Bridging the Local Persistence and Long-Range Dispersal of Highly Pathogenic Avian Influenza Virus (HPAIv): A Case Study of HPAIv-Infected Sedentary and Migratory Wildfowls Inhabiting Infected Premises
Abstract
:1. Introduction
2. Materials and Methods
2.1. Avian Influenza Outbreak Data
2.2. Telemetry Records for Wild Waterfowls
3. Methods
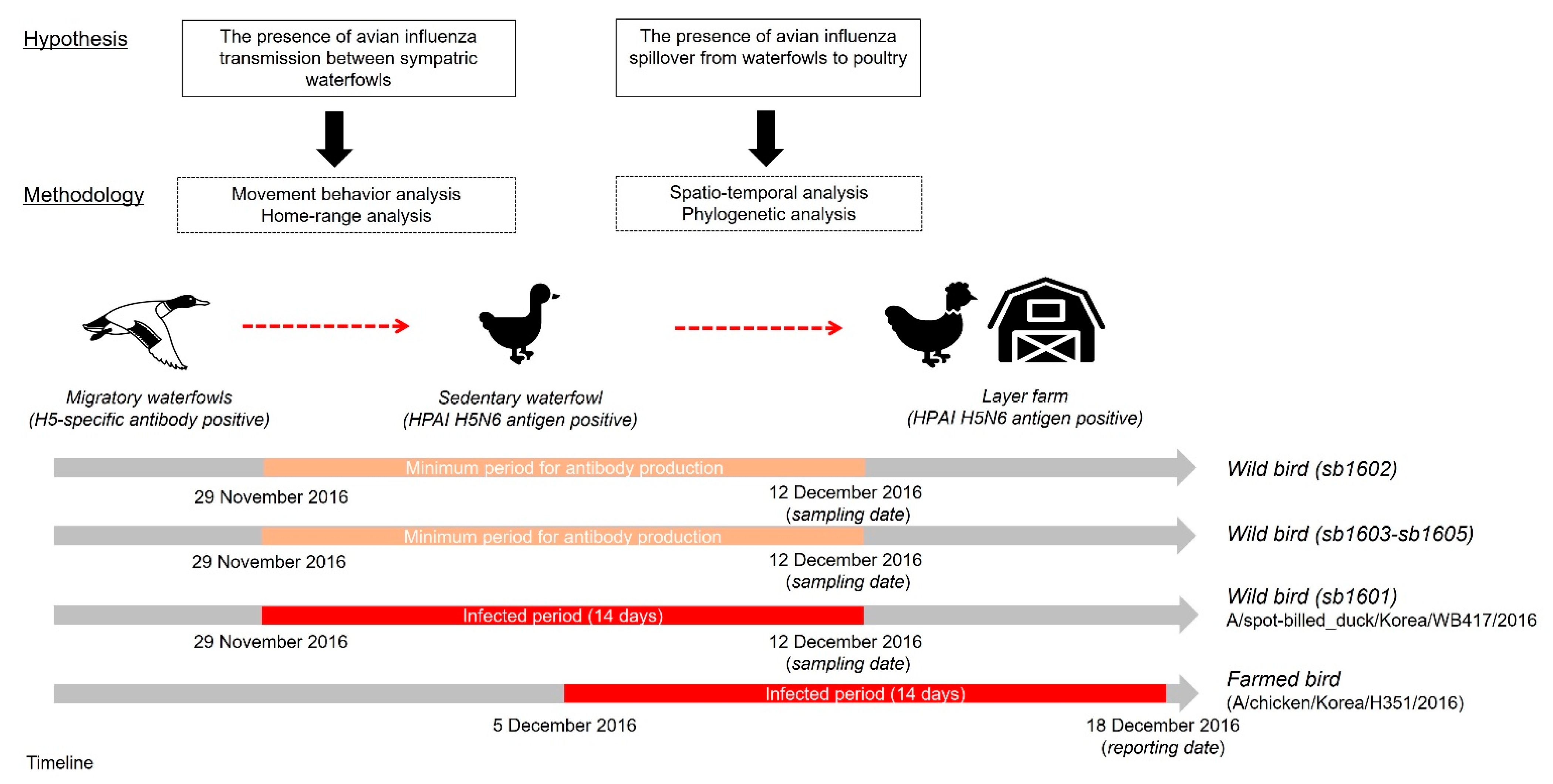
3.1. Study Framework
3.2. Movement Characteristics of Avian Influenza-Infected Wild Birds
3.3. Interaction Analyses between Poultry and Wild Birds
4. Results
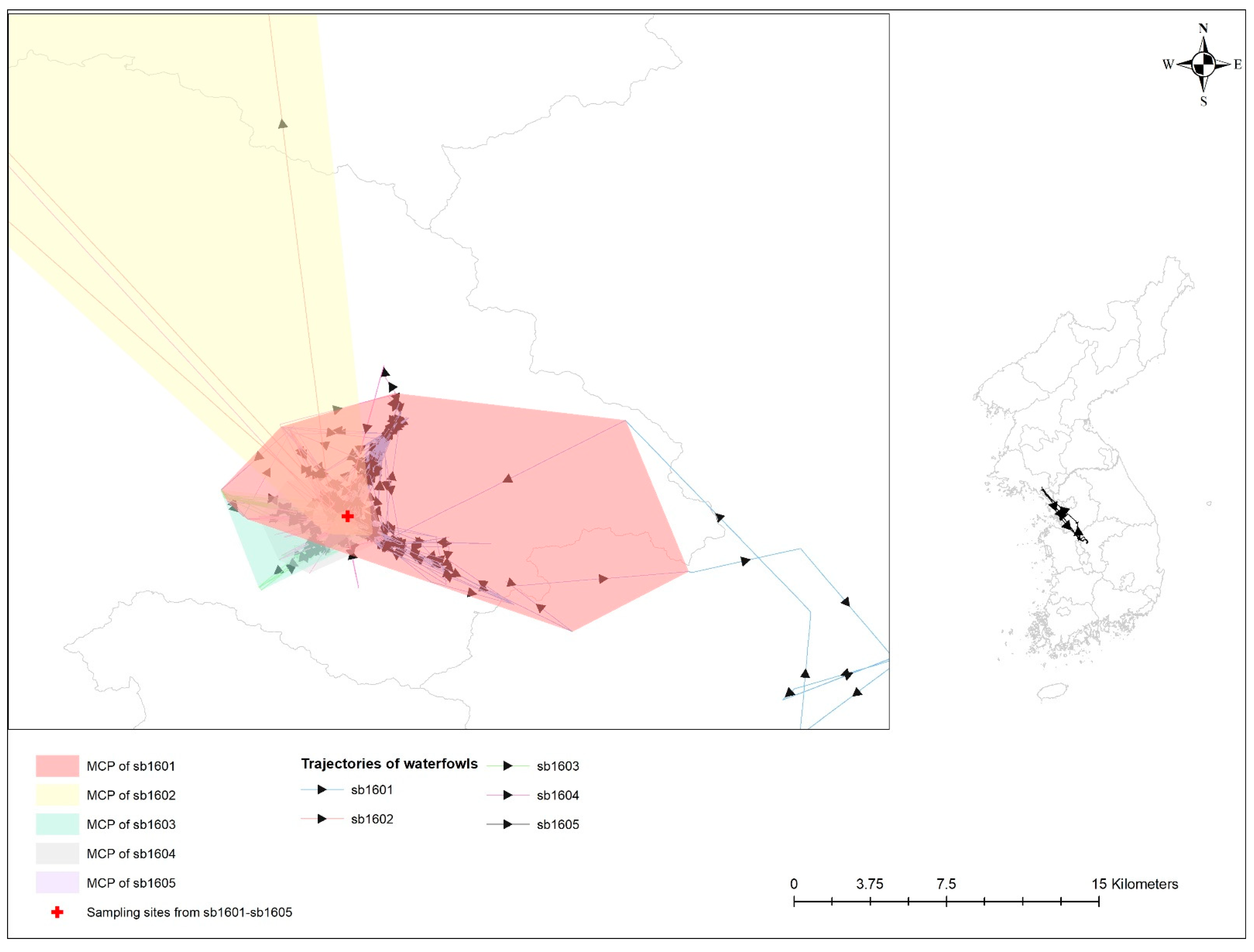
4.1. Movement Characteristics
4.2. Geographical Relationship of Avian Influenza-Infected Wild Birds
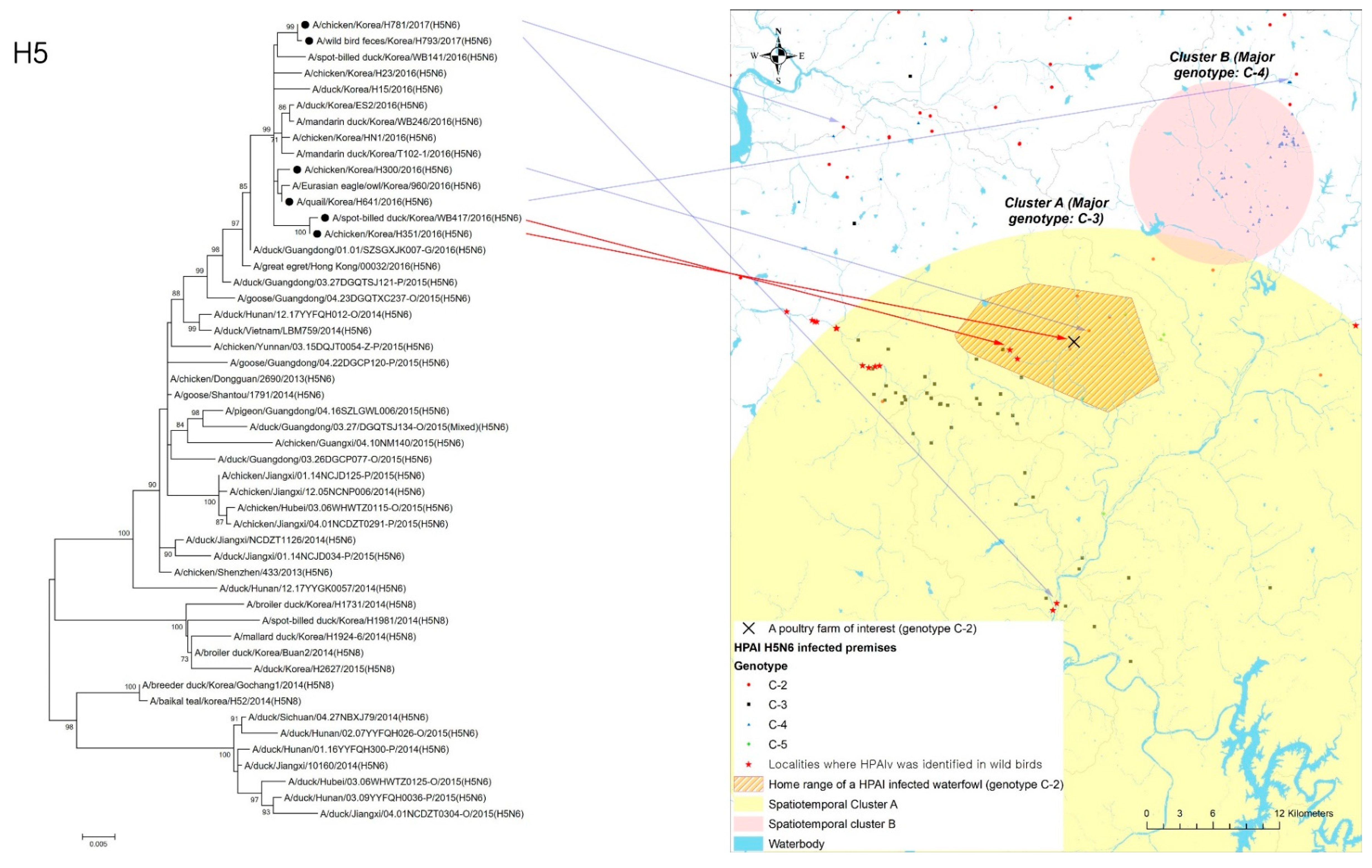
4.3. Geographical and Phylogenetic Relationship of Wild Birds to Infected Poultry Premises
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaurock, C.; Breithaupt, A.; Scheibner, D.; Bagato, O.; Karger, A.; Mettenleiter, T.C.; Abdelwhab, E.M. Preferential selection and contribution of non-structural protein 1 (NS1) to the efficient transmission of panzootic avian influenza H5N8 virus clades 2.3.4.4A and B in chickens and ducks. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Verhagen, J.H.; Eriksson, P.; Leijten, L.; Blixt, O.; Olsen, B.; Waldenström, J.; Ellström, P.; Kuiken, T. Host range of influenza a virus H1 to H16 in Eurasian ducks based on tissue and receptor binding studies. J. Virol. 2021, 95, e01873-20. [Google Scholar] [CrossRef]
- Gilbert, M.; Newman, S.H.; Takekawa, J.Y.; Loth, L.; Biradar, C.; Prosser, D.J.; Balachandran, S.; Rao, M.V.S.; Mundkur, T.; Yan, B. Flying over an infected landscape: Distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. Ecohealth 2010, 7, 448–458. [Google Scholar] [CrossRef] [Green Version]
- Gaidet, N.; Cappelle, J.; Takekawa, J.Y.; Prosser, D.J.; Iverson, S.A.; Douglas, D.C.; Perry, W.M.; Mundkur, T.; Newman, S.H. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: Dispersal ranges and rates determined from large-scale satellite telemetry. J. Appl. Ecol. 2010, 47, 1147–1157. [Google Scholar] [CrossRef]
- Yoo, D.S.; Lee, K.; Beatriz, M.L.; Chun, B.C.; Belkhiria, J.; Lee, K.N. Spatio-temporal risk assessment for avian influenza outbreak based on the dynamics of habitat suitability for wild birds. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Animal and Plant Quarantine Agency. Epidemiology Report for 2016–2017 Outbreak of Highly Pathogenic Avian Influenza (HPAI) in Republic of Korea; Ministry of Agriculture, Food and Rural Affairs: Gimcheon, Korea, 2018; pp. 42–43.
- Animal and Plant Quarantine Agency. Project Report on Wild Bird Capture, Sampling and Telemetery Transmitter Attachment for Highly Pathogenic Avian Influenza Surveillance; 2014 in Republic of Korea; Division of Veterinary Epidemiology, Ed.; Ministry of Agriculture, Food and Rural Affairs: Gimcheon, Korea, 2014.
- Animal and Plant Quarantine Agency. Epidemiology Report for 2017–2018 Outbreak of Highly Pathogenic Avian Influenza (HPAI) in Republic of Korea; Division of Veterinary Epidemiology, Ed.; Ministry of Agriculture, Food and Rural Affairs: Gimcheon, Korea, 2018; p. 31.
- Lee, E.K.; Kang, H.M.; Song, B.M.; Lee, Y.N.; Heo, G.B.; Lee, H.S.; Lee, Y.J.; Kim, J.H. Surveillance of avian influenza viruses in South Korea between 2012 and 2014. Virol. J. 2017, 14, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Yu, D.; Martínez-López, B.; Yoon, H.; Kang, S.-I.; Hong, S.-K.; Lee, I.; Kang, Y.; Jeong, W.; Lee, E. Fine-scale tracking of wild waterfowl and their impact on highly pathogenic avian influenza outbreaks in the Republic of Korea, 2014–2015. Sci. Rep. 2020, 10, 18631. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.U.; Shin, M.S.; Lee, H.S.; Kang, Y.M.; Jeong, W.S.; Choi, J.D.; Yoon, H.C.; Oh, H.S.K. Home-Range of Mallard and Spot-billed Duck in Korea1a. Korean J. Environ. Ecol. 2016, 30, 165–172. [Google Scholar] [CrossRef]
- Bunnefeld, N.; Borger, L.; van Moorter, B.; Rolandsen, C.M.; Dettki, H.; Solberg, E.J.; Ericsson, G. A model-driven approach to quantify migration patterns: Individual, regional and yearly differences. J. Anim. Ecol. 2011, 80, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.J.; Allen, A.M.; Ericsson, G. Quantifying Migration Behaviour Using Net Squared Displacement Approach: Clarifications and Caveats. PLoS ONE 2016, 11, e0149594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranstauber, B.; Kays, R.; LaPoint, S.D.; Wikelski, M.; Safi, K. A dynamic Brownian bridge movement model to estimate utilization distributions for heterogeneous animal movement. J. Anim. Ecol. 2012, 81, 738–746. [Google Scholar] [CrossRef]
- Fieberg, J.; Kochanny, C.O. Quantifying home-range overlap: The importance of the utilization distribution. J. Wildl. Manag. 2005, 69, 1346–1359. [Google Scholar] [CrossRef]
- Calenge, C. The package adehabitatHR for the R software: A tool for the analysis of space and habitat use by animals. Ecol. Model. 2006, 197, 516–519. [Google Scholar] [CrossRef]
- Calenge, C. The package adehabitatLT for the R software: A tool for the analysis of space and habitat use by animals. Ecol. Model. 2006, 197, 1035. [Google Scholar] [CrossRef]
- Yoo, D.-S.; Chun, B.; Min, K.-D.; Lim, J.-S.; Moon, O.-K.; Lee, K.-N. Elucidating the Local Transmission Dynamics of Highly Pathogenic Avian Influenza H5N6 in the Republic of Korea by Integrating Phylogenetic Information. Pathogens 2021, 10, 691. [Google Scholar] [CrossRef]
- Si, Y.J.; Lee, I.W.; Kim, E.H.; Kim, Y.I.; Kwon, H.I.; Park, S.J.; Nguyen, H.D.; Kim, S.M.; Kwon, J.J.; Choi, W.S.; et al. Genetic characterisation of novel, highly pathogenic avian influenza (HPAI) H5N6 viruses isolated in birds, South Korea, November 2016. Euro Surveill 2017, 22, 30434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.K.; Song, B.M.; Lee, Y.N.; Heo, G.B.; Bae, Y.C.; Joh, S.J.; Park, S.C.; Choi, K.S.; Lee, H.J.; Jang, I.; et al. Multiple novel H5N6 highly pathogenic avian influenza viruses, South Korea, 2016. Infect. Genet. Evol. 2017, 51, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Emerging Infectious Diseases of Wildlife—Threats to Biodiversity and Human Health. Science 2000, 287, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Siembieda, J.; Johnson, C.K.; Boyce, W.; Sandrock, C.; Cardona, C. Risk for avian influenza virus exposure at human–wildlife interface. Emerg. Infect. Dis. 2008, 14, 1151. [Google Scholar] [CrossRef]
- Verhagen, J.H.; Fouchier, R.A.M.; Lewis, N. Highly pathogenic avian influenza viruses at the wild–domestic bird interface in europe: Future directions for research and surveillance. Viruses 2021, 13, 212. [Google Scholar] [CrossRef]
- Ellis, J.W.; Root, J.J.; McCurdy, L.M.; Bentler, K.T.; Barrett, N.L.; VanDalen, K.K.; Dirsmith, K.L.; Shriner, S.A. Avian influenza A virus susceptibility, infection, transmission, and antibody kinetics in European starlings. PLoS Pathog. 2021, 17, e1009879. [Google Scholar] [CrossRef]
- Gilbert, M.; Prosser, D.J.; Zhang, G.; Artois, J.; Dhingra, M.S.; Tildesley, M.; Newman, S.H.; Guo, F.; Black, P.; Claes, F.; et al. Could Changes in the Agricultural Landscape of Northeastern China Have Influenced the Long-Distance Transmission of Highly Pathogenic Avian Influenza H5Nx Viruses? Front. Vet. Sci. 2017, 4, 225. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Zhou, H.; Fan, L.; Zhu, G.; Li, Y.; Chen, G.; Zhang, J.; Li, J.; Zheng, H.; Feng, W.; et al. Emerging highly pathogenic avian influenza (H5N8) virus in migratory birds in Central China, 2020. Emerg. Microbes Infect. 2021, 10, 1503–1506. [Google Scholar] [CrossRef]
- Kwon, Y.; Thomas, C.; Swayne, D. Variability in pathobiology of South Korean H5N1 high-pathogenicity avian influenza virus infection for 5 species of migratory waterfowl. Vet. Pathol. 2010, 47, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Jin, S.; Wang, T.; Sun, W.; Zhang, Y.; Li, F.; Zhao, M.; Sun, L.; Hu, X.; et al. H9N2 influenza virus spillover into wild birds from poultry in China bind to human-type receptors and transmit in mammals via respiratory droplets. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, J.G.; Kleyheeg, E.; Soons, M.B.; Nolet, B.A.; Fouchier, R.A.; Klaassen, M. Weak negative associations between avian influenza virus infection and movement behaviour in a key host species, the mallard Anas platyrhynchos. Oikos 2015, 124, 1293–1303. [Google Scholar] [CrossRef] [Green Version]
- Keawcharoen, J.; van den Broek, J.; Bouma, A.; Tiensin, T.; Osterhaus, A.D.M.E.; Heesterbeek, H. Wild birds and increased transmission of highly pathogenic avian influenza (H5N1) among poultry, Thailand. Emerg. Infect. Dis. 2011, 17, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Department of Biological Resource Department. Wildlife Survey; National Institue of Biological Resources: Incheon, Korea, 2016. [Google Scholar]
- De Maio, N.; Wu, C.H.; Wilson, D.J. SCOTTI: Efficient reconstruction of transmission within outbreaks with the structured coalescent. PLoS Comput. Biol. 2016, 12, e1005130. [Google Scholar] [CrossRef]


| Virus Name | Samples | Collection Date | Latitude/Longitude | GISAID Isolate ID |
|---|---|---|---|---|
| A/spot-billed_duck/Korea/WB417/2016 † | Oral and Cloacal swab | 12 December 2016 | 36°45′9.550″ N/127°7′0.5052″ E | EPI_ISL_7412777 |
| A/chicken/Korea/H300/2016 | Oral and Cloacal swab | 15 December 2016 | 37°47′35.448″ N/127°21′16.6968″ E | EPI_ISL_7412755 |
| A/chicken/Korea/H351/2016 ‡ | Oral and Cloacal swab | 18 December 2016 | 36°46′30.234″ N/127°19′12.684″ E | EPI_ISL_7412792 |
| A/quail/Korea/H641/2016 | Oral and Cloacal swab | 29 December 2016 | 36°59′30.3432″ N/127°38′9292″ E | EPI_ISL_7413688 |
| A/wild_duck/Korea/H793–1/2017 | Feces | 4 January 2017 | 36°33′17.1972″ N/127°17′51.468″ E | EPI_ISL_7413520 |
| A/chicken/Korea/H781/2017 | Oral and Cloacal swab | 7 January 2017 | 36°55′1.9668″ N/127°4′10.5564″ E | EPI_ISL_7412735 |
| ID | Test Result | Movement Mode | dBBMM (Unit: km2) | MCP (Unit: km2) | Date of the First Signal | Date of the Last Signal | |
|---|---|---|---|---|---|---|---|
| 50% | 95% | 99% | |||||
| sb1601 | HPAIV H5N6 antigen | Sedentary | 2.40 | 46.46 | 13.77 | 17 December 2016 | 10 August 2017 |
| sb1602 | AIV H5 antibody | Migratory | 13.46 | 1313.11 | 259.75 | 26 March 2017 | |
| sb1603 | AIV H5 antibody | Sedentary | 0.18 | 16.30 | 1.56 | 24 April 2017 | |
| sb1604 | AIV H5 antibody | Sedentary | 0.01 | 7.55 | 1.67 | 15 July 2017 | |
| sb1605 | AIV H5 antibody | Sedentary | 0.18 | 3.06 | 140.94 | 23 September 2017 | |
| ID | ID (Movement Mode, Type of Positive Test) | ||||
|---|---|---|---|---|---|
| sb1601 (Sedentary, Antigen) | sb1602 (Migratory, Antibody) | sb1603 (Sedentary, Antibody) | sb1604 (Sedentary, Antibody) | sb1605 (Sedentary, Antibody) | |
| sb1601 | 1.000 | 0.032 | 0.279 | 0.173 | 0.430 |
| sb1602 | 1.000 | 0.024 | 0.021 | 0.006 | |
| sb1603 | 1.000 | 0.164 | 0.253 | ||
| sb1604 | 1.000 | 0.151 | |||
| sb1605 | 1.000 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, D.-s.; Kang, S.-I.; Lee, Y.-N.; Lee, E.-K.; Kim, W.-y.; Lee, Y.-J. Bridging the Local Persistence and Long-Range Dispersal of Highly Pathogenic Avian Influenza Virus (HPAIv): A Case Study of HPAIv-Infected Sedentary and Migratory Wildfowls Inhabiting Infected Premises. Viruses 2022, 14, 116. https://doi.org/10.3390/v14010116
Yoo D-s, Kang S-I, Lee Y-N, Lee E-K, Kim W-y, Lee Y-J. Bridging the Local Persistence and Long-Range Dispersal of Highly Pathogenic Avian Influenza Virus (HPAIv): A Case Study of HPAIv-Infected Sedentary and Migratory Wildfowls Inhabiting Infected Premises. Viruses. 2022; 14(1):116. https://doi.org/10.3390/v14010116
Chicago/Turabian StyleYoo, Dae-sung, Sung-Il Kang, Yu-Na Lee, Eun-Kyoung Lee, Woo-yuel Kim, and Youn-Jeong Lee. 2022. "Bridging the Local Persistence and Long-Range Dispersal of Highly Pathogenic Avian Influenza Virus (HPAIv): A Case Study of HPAIv-Infected Sedentary and Migratory Wildfowls Inhabiting Infected Premises" Viruses 14, no. 1: 116. https://doi.org/10.3390/v14010116
APA StyleYoo, D.-s., Kang, S.-I., Lee, Y.-N., Lee, E.-K., Kim, W.-y., & Lee, Y.-J. (2022). Bridging the Local Persistence and Long-Range Dispersal of Highly Pathogenic Avian Influenza Virus (HPAIv): A Case Study of HPAIv-Infected Sedentary and Migratory Wildfowls Inhabiting Infected Premises. Viruses, 14(1), 116. https://doi.org/10.3390/v14010116






