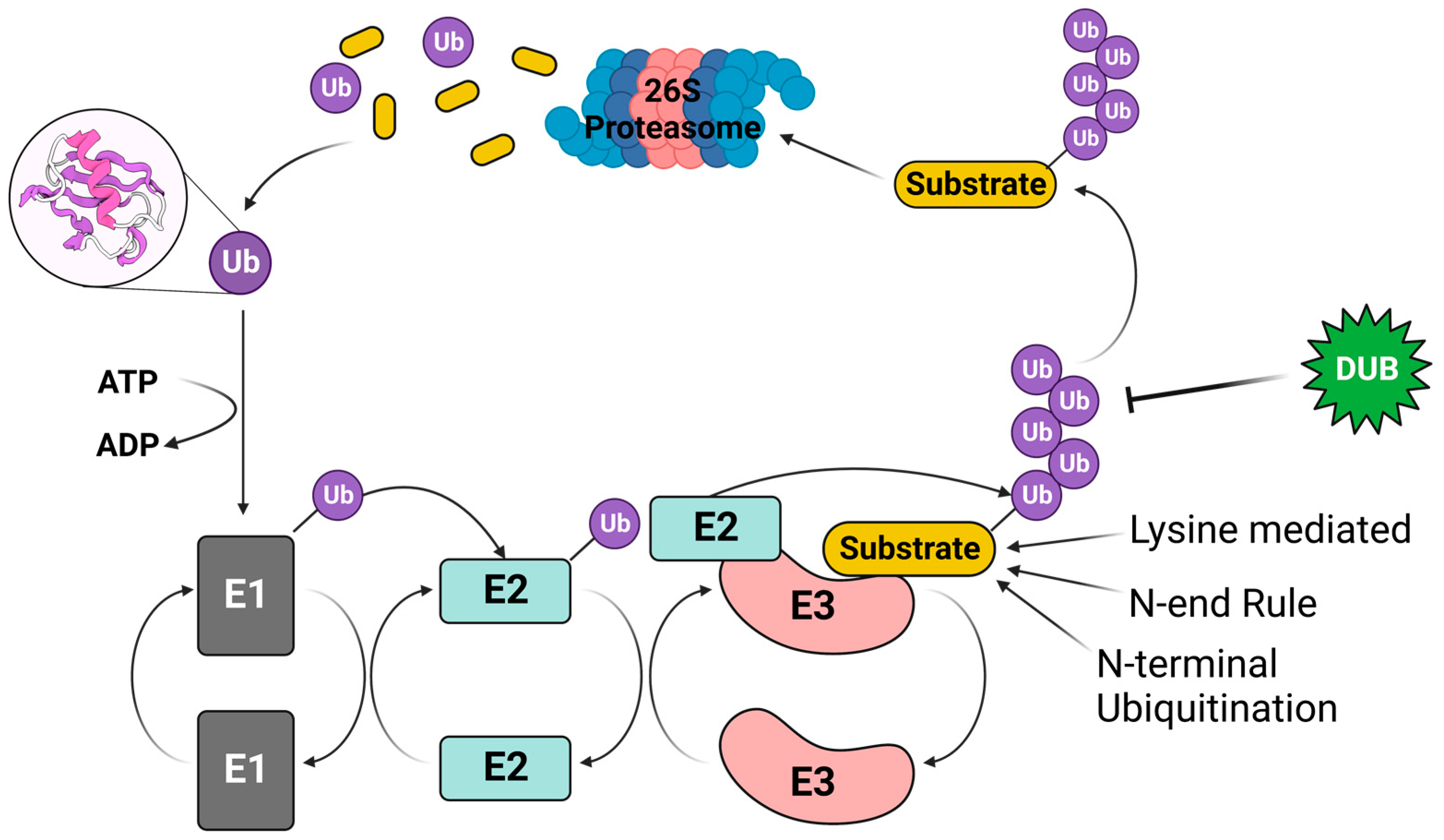
2. The Ubiquitin–Proteasome Pathway (UPP)
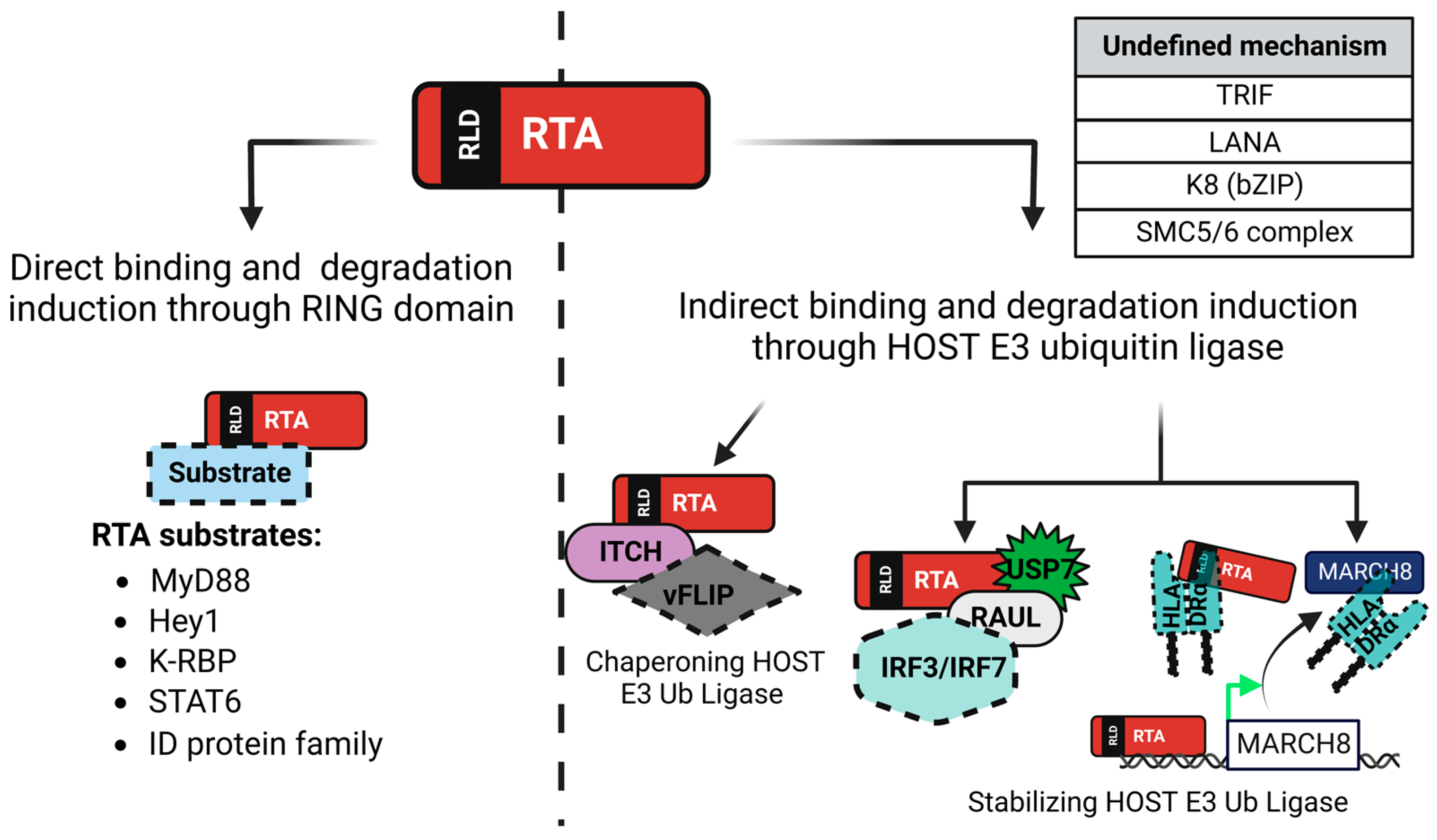
3. KSHV-Encoded Viral Factors Inducing Protein Degradation
4. RTA Suppresses Innate and Adaptive Immune Responses Induced by KSHV Infection
4.1. Toll-Interleukin-1 Receptor Domain-Containing Adaptor Protein-Inducing Interferon β (TRIF)
4.2. Myeloid Differentiation Factor 88 (MyD88)
4.3. Interferon Regulatory Factors 3 and 7 (IRF3 and IRF7)
4.4. Signal Transducer and Activator of Transcription 6 (STAT6)
4.5. Major Histocompatibility Complex, Class II, DR Alpha (HLA-DRα)
5. RTA-Mediated Degradation of Cellular Repressors of RTA
5.1. Inhibitor of DNA Binding Protein (ID) Family
5.2. Hairy/Enhancer-of-Split Related with YRPW Motif Protein 1 (Hey1)
5.3. KSHV-RTA Binding Protein (K-RBP)
5.4. Structural Maintenance of Chromosome 5 and 6 (SMC5/6) Complex
6. Viral Targets of KSHV RTA for Protein Degradation
6.1. K-bZIP (K8)
6.2. Latency-Associated Nuclear Antigen (LANA)
6.3. vFLIP
7. Gammaherpesvirus RTA Homologs
7.1. The Lymphocryptovirus: Epstein–Barr Virus (EBV)
7.2. The Rhadinoviruses: Herpesvirus Saimiri (HVS), Rhesus Monkey Rhadinovirus (RRV), and Murine Gammaherpesvirus 68 (MHV-68)
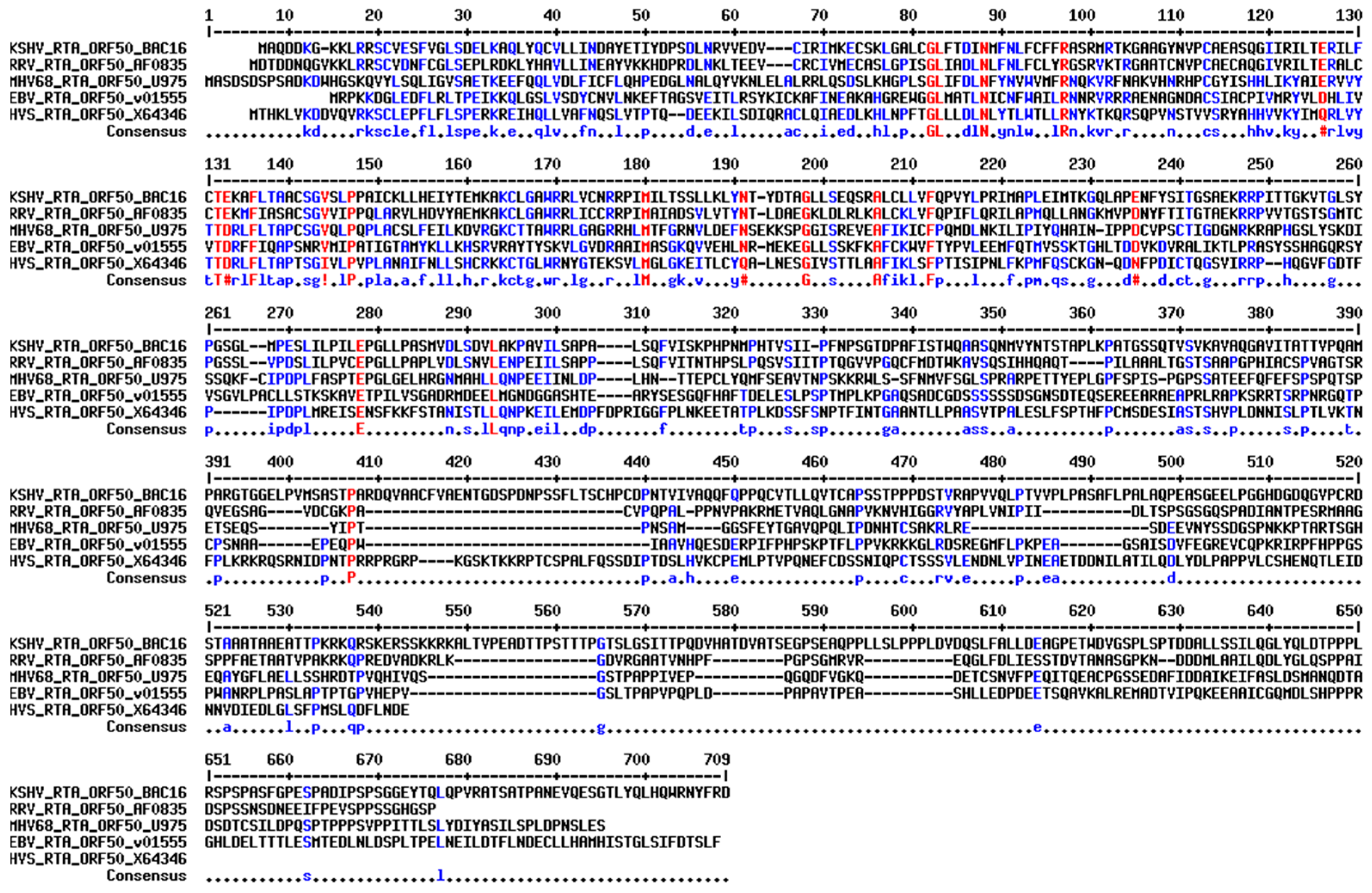
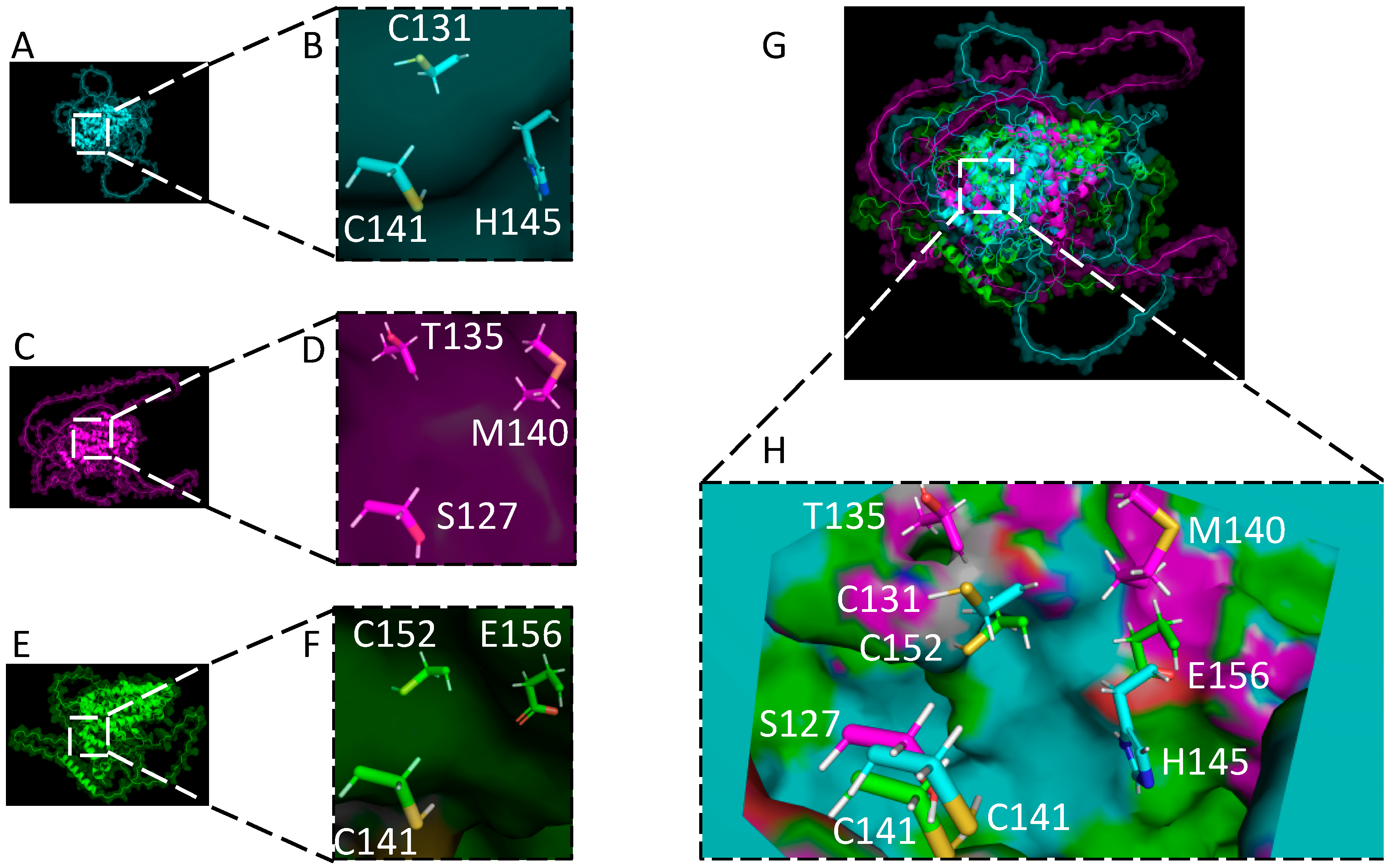
8. Structural Comparison of Gammaherpesvirus RTAs
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Vossen, M.T.; Westerhout, E.M.; Söderberg-Nauclér, C.; Wiertz, E.J. Viral immune evasion: A masterpiece of evolution. Immunogenetics 2002, 54, 527–542. [Google Scholar] [CrossRef]
- Sun, R.; Lin, S.F.; Gradoville, L.; Yuan, Y.; Zhu, F.; Miller, G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1998, 95, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Dupin, N.; Grandadam, M.; Calvez, V.; Gorin, I.; Aubin, J.T.; Havard, S.; Lamy, F.; Leibowitch, M.; Huraux, J.M.; Escande, J.P.; et al. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi’s sarcoma. Lancet 1995, 345, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef]
- Polizzotto, M.N.; Uldrick, T.S.; Wyvill, K.M.; Aleman, K.; Marshall, V.; Wang, V.; Whitby, D.; Pittaluga, S.; Jaffe, E.S.; Millo, C.; et al. Clinical Features and Outcomes of Patients With Symptomatic Kaposi Sarcoma Herpesvirus (KSHV)-associated Inflammation: Prospective Characterization of KSHV Inflammatory Cytokine Syndrome (KICS). Clin. Infect. Dis. 2015, 62, 730–738. [Google Scholar] [CrossRef]
- Cesarman, E. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 2011, 305, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of Herpesvirus-Like DNA Sequences in AIDS-Sssociated Kaposi’s Sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Greene, W.; Zhang, W.; He, M.; Witt, C.; Ye, F.; Gao, S.-J. The Ubiquitin/Proteasome System Mediates Entry and Endosomal Trafficking of Kaposi’s Sarcoma-Associated Herpesvirus in Endothelial Cells. PLoS Pathog. 2012, 8, e1002703. [Google Scholar] [CrossRef]
- Papp, B.; Motlagh, N.; Smindak, R.J.; Jin Jang, S.; Sharma, A.; Alonso, J.D.; Toth, Z. Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi’s Sarcoma-Associated Herpesvirus Lytic Reactivation. J. Virol. 2019, 93, e01978-18. [Google Scholar] [CrossRef]
- Gonzalez-Lopez, O.; DeCotiis, J.; Goyeneche, C.; Mello, H.; Vicente-Ortiz, B.A.; Shin, H.J.; Driscoll, K.E.; Du, P.; Palmeri, D.; Lukac, D.M. A herpesvirus transactivator and cellular POU proteins extensively regulate DNA binding of the host Notch signaling protein RBP-Jκ to the virus genome. J. Biol. Chem. 2019, 294, 13073–13092. [Google Scholar] [CrossRef]
- Ellison, T.J.; Izumiya, Y.; Izumiya, C.; Luciw, P.A.; Kung, H.J. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi’s sarcoma-associated herpesvirus. Virology 2009, 387, 76–88. [Google Scholar] [CrossRef]
- Campbell, M.; Watanabe, T.; Nakano, K.; Davis, R.R.; Lyu, Y.; Tepper, C.G.; Durbin-Johnson, B.; Fujimuro, M.; Izumiya, Y. KSHV episomes reveal dynamic chromatin loop formation with domain-specific gene regulation. Nat. Commun. 2018, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Kaul, R.; Purushothaman, P.; Uppal, T.; Verma, S.C. KSHV lytic proteins K-RTA and K8 bind to cellular and viral chromatin to modulate gene expression. PLoS ONE 2019, 14, e0215394. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, S.E.; Hayward, G.S. The KSHV Immediate-Early Transcription Factor RTA Encodes Ubiquitin E3 Ligase Activity that Targets IRF7 for Proteosome-Mediated Degradation. Immunity 2005, 22, 59–70. [Google Scholar] [CrossRef]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D.; Audhya, T.K. Stimulation of ATP-dependent proteolysis requires ubiquitin with the COOH-terminal sequence Arg-Gly-Gly. J. Biol. Chem. 1981, 256, 9235–9241. [Google Scholar] [CrossRef] [PubMed]
- Vijay-Kumar, S.; Bugg, C.E.; Cook, W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987, 194, 531–544. [Google Scholar] [CrossRef]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef]
- Schwarz, L.A.; Patrick, G.N. Ubiquitin-dependent endocytosis, trafficking and turnover of neuronal membrane proteins. Mol. Cell Neurosci. 2012, 49, 387–393. [Google Scholar] [CrossRef]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, K.; Schell, M.; Hoppe, T.; Kashkar, H. Regulation of the DNA damage response by ubiquitin conjugation. Front. Genet. 2015, 6, 98. [Google Scholar] [CrossRef]
- Dang, F.; Nie, L.; Wei, W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021, 28, 427–438. [Google Scholar] [CrossRef]
- Haas, A.L.; Rose, I.A. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 1982, 257, 10329–10337. [Google Scholar] [CrossRef]
- Pickart, C.M.; Rose, I.A. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 1985, 260, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Jin, J.; Li, X.; Gygi, S.P.; Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007, 447, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Nakayama, K.I.; Nakayama, K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 2006, 6, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, S. The ubiquitin-conjugation system. Annu. Rev. Genet. 1992, 26, 179–207. [Google Scholar] [CrossRef]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. Types of Ubiquitin Ligases. Cell 2016, 165, 248.e1. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Dunn, R. Regulation of Membrane Protein Transport by Ubiquitin and Ubiquitin-Binding Proteins. Annu. Rev. Cell Dev. Biol. 2003, 19, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef]
- Behrends, C.; Harper, J.W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 2011, 18, 520–528. [Google Scholar] [CrossRef]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576. [Google Scholar] [CrossRef] [PubMed]
- Haglund, K.; Dikic, I. Ubiquitylation and cell signaling. EMBO J. 2005, 24, 3353–3359. [Google Scholar] [CrossRef] [PubMed]
- McDowell, G.S.; Philpott, A. Non-canonical ubiquitylation: Mechanisms and consequences. Int. J. Biochem. Cell Biol. 2013, 45, 1833–1842. [Google Scholar] [CrossRef]
- Geimonen, E.; Fernandez, I.; Gavrilovskaya, I.N.; Mackow, E.R. Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail. J. Virol. 2003, 77, 10760–10868. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 1996, 93, 12142–12149. [Google Scholar] [CrossRef]
- Breitschopf, K.; Bengal, E.; Ziv, T.; Admon, A.; Ciechanover, A. A novel site for ubiquitination: The N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998, 17, 5964–5973. [Google Scholar] [CrossRef] [PubMed]
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef]
- Hu, M.; Li, P.; Li, M.; Li, W.; Yao, T.; Wu, J.-W.; Gu, W.; Cohen, R.E.; Shi, Y. Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex with Ubiquitin Aldehyde. Cell 2002, 111, 1041–1054. [Google Scholar] [CrossRef]
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 114–128. [Google Scholar] [CrossRef]
- Li, Y.; Shi, F.; Hu, J.; Xie, L.; Bode, A.M.; Cao, Y. The Role of Deubiquitinases in Oncovirus and Host Interactions. J. Oncol. 2019, 2019, 2128410. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, D.; Lin, X.; Robertson, E.S.; Lan, K. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen reduces interleukin-8 expression in endothelial cells and impairs neutrophil chemotaxis by degrading nuclear p65. J. Virol. 2011, 85, 8606–8615. [Google Scholar] [CrossRef]
- Tadmor, H.; Greenway, M.; Ahuja, A.; Orgil, O.; Liao, G.; Ambinder, R.F.; Hayward, S.D.; Shamay, M. Kaposi’s Sarcoma-Associated Herpesvirus LANA Modulates the Stability of the E3 Ubiquitin Ligase RLIM. J. Virol. 2020, 94, e01578-19. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.; Toth, Z.; Wong, L.-Y.; Feng, P.; Gao, S.-J.; Ensser, A.; Jung Jae, U.; Longnecker, R.M. Kaposi’s Sarcoma-Associated Herpesvirus K3 and K5 Ubiquitin E3 Ligases Have Stage-Specific Immune Evasion Roles during Lytic Replication. J. Virol. 2014, 88, 9335–9349. [Google Scholar] [CrossRef]
- Kajikawa, M.; Li, P.C.; Goto, E.; Miyashita, N.; Aoki-Kawasumi, M.; Mito-Yoshida, M.; Ikegaya, M.; Sugita, Y.; Ishido, S. The intertransmembrane region of Kaposi’s sarcoma-associated herpesvirus modulator of immune recognition 2 contributes to B7-2 downregulation. J. Virol. 2012, 86, 5288–5296. [Google Scholar] [CrossRef]
- Chung, W.-C.; Lee, S.; Kim, Y.; Seo, J.B.; Song, M.J. Kaposi’s sarcoma-associated herpesvirus processivity factor (PF-8) recruits cellular E3 ubiquitin ligase CHFR to promote PARP1 degradation and lytic replication. PLoS Pathog. 2021, 17, e1009261. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Kousoulas, K.G. The Kaposi’s sarcoma-associated herpesvirus ORF34 protein binds to HIF-1α and causes its degradation via the proteasome pathway. J. Virol. 2013, 87, 2164–2173. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Broussard, G.; Damania, B. KSHV: Immune Modulation and Immunotherapy. Front. Immunol. 2020, 10, 3084. [Google Scholar] [CrossRef] [PubMed]
- Ashizawa, A.; Higashi, C.; Masuda, K.; Ohga, R.; Taira, T.; Fujimuro, M. The Ubiquitin System and Kaposi’s Sarcoma-Associated Herpesvirus. Front. Microbiol. 2012, 3, 66. [Google Scholar] [CrossRef]
- West, J.T.; Wood, C. The role of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 2003, 22, 5150–5163. [Google Scholar] [CrossRef]
- Izumiya, Y.; Kobayashi, K.; Kim, K.Y.; Pochampalli, M.; Izumiya, C.; Shevchenko, B.; Wang, D.-H.; Huerta, S.B.; Martinez, A.; Campbell, M.; et al. Kaposi’s sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog. 2013, 9, e1003506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Mohan, C. Toll-Like Receptor Signaling Pathways—Therapeutic Opportunities. Mediat. Inflamm. 2010, 2010, 781235. [Google Scholar] [CrossRef]
- Yu, Y.; Hayward, G.S. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 2010, 33, 863–877. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Mori, K.; Hoshino, K.; Takeuchi, O.; Takeda, K.; Akira, S. Cutting Edge: A Novel Toll/IL-1 Receptor Domain-Containing Adapter That Preferentially Activates the IFN-β Promoter in the Toll-Like Receptor Signaling. J. Immunol. 2002, 169, 6668–6672. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of Adaptor TRIF in the MyD88-Independent Toll-Like Receptor Signaling Pathway. Science 2003, 301, 640–643. [Google Scholar] [CrossRef]
- Ahmad, H.; Gubbels, R.; Ehlers, E.; Meyer, F.; Waterbury, T.; Lin, R.; Zhang, L. Kaposi Sarcoma-associated Herpesvirus Degrades Cellular Toll-Interleukin-1 Receptor Domain-containing Adaptor-inducing β-Interferon (TRIF). J. Biol. Chem. 2011, 286, 7865–7872. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Ehlers, E.; Steadman, A.; Waterbury, T.; Cao, M.; Zhang, L. TLR-TRIF pathway enhances the expression of KSHV replication and transcription activator. J. Biol. Chem. 2013, 288, 20435–20442. [Google Scholar] [CrossRef] [PubMed]
- Wesche, H.; Henzel, W.J.; Shillinglaw, W.; Li, S.; Cao, Z. MyD88: An Adapter That Recruits IRAK to the IL-1 Receptor Complex. Immunity 1997, 7, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.; Martinon, F.; Esslinger, C.; Pahl, H.; Schneider, P.; Bodmer, J.-L.; Di Marco, F.; French, L.; Tschopp, J. MyD88, an Adapter Protein Involved in Interleukin-1 Signaling. J. Biol. Chem. 1998, 273, 12203–12209. [Google Scholar] [CrossRef]
- Adachi, O.; Kawai, T.; Takeda, K.; Matsumoto, M.; Tsutsui, H.; Sakagami, M.; Nakanishi, K.; Akira, S. Targeted Disruption of the MyD88 Gene Results in Loss of IL-1- and IL-18-Mediated Function. Immunity 1998, 9, 143–150. [Google Scholar] [CrossRef]
- West, A.P.; Koblansky, A.A.; Ghosh, S. Recognition and Signaling by Toll-Like Receptors. Annu. Rev. Cell Dev. Biol. 2006, 22, 409–437. [Google Scholar] [CrossRef]
- Zhao, Q.; Liang, D.; Sun, R.; Jia, B.; Xia, T.; Xiao, H.; Lan, K. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded Replication and Transcription Activator Impairs Innate Immunity via Ubiquitin-Mediated Degradation of Myeloid Differentiation Factor 88. J. Virol. 2015, 89, 415–427. [Google Scholar] [CrossRef]
- Katze, M.G.; He, Y.; Gale, M. Viruses and interferon: A fight for supremacy. Nat. Rev. Immunol. 2002, 2, 675–687. [Google Scholar] [CrossRef]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef]
- Hwang, S.-W.; Kim, D.; Jung, J.U.; Lee, H.-R. KSHV-encoded viral interferon regulatory factor 4 (vIRF4) interacts with IRF7 and inhibits interferon alpha production. Biochem. Biophys. Res. Commun. 2017, 486, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-j.; Li, W.; Shao, Y.; Avey, D.; Fu, B.; Gillen, J.; Hand, T.; Ma, S.; Liu, X.; Miley, W.; et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 2015, 18, 333–344. [Google Scholar] [CrossRef]
- Jacobs, S.R.; Damania, B. The viral interferon regulatory factors of KSHV: Immunosuppressors or oncogenes? Front. Immunol. 2011, 2, 19. [Google Scholar] [CrossRef]
- Bisson, S.A.; Page, A.L.; Ganem, D. A Kaposi’s sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. J. Virol. 2009, 83, 5056–5066. [Google Scholar] [CrossRef]
- Lefort, S.; Soucy-Faulkner, A.; Grandvaux, N.; Flamand, L. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 2007, 81, 10950–10960. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.X.; King, S.M.; Smith, E.J.; Levy, D.E.; Yuan, Y. A Kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 2002, 99, 5573–5578. [Google Scholar] [CrossRef]
- Cloutier, N.; Flamand, L. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J. Biol. Chem. 2010, 285, 7208–7221. [Google Scholar] [CrossRef]
- Zhang, G.; Chan, B.; Samarina, N.; Abere, B.; Weidner-Glunde, M.; Buch, A.; Pich, A.; Brinkmann, M.M.; Schulz, T.F. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. USA 2016, 113, E1034–E1043. [Google Scholar] [CrossRef]
- Goenka, S.; Kaplan, M.H. Transcriptional regulation by STAT6. Immunol. Res. 2011, 50, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Karpathiou, G.; Papoudou-Bai, A.; Ferrand, E.; Dumollard, J.M.; Peoc’h, M. STAT6: A review of a signaling pathway implicated in various diseases with a special emphasis in its usefulness in pathology. Pathol.-Res. Pract. 2021, 223, 153477. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, C.; Wei, F.; Gao, S.; Zhang, L.; Li, Y.; Feng, Y.; Tong, Y.; Xu, J.; Wang, B.; et al. Nuclear Localization and Cleavage of STAT6 Is Induced by Kaposi’s Sarcoma-Associated Herpesvirus for Viral Latency. PLoS Pathog. 2017, 13, e1006124. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, C.; Wei, F.; Zhang, L.; Mo, X.; Feng, Y.; Xu, J.; Yuan, Z.; Robertson, E.; Cai, Q. Constitutive Activation of Interleukin-13/STAT6 Contributes to Kaposi’s Sarcoma-Associated Herpesvirus-Related Primary Effusion Lymphoma Cell Proliferation and Survival. J. Virol. 2015, 89, 10416–10426. [Google Scholar] [CrossRef]
- Cai, Q.; Verma, S.C.; Choi, J.Y.; Ma, M.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus inhibits interleukin-4-mediated STAT6 phosphorylation to regulate apoptosis and maintain latency. J. Virol. 2010, 84, 11134–11144. [Google Scholar] [CrossRef]
- Gu, F.; Wang, C.; Wei, F.; Wang, Y.; Zhu, Q.; Ding, L.; Xu, W.; Zhu, C.; Cai, C.; Qian, Z.; et al. STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus. PLoS Pathog. 2018, 14, e1007416. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shao, Y.; Fu, R. Current research status of HLA in immune-related diseases. Immun. Inflamm. Dis. 2021, 9, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Jha Hem, C.; Pei, Y.-g.; Robertson, E.S.; Longnecker, R.M. Major Histocompatibility Complex Class II HLA-DRα Is Downregulated by Kaposi’s Sarcoma-Associated Herpesvirus-Encoded Lytic Transactivator RTA and MARCH8. J. Virol. 2016, 90, 8047–8058. [Google Scholar] [CrossRef] [PubMed]
- Lapaque, N.; Jahnke, M.; Trowsdale, J.; Kelly, A.P. The HLA-DRα Chain Is Modified by Polyubiquitination. J. Biol. Chem. 2009, 284, 7007–7016. [Google Scholar] [CrossRef]
- Coscoy, L.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 8051–8056. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Kang, B.; Sun, X.-H. Chapter Five-Id Proteins: Small Molecules, Mighty Regulators. Curr. Top. Dev. Biol. 2014, 110, 189–216. [Google Scholar]
- Liang, D. Oncogenic Herpesvirus KSHV Hijacks BMP-Smad1-Id Signaling to Promote Tumorigenesis. PLoS Pathog. 2014, 10, e1004253. [Google Scholar] [CrossRef]
- Combs Lauren, R.; Spires Lauren, M.; Alonso Juan, D.; Papp, B.; Toth, Z.; Goodrum, F. KSHV RTA Induces Degradation of the Host Transcription Repressor ID2 To Promote the Viral Lytic Cycle. J. Virol. 2022, 96, e00101-22. [Google Scholar] [CrossRef]
- Wang, X.; He, Z.; Xia, T.; Li, X.; Liang, D.; Lin, X.; Wen, H.; Lan, K. Latency-Associated Nuclear Antigen of Kaposi Sarcoma–Associated Herpesvirus Promotes Angiogenesis through Targeting Notch Signaling Effector Hey1. Cancer Res. 2014, 74, 2026. [Google Scholar] [CrossRef]
- Iso, T.; Sartorelli, V.; Poizat, C.; Iezzi, S.; Wu, H.-Y.; Chung, G.; Kedes, L.; Hamamori, Y. HERP, a Novel Heterodimer Partner of HES/E(spl) in Notch Signaling. Mol. Cell. Biol. 2001, 21, 6080–6089. [Google Scholar] [CrossRef] [PubMed]
- Gould, F.; Harrison, S.M.; Hewitt, E.W.; Whitehouse, A. Kaposi’s Sarcoma-Associated Herpesvirus RTA Promotes Degradation of the Hey1 Repressor Protein through the Ubiquitin Proteasome Pathway. J. Virol. 2009, 83, 6727–6738. [Google Scholar] [CrossRef]
- Yada, K.; Do, E.; Sakakibara, S.; Ohsaki, E.; Ito, E.; Watanabe, S.; Ueda, K. KSHV RTA induces a transcriptional repressor, HEY1 that represses rta promoter. Biochem. Biophys. Res. Commun. 2006, 345, 410–418. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Wu, M.-H.; Geng, Y.; Wood, C. Identification of a Cellular Protein That Interacts and Synergizes with the RTA (ORF50) Protein of Kaposi’s Sarcoma-Associated Herpesvirus in Transcriptional Activation. J. Virol. 2001, 75, 11961–11973. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wood, C. The transcriptional repressor K-RBP modulates RTA-mediated transactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2007, 81, 6294–6306. [Google Scholar] [CrossRef]
- Yang, Z.; Wen, H.-J.; Minhas, V.; Wood, C. The zinc finger DNA-binding domain of K-RBP plays an important role in regulating Kaposi’s sarcoma-associated herpesvirus RTA-mediated gene expression. Virology 2009, 391, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yan, Z.; Wood, C. Kaposi’s Sarcoma-Associated Herpesvirus Transactivator RTA Promotes Degradation of the Repressors To Regulate Viral Lytic Replication. J. Virol. 2008, 82, 3590–3603. [Google Scholar] [CrossRef] [PubMed]
- Aragón, L. The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative. Annu. Rev. Genet. 2018, 52, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Serrano, D.; Cordero, G.; Kawamura, R.; Sverzhinsky, A.; Sarker, M.; Roy, S.; Malo, C.; Pascal, J.M.; Marko, J.F.; D’Amours, D. The Smc5/6 Core Complex Is a Structure-Specific DNA Binding and Compacting Machine. Mol. Cell 2020, 80, 1025–1038.e5. [Google Scholar] [CrossRef] [PubMed]
- Irwan, I.D.; Cullen, B.R. The SMC5/6 complex: An emerging antiviral restriction factor that can silence episomal DNA. PLoS Pathog. 2023, 19, e1011180. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Zhang, D.; Gui, C.; Huang, L.; Chang, S.; Dong, L.; Bai, L.; Wu, S.; Lan, K. KSHV RTA antagonizes SMC5/6 complex-induced viral chromatin compaction by hijacking the ubiquitin-proteasome system. PLoS Pathog. 2022, 18, e1010744. [Google Scholar] [CrossRef]
- Lin, S.-F.; Robinson Dan, R.; Miller, G.; Kung, H.-J. Kaposi’s Sarcoma-Associated Herpesvirus Encodes a bZIP Protein with Homology to BZLF1 of Epstein-Barr Virus. J. Virol. 1999, 73, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sathish, N.; Hollow, C.; Yuan, Y. Functional characterization of Kaposi’s sarcoma-associated herpesvirus open reading frame K8 by bacterial artificial chromosome-based mutagenesis. J. Virol. 2011, 85, 1943–1957. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, Q.; Maul, G.G.; Yuan, Y. Kaposi’s sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: Dual role of replication and transcription activator. J. Virol. 2006, 80, 12171–12186. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Tang, Q.; Maul, G.G.; Yuan, Y. Kaposi’s sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: Involvement of host cellular factors. J. Virol. 2008, 82, 2867–2882. [Google Scholar] [CrossRef]
- Izumiya, Y.; Lin, S.-F.; Ellison, T.; Chen, L.-Y.; Izumiya, C.; Luciw, P.; Kung, H.-J. Kaposi’s Sarcoma-Associated Herpesvirus K-bZIP Is a Coregulator of K-Rta: Physical Association and Promoter-Dependent Transcriptional Repression. J. Virol. 2003, 77, 1441–1451. [Google Scholar] [CrossRef]
- Liao, W.; Tang, Y.; Lin, S.-F.; Kung, H.-J.; Giam, C.-Z. K-bZIP of Kaposi’s Sarcoma-Associated Herpesvirus/Human Herpesvirus 8 (KSHV/HHV-8) Binds KSHV/HHV-8 Rta and Represses Rta-Mediated Transactivation. J. Virol. 2003, 77, 3809–3815. [Google Scholar] [CrossRef]
- Kato-Noah, T.; Xu, Y.; Rossetto, C.C.; Colletti, K.; Papousková, I.; Pari, G.S. Overexpression of the kaposi’s sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion BACmid and yields an enhanced growth phenotype. J. Virol. 2007, 81, 13519–13532. [Google Scholar] [CrossRef] [PubMed]
- Uppal, T.; Banerjee, S.; Sun, Z.; Verma, C.S.; Robertson, S.E. KSHV LANA—The Master Regulator of KSHV Latency. Viruses 2014, 6, 4961–4998. [Google Scholar] [CrossRef] [PubMed]
- Lan, K.; Kuppers, D.A.; Verma, S.C.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: A potential mechanism for virus-mediated control of latency. J. Virol. 2004, 78, 6585–6594. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Liang, D.; Gao, Y.; Lan, K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J. Virol. 2014, 88, 7331–7344. [Google Scholar] [CrossRef]
- Lan, K.; Kuppers Daniel, A.; Robertson Erle, S. Kaposi’s Sarcoma-Associated Herpesvirus Reactivation Is Regulated by Interaction of Latency-Associated Nuclear Antigen with Recombination Signal Sequence-Binding Protein Jκ, the Major Downstream Effector of the Notch Signaling Pathway. J. Virol. 2005, 79, 3468–3478. [Google Scholar] [CrossRef]
- Jin, Y.; He, Z.; Liang, D.; Zhang, Q.; Zhang, H.; Deng, Q.; Robertson, E.S.; Lan, K. Carboxyl-terminal amino acids 1052 to 1082 of the latency-associated nuclear antigen (LANA) interact with RBP-Jκ and are responsible for LANA-mediated RTA repression. J. Virol. 2012, 86, 4956–4969. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Verma, S.C.; Cai, Q.; Saha, A.; Dzeng, R.K.; Robertson, E.S. The RBP-Jκ binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation. PLoS Pathog. 2012, 8, e1002479. [Google Scholar] [CrossRef]
- Lan, K.; Kuppers, D.A.; Verma, S.C.; Sharma, N.; Murakami, M.; Robertson, E.S. Induction of Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: A novel mechanism for establishment of latency. J. Virol. 2005, 79, 7453–7465. [Google Scholar] [CrossRef]
- Ye, F.C.; Zhou, F.C.; Xie, J.P.; Kang, T.; Greene, W.; Kuhne, K.; Lei, X.F.; Li, Q.H.; Gao, S.J. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: A novel mechanism of virus control of latency. J. Virol. 2008, 82, 4235–4249. [Google Scholar] [CrossRef]
- Field, N.; Low, W.; Daniels, M.; Howell, S.; Daviet, L.; Boshoff, C.; Collins, M. KSHV vFLIP binds to IKK-γ to activate IKK. J. Cell Sci. 2003, 116, 3721–3728. [Google Scholar] [CrossRef]
- Grossmann, C.; Ganem, D. Effects of NFκB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology 2008, 375, 94–102. [Google Scholar] [CrossRef]
- Izumiya, Y.; Izumiya, C.; Hsia, D.; Ellison, T.J.; Luciw, P.A.; Kung, H.J. NF-kappaB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J. Virol. 2009, 83, 4435–4446. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Matta, H.; Chaudhary, P.M. The human herpes virus 8–encoded viral FLICE inhibitory protein protects against growth factor withdrawal–induced apoptosis via NF-κB activation. Blood 2003, 101, 1956–1961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guasparri, I.; Keller, S.A.; Cesarman, E. KSHV vFLIP Is Essential for the Survival of Infected Lymphoma Cells. J. Exp. Med. 2004, 199, 993–1003. [Google Scholar] [CrossRef]
- Ehrlich, E.S.; Chmura, J.C.; Smith, J.C.; Kalu, N.N.; Hayward, G.S. KSHV RTA abolishes NFκB responsive gene expression during lytic reactivation by targeting vFLIP for degradation via the proteasome. PLoS ONE 2014, 9, e91359. [Google Scholar] [CrossRef] [PubMed]
- Chmura, J.C.; Herold, K.; Ruffin, A.; Atuobi, T.; Fabiyi, Y.; Mitchell, A.E.; Choi, Y.B.; Ehrlich, E.S. The Itch ubiquitin ligase is required for KSHV RTA induced vFLIP degradation. Virology 2017, 501, 119–126. [Google Scholar] [CrossRef]
- Damania, B.; Jeong, J.H.; Bowser, B.S.; DeWire, S.M.; Staudt, M.R.; Dittmer, D.P. Comparison of the Rta/Orf50 Transactivator Proteins of Gamma-2-Herpesviruses. J. Virol. 2004, 78, 5491–5499. [Google Scholar] [CrossRef]
- Staudt, M.R.; Dittmer, D.P. The Rta/Orf50 Transactivator Proteins of the Gamma-Herpesviridae. In Kaposi Sarcoma Herpesvirus: New Perspectives; Boshoff, C., Weiss, R.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 71–100. [Google Scholar] [CrossRef]
- Damania, B.; Kenney, S.C.; Raab-Traub, N. Epstein-Barr virus: Biology and clinical disease. Cell 2022, 185, 3652–3670. [Google Scholar] [CrossRef]
- Ragoczy, T.; Heston, L.; Miller, G. The Epstein-Barr Virus Rta Protein Activates Lytic Cycle Genes and Can Disrupt Latency in B Lymphocytes. J. Virol. 1998, 72, 7978–7984. [Google Scholar] [CrossRef]
- Feederle, R.; Kost, M.; Baumann, M.; Janz, A.; Drouet, E.; Hammerschmidt, W.; Delecluse, H.-J. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000, 19, 3080–3089. [Google Scholar] [CrossRef]
- Bentz, G.L.; Liu, R.; Hahn, A.M.; Shackelford, J.; Pagano, J.S. Epstein-Barr virus BRLF1 inhibits transcription of IRF3 and IRF7 and suppresses induction of interferon-beta. Virology 2010, 402, 121–128. [Google Scholar] [CrossRef]
- De La Cruz-Herrera, C.F.; Shire, K.; Siddiqi, U.Z.; Frappier, L. A genome-wide screen of Epstein-Barr virus proteins that modulate host SUMOylation identifies a SUMO E3 ligase conserved in herpesviruses. PLoS Pathog. 2018, 14, e1007176. [Google Scholar] [CrossRef] [PubMed]
- Fickenscher, H.; Fleckenstein, B. Herpesvirus saimiri. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001, 356, 545–567. [Google Scholar] [CrossRef]
- Daniel, M.D.; Meléndez, L.V. Herpes T Virus Variants: Isolation and Characterization. Arch. Virol. 1968, 25, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Meléndez, L.V.; Hunt, R.D.; Daniel, M.D.; Fraser, C.E.O.; García, F.G.; Williamson, M.E. Lethal reticuloproliferative disease induced in Cebus albifrons monkeys by Herpesvirus saimiri. Int. J. Cancer 1970, 6, 431–435. [Google Scholar] [CrossRef]
- Falk, L.; Wright, J.; Deinhardt, F.; Wolfe, L.; Schaffer, P.; Benyesh-Melnick, M. Experimental infection of squirrel and marmoset monkeys with attenuated Herpesvirus saimiri. Cancer Res. 1976, 36, 707. [Google Scholar]
- Rabin, H.; Adamson, R.H.; Neubauer, R.H.; Cicmanec, J.L.; Wallen, W.C. Pilot Studies with Human Interferon in Herpesvirus saimiri-induced Lymphoma in Owl Monkeys. Cancer Res. 1976, 36, 715. [Google Scholar]
- Nicholas, J.; Coles, L.S.; Newman, C.; Honess, R.W. Regulation of the herpesvirus saimiri (HVS) delayed-early 110-kilodalton promoter by HVS immediate-early gene products and a homolog of the Epstein-Barr virus R trans activator. J. Virol. 1991, 65, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Goodwin Delyth, J.; Walters Matthew, S.; Smith Peter, G.; Thurau, M.; Fickenscher, H.; Whitehouse, A. Herpesvirus Saimiri Open Reading Frame 50 (Rta) Protein Reactivates the Lytic Replication Cycle in a Persistently Infected A549 Cell Line. J. Virol. 2001, 75, 4008–4013. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Walters, M.S.; Hall, K.T.; Whitehouse, A. The herpesvirus saimiri open reading frame (ORF) 50 (Rta) protein contains an at hook required for binding to the ORF 50 response element in delayed-early promoters. J. Virol. 2004, 78, 4936–4942. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.C.; Sasseville, V.G.; Czajak, S.C.; Zhang, X.; Mansfield, K.G.; Kaur, A.; Johnson, R.P.; Lackner, A.A.; Jung, J.U. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1997, 71, 9764–9769. [Google Scholar] [CrossRef]
- Searles Robert, P.; Bergquam Eric, P.; Axthelm Michael, K.; Wong Scott, W. Sequence and Genomic Analysis of a Rhesus Macaque Rhadinovirus with Similarity to Kaposi’s Sarcoma-Associated Herpesvirus/Human Herpesvirus 8. J. Virol. 1999, 73, 3040–3053. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.; Denekamp, L.; Knapp, A.; Auerbach, M.R.; Damania, B.; Desrosiers, R.C. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: Sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 2000, 74, 3388–3398. [Google Scholar] [CrossRef] [PubMed]
- Greensill, J.; Sheldon, J.A.; Renwick, N.M.; Beer, B.E.; Norley, S.; Goudsmit, J.; Schulz, T.F. Two distinct gamma-2 herpesviruses in African green monkeys: A second gamma-2 herpesvirus lineage among old world primates? J. Virol. 2000, 74, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.R.; Rankin, G.W., Jr.; Blanc, M.P.; Raden, B.W.; Tsai, C.C.; Rose, T.M. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2000, 74, 4919–4928. [Google Scholar] [CrossRef][Green Version]
- Chalifoux, L.V.; King, N.W.; Daniel, M.D.; Kannagi, M.; Desrosiers, R.C.; Sehgal, P.K.; Waldron, L.M.; Hunt, R.D.; Letvin, N.L. Lymphoproliferative syndrome in an immunodeficient rhesus monkey naturally infected with an HTLV-III-like virus (STLV-III). Lab Investig. 1986, 55, 43–50. [Google Scholar] [PubMed]
- Baskin, G.B.; Martin, L.N.; Rangan, S.R.; Gormus, B.J.; Murphey-Corb, M.; Wolf, R.H.; Soike, K.F. Transmissible lymphoma and simian acquired immunodeficiency syndrome in rhesus monkeys. J. Natl. Cancer Inst. 1986, 77, 127–139. [Google Scholar]
- Tsai, C.C.; Tsai, C.C.; Roodman, S.T.; Woon, M.D. Mesenchymoproliferative disorders (MPD) in simian AIDS associated with SRV-2 infection. J. Med. Primatol. 1990, 19, 189–202. [Google Scholar] [CrossRef]
- Wong, S.W.; Bergquam, E.P.; Swanson, R.M.; Lee, F.W.; Shiigi, S.M.; Avery, N.A.; Fanton, J.W.; Axthelm, M.K. Induction of B Cell Hyperplasia in Simian Immunodeficiency Virus–Infected Rhesus Macaques with the Simian Homologue of Kaposi’s Sarcoma–Associated Herpesvirus. J. Exp. Med. 1999, 190, 827–840. [Google Scholar] [CrossRef]
- Lin, S.-F.; Robinson, D.R.; Oh, J.; Jung, J.U.; Luciw, P.A.; Kung, H.-J. Identification of the bZIP and Rta Homologues in the Genome of Rhesus Monkey Rhadinovirus. Virology 2002, 298, 181–188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- DeWire Scott, M.; McVoy Michael, A.; Damania, B. Kinetics of Expression of Rhesus Monkey Rhadinovirus (RRV) and Identification and Characterization of a Polycistronic Transcript Encoding the RRV Orf50/Rta, RRV R8, and R8.1 Genes. J. Virol. 2002, 76, 9819–9831. [Google Scholar] [CrossRef]
- Dittmer, D.P.; Gonzalez, C.M.; Vahrson, W.; DeWire, S.M.; Hines-Boykin, R.; Damania, B. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J. Virol. 2005, 79, 8637–8650. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sunil-Chandra, N.P.; Efstathiou, S.; Arno, J.; Nash, A.A. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 1992, 73 Pt 9, 2347–2356. [Google Scholar] [CrossRef]
- Blaskovic, D.; Stanceková, M.; Svobodová, J.; Mistríková, J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980, 24, 468. [Google Scholar] [PubMed]
- Virgin, H.W.; Latreille, P.; Wamsley, P.; Hallsworth, K.; Weck, K.E.; Dal Canto, A.J.; Speck, S.H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 1997, 71, 5894–5904. [Google Scholar] [CrossRef]
- Sunil-Chandra, N.P.; Efstathiou, S.; Nash, A.A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 1992, 73 Pt 12, 3275–3279. [Google Scholar] [CrossRef] [PubMed]
- Ehtisham, S.; Sunil-Chandra, N.P.; Nash, A.A. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 1993, 67, 5247–5252. [Google Scholar] [CrossRef]
- Dutia, B.M.; Stewart, J.P.; Clayton, R.A.E.; Dyson, H.; Nash, A.A. Kinetic and phenotypic changes in murine lymphocytes infected with murine gammaherpesvirus-68 in vitro. J. Gen. Virol. 1999, 80, 2729–2736. [Google Scholar] [CrossRef]
- Usherwood, E.J.; Stewart, J.P.; Robertson, K.; Allen, D.J.; Nash, A.A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 1996, 77, 2819–2825. [Google Scholar] [CrossRef] [PubMed]
- Flaño, E.; Husain, S.M.; Sample, J.T.; Woodland, D.L.; Blackman, M.A. Latent Murine γ-Herpesvirus Infection Is Established in Activated B Cells, Dendritic Cells, and Macrophages. J. Immunol. 2000, 165, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-T.; Usherwood Edward, J.; Stewart James, P.; Nash Anthony, A.; Sun, R. Rta of Murine Gammaherpesvirus 68 Reactivates the Complete Lytic Cycle from Latency. J. Virol. 2000, 74, 3659–3667. [Google Scholar] [CrossRef]
- Wu, T.-T.; Tong, L.; Rickabaugh, T.; Speck, S.; Sun, R. Function of Rta Is Essential for Lytic Replication of Murine Gammaherpesvirus 68. J. Virol. 2001, 75, 9262–9273. [Google Scholar] [CrossRef]
- Dong, X.; He, Z.; Durakoglugil, D.; Arneson, L.; Shen, Y.; Feng, P. Murine Gammaherpesvirus 68 Evades Host Cytokine Production via Replication Transactivator-Induced RelA Degradation. J. Virol. 2012, 86, 1930–1941. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Laitaoja, M.; Valjakka, J.; Jänis, J. Zinc Coordination Spheres in Protein Structures. Inorg. Chem. 2013, 52, 10983–10991. [Google Scholar] [CrossRef]
- Boles, G.C.; Hightower, R.L.; Berden, G.; Oomens, J.; Armentrout, P.B. Zinc and Cadmium Complexation of l-Threonine: An Infrared Multiple Photon Dissociation Spectroscopy and Theoretical Study. J. Phys. Chem. B 2019, 123, 9343–9354. [Google Scholar] [CrossRef]
- Ataie, N.J.; Hoang, Q.Q.; Zahniser, M.P.D.; Tu, Y.; Milne, A.; Petsko, G.A.; Ringe, D. Zinc Coordination Geometry and Ligand Binding Affinity: The Structural and Kinetic Analysis of the Second-Shell Serine 228 Residue and the Methionine 180 Residue of the Aminopeptidase from Vibrio proteolyticus. Biochemistry 2008, 47, 7673–7683. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Combs, L.R.; Combs, J.; McKenna, R.; Toth, Z. Protein Degradation by Gammaherpesvirus RTAs: More Than Just Viral Transactivators. Viruses 2023, 15, 730. https://doi.org/10.3390/v15030730
Combs LR, Combs J, McKenna R, Toth Z. Protein Degradation by Gammaherpesvirus RTAs: More Than Just Viral Transactivators. Viruses. 2023; 15(3):730. https://doi.org/10.3390/v15030730
Chicago/Turabian StyleCombs, Lauren R., Jacob Combs, Robert McKenna, and Zsolt Toth. 2023. "Protein Degradation by Gammaherpesvirus RTAs: More Than Just Viral Transactivators" Viruses 15, no. 3: 730. https://doi.org/10.3390/v15030730
APA StyleCombs, L. R., Combs, J., McKenna, R., & Toth, Z. (2023). Protein Degradation by Gammaherpesvirus RTAs: More Than Just Viral Transactivators. Viruses, 15(3), 730. https://doi.org/10.3390/v15030730





