Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of DZIF-10c
2.2. Cryoelectron Microscopy
2.3. Analysis of the Pharmacokinetic Profile of DZIF-10c in NRG and FcRn Mice
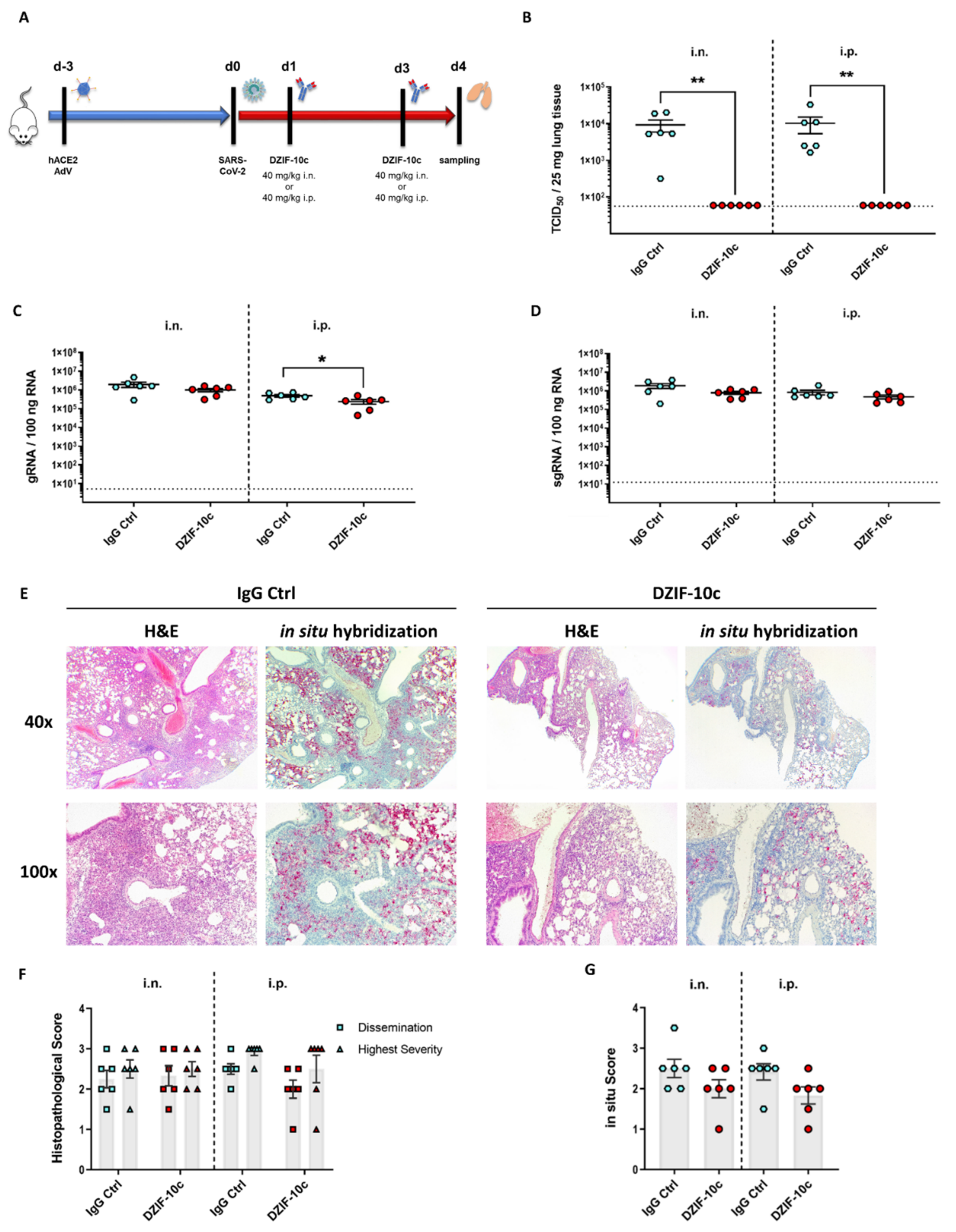
2.4. Analysis of DZIF-10c In Vivo Efficacy after the SARS-CoV-2 Challenge
2.5. Histopathological Examination of Lung Tissue
2.6. Statistical Analysis
3. Results
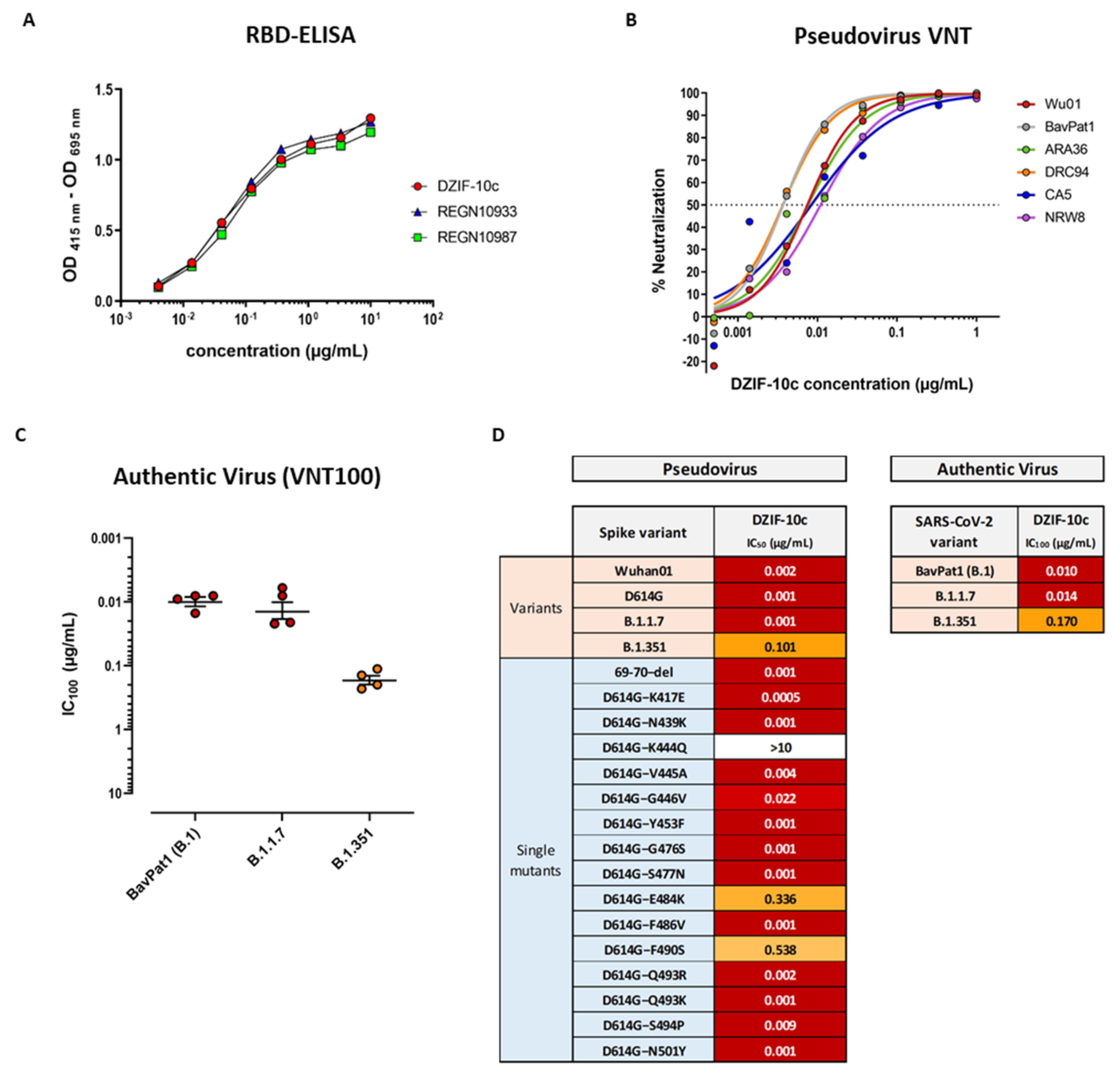
3.1. DZIF-10c Displays an Extraordinary Neutralizing Capacity against SARS-CoV-2 and Remains Active against SARS-CoV-2 Variants B.1.1.7 and B.1.351
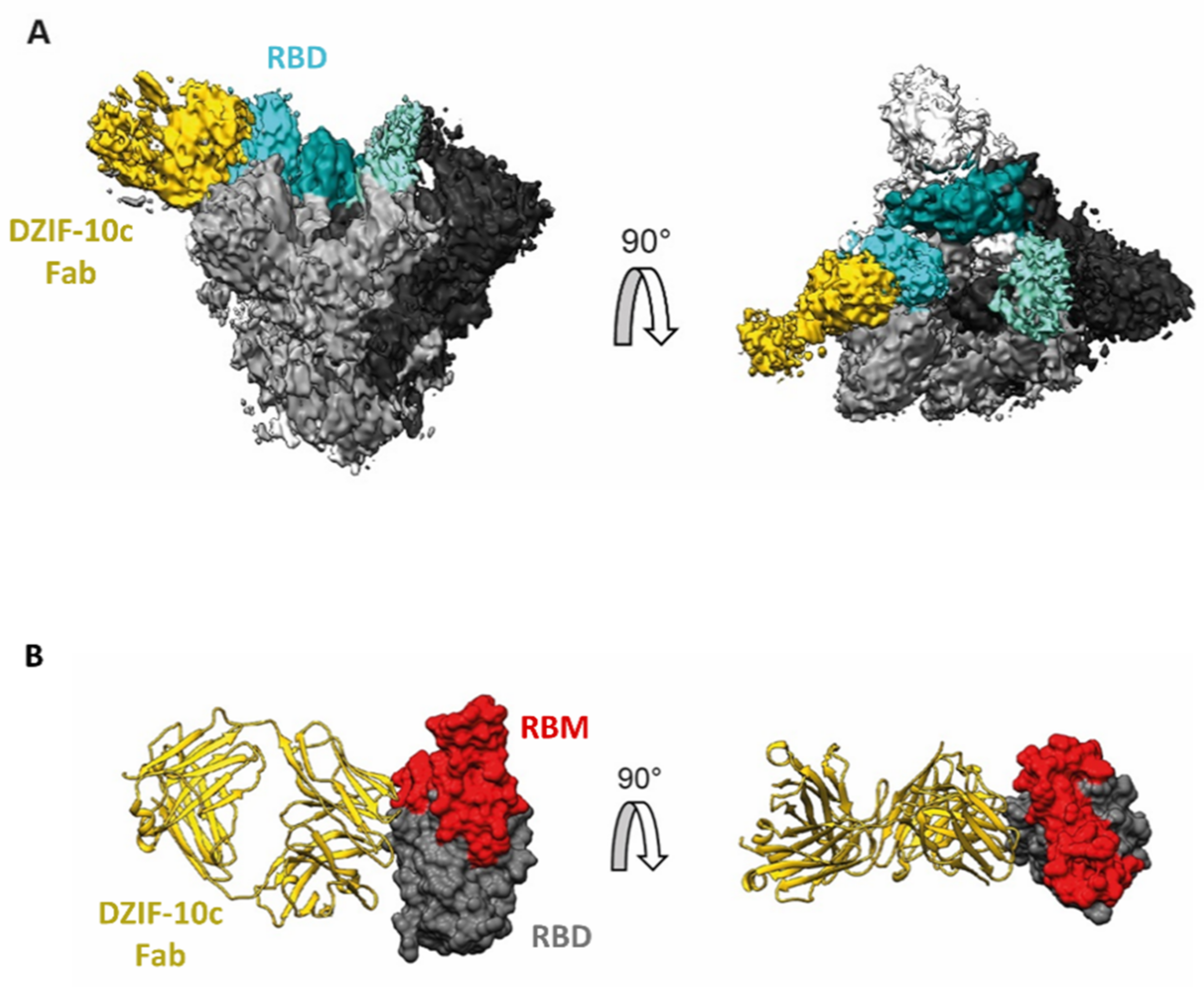
3.2. Structural Analysis Indicates Binding of DZIF-10c to the Prefusion Conformation of S Adjacent to the Receptor Binding Motif
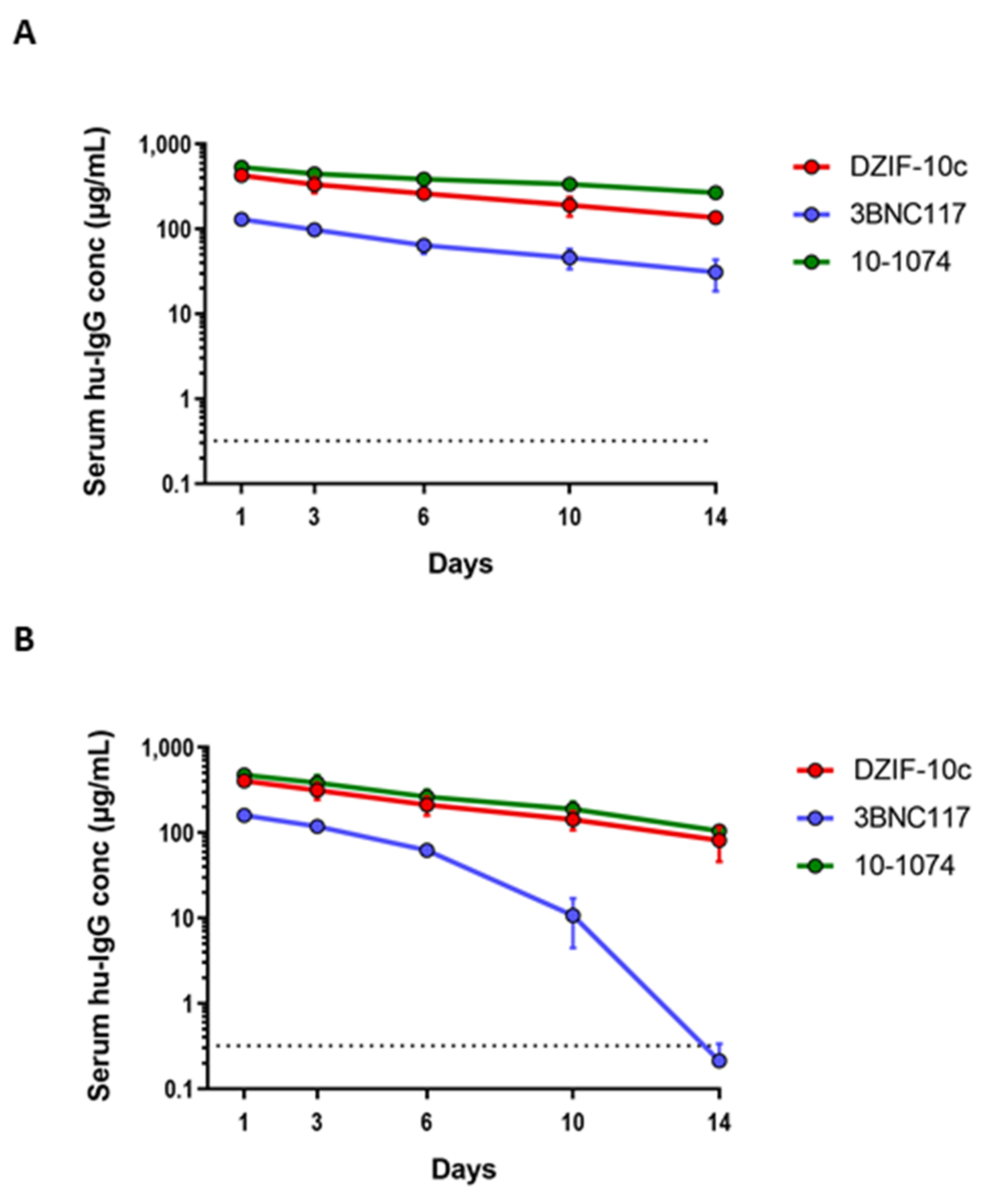
3.3. DZIF-10c Shows a Favorable Pharmacokinetic Profile In Vivo
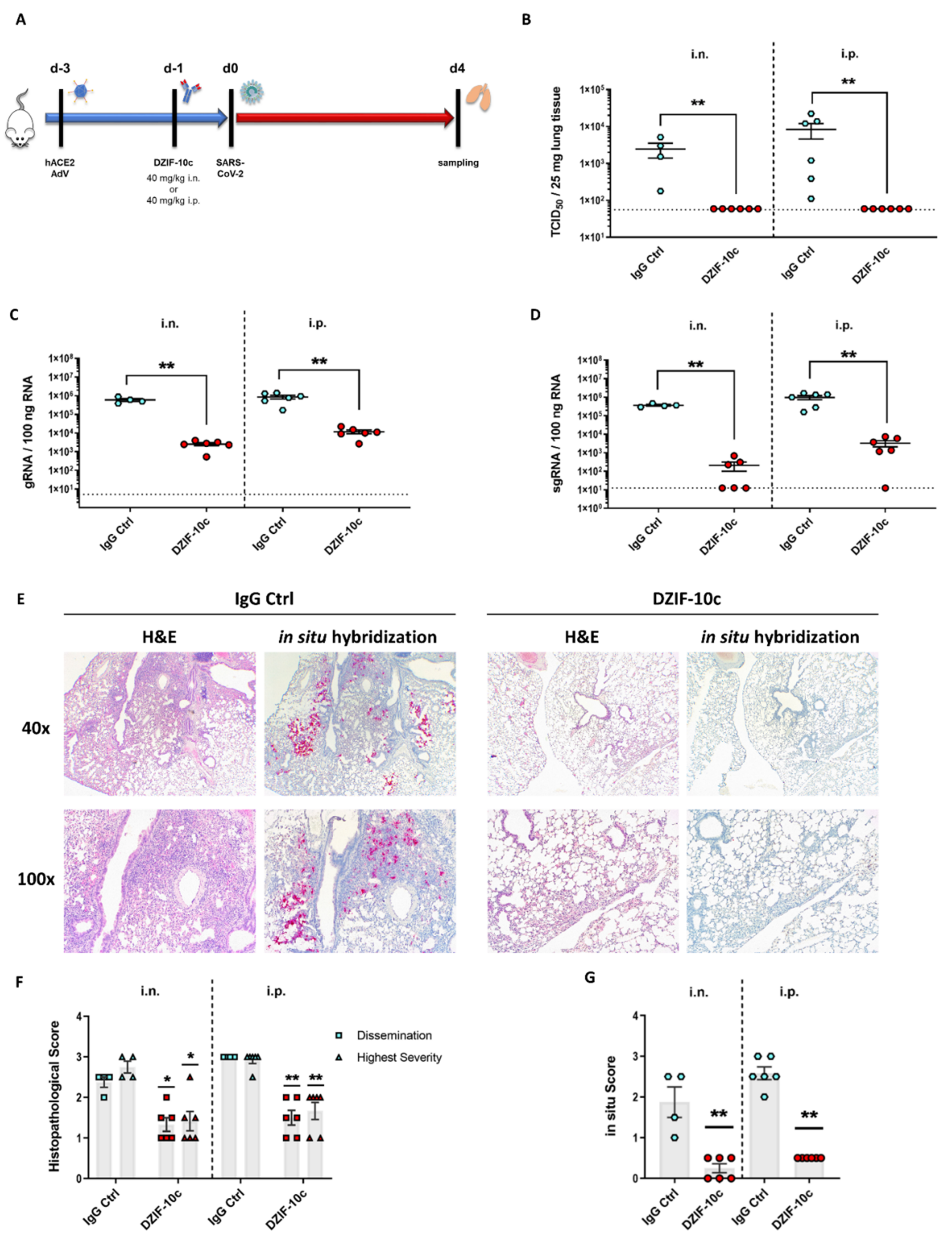
3.4. DZIF-10c Efficiently Protects hACE2-Transduced Mice from Infection with SARS-CoV-2
3.5. DZIF-10c Shows No Evidence of Enhanced Infection in an In Vitro ADE Assay
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johns Hopkins Coronavirus Resource Center. COVID-19 United States Cases by County—Johns Hopkins Coronavirus Resource Center. Available online: https://coronavirus.jhu.edu/map (accessed on 6 July 2021).
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Malley, R.; DeVincenzo, J.; Ramilo, O.; Dennehy, P.H.; Meissner, H.C.; Gruber, W.C.; Sanchez, P.J.; Jafri, H.; Balsley, J.; Carlin, D.; et al. Reduction of Respiratory Syncytial Virus (RSV) in Tracheal Aspirates in Intubated Infants by Use of Humanized Monoclonal Antibody to RSV F Protein. J. Infect. Dis. 1998, 178, 1555–1561. [Google Scholar] [CrossRef] [Green Version]
- Klein, F.; Halper-Stromberg, A.; Horwitz, J.A.; Scheid, J.F.; Bournazos, S.; Mouquet, H.; Spatz, L.A.; Abadir, A.; Zang, T.; Dorner, M.; et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 2012, 492, 118–122. [Google Scholar] [CrossRef] [Green Version]
- Caskey, M.; Klein, F.; Lorenzi, J.C.C.; Seaman, M.S.; West, A.P.; Buckley, N.; Kremer, G.; Nogueira, L.; Braunschweig, M.; Scheid, J.F.; et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 2015, 522, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Wong, G.; Audet, J.; Bello, A.; Fernando, L.; Alimonti, J.B.; Fausther-Bovendo, H.; Wei, H.; Aviles, J.; Hiatt, E.; et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Mulangu, S.; Dodd, L.E.; Davey, R.T.; Tshiani Mbaya, O.; Proschan, M.; Mukadi, D.; Lusakibanza Manzo, M.; Nzolo, D.; Tshomba Oloma, A.; Ibanda, A.; et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N. Engl. J. Med. 2019, 381, 2293–2303. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, H.; Yu, H.; Li, S.; Zheng, Q.; Xia, N. Neutralizing antibodies against SARS-CoV-2: Current understanding, challenge and perspective. Antib. Ther. 2020, 3, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.-W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Baum, A.; Pascal, K.E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B.O.; Yan, Y.; Koon, K.; Patel, K.; et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020, 369, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Nirula, A.; Heller, B.; Gottlieb, R.L.; Boscia, J.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021, 384, 229–237. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Nirula, A.; Chen, P.; Boscia, J.; Heller, B.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021, 325, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Ryu, D.-K.; Lee, J.; Kim, Y.-I.; Seo, J.-M.; Kim, Y.-G.; Jeong, J.-H.; Kim, M.; Kim, J.-I.; Kim, P.; et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182, 744–753.e4. [Google Scholar] [CrossRef] [PubMed]
- Tuccori, M.; Ferraro, S.; Convertino, I.; Cappello, E.; Valdiserra, G.; Blandizzi, C.; Maggi, F.; Focosi, D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: Clinical pipeline. mAbs 2020, 12, 1854149. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Teran, C.; Tiruthani, K.; McSweeney, M.; Ma, A.; Pickles, R.; Lai, S.K. Challenges and opportunities for antiviral monoclonal antibodies as COVID-19 therapy. Adv. Drug Deliv. Rev. 2021, 169, 100–117. [Google Scholar] [CrossRef]
- Kreer, C.; Zehner, M.; Weber, T.; Ercanoglu, M.S.; Gieselmann, L.; Rohde, C.; Halwe, S.; Korenkov, M.; Schommers, P.; Vanshylla, K.; et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell 2020, 182, 843–854.e12. [Google Scholar] [CrossRef] [PubMed]
- Khawli, L.A.; Goswami, S.; Hutchinson, R.; Kwong, Z.W.; Yang, J.; Wang, X.; Yao, Z.; Sreedhara, A.; Cano, T.; Tesar, D.; et al. Charge variants in IgG1: Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs 2010, 2, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Yu, C.; Yadav, D.B.; Hu, Z.; Amurao, A.; Duenas, E.; Wong, M.; Iverson, M.; Zheng, K.; Lam, X.; et al. Evaluation of Heavy-Chain C-Terminal Deletion on Product Quality and Pharmacokinetics of Monoclonal Antibodies. J. Pharm. Sci. 2016, 105, 2066–2072. [Google Scholar] [CrossRef]
- Schommers, P.; Gruell, H.; Abernathy, M.E.; Tran, M.-K.; Dingens, A.S.; Gristick, H.B.; Barnes, C.O.; Schoofs, T.; Schlotz, M.; Vanshylla, K.; et al. Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody. Cell 2020, 180, 471–489.e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malczyk, A.H.; Kupke, A.; Prüfer, S.; Scheuplein, V.A.; Hutzler, S.; Kreuz, D.; Beissert, T.; Bauer, S.; Hubich-Rau, S.; Tondera, C.; et al. A Highly Immunogenic and Protective Middle East Respiratory Syndrome Coronavirus Vaccine Based on a Recombinant Measles Virus Vaccine Platform. J. Virol. 2015, 89, 11654–11667. [Google Scholar] [CrossRef] [Green Version]
- Volz, A.; Kupke, A.; Song, F.; Jany, S.; Fux, R.; Shams-Eldin, H.; Schmidt, J.; Becker, C.; Eickmann, M.; Becker, S.; et al. Protective Efficacy of Recombinant Modified Vaccinia Virus Ankara Delivering Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. J. Virol. 2015, 89, 8651–8656. [Google Scholar] [CrossRef] [Green Version]
- Sauerhering, L.; Kupke, A.; Meier, L.; Dietzel, E.; Hoppe, J.; Gruber, A.D.; Gattenloehner, S.; Witte, B.; Fink, L.; Hofmann, N.; et al. Cyclophilin inhibitors restrict Middle East respiratory syndrome coronavirus via interferon-λ in vitro and in mice. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef]
- Tscherne, A.; Schwarz, J.H.; Rohde, C.; Kupke, A.; Kalodimou, G.; Limpinsel, L.; Okba, N.M.; Bošnjak, B.; Sandrock, I.; Odak, I.; et al. Immunogenicity and efficacy of the COVID-19 candidate vector vaccine MVA-SARS-2-S in preclinical vaccination. Proc. Natl. Acad. Sci. USA 2021, 118(28), e2026207118. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. [Google Scholar] [CrossRef]
- Vanshylla, K.; Di Cristanziano, V.; Kleipass, F.; Dewald, F.; Schommers, P.; Gieselmann, L.; Gruell, H.; Schlotz, M.; Ercanoglu, M.S.; Stumpf, R.; et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe 2021, 29, 917–929.e4. [Google Scholar] [CrossRef]
- Collier, D.A.; de Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.; Walls, A.C.; Kemp, S.S.A.; Bassi, J.; Pinto, D.; Fregni, C.S.; et al. SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv 2021. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Widera, M.; Wilhelm, A.; Hoehl, S.; Pallas, C.; Kohmer, N.; Wolf, T.; Rabenau, H.F.; Corman, V.; Drosten, C.; Vehreschild, M.J.; et al. Bamlanivimab does not neutralize two SARS-CoV-2 variants carrying E484K in vitro. medRxiv 2021. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, L.; Kuwahara, K.; Li, L.; Liu, Z.; Li, T.; Zhu, H.; Liu, J.; Xu, Y.; Xie, J.; et al. Immunodominant SARS Coronavirus Epitopes in Humans Elicited both Enhancing and Neutralizing Effects on Infection in Non-human Primates. ACS Infect. Dis. 2016, 2, 361–376. [Google Scholar] [CrossRef]
- Sjors, H.W.S.; Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 2012, 9, 853–854. [Google Scholar] [CrossRef]
- Proetzel, G.; Roopenian, D.C. Humanized FcRn mouse models for evaluating pharmacokinetics of human IgG antibodies. Methods 2014, 65, 148–153. [Google Scholar] [CrossRef] [Green Version]
- Caskey, M.; Schoofs, T.; Settler, A.; Karagounis, T.; Kreider, E.F.; Murrell, B.; Nogueira, L.; Oliveira, T.Y.; Learn, G.H.; Cohen, Y.Z.; et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017, 23, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, J.A.; Halper-Stromberg, A.; Mouquet, H.; Gitlin, A.D.; Tretiakova, A.; Eisenreich, T.R.; Malbec, M.; Gravemann, S.; Billerbeck, E.; Dorner, M.; et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. USA 2013, 110, 16538–16543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Alsoussi, W.B.; Turner, J.S.; Case, J.B.; Zhao, H.; Schmitz, A.J.; Zhou, J.Q.; Chen, R.E.; Lei, T.; Rizk, A.A.; McIntire, K.M.; et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020, 205, 915–922. [Google Scholar] [CrossRef]
- Sun, J.; Zhuang, Z.; Zheng, J.; Li, K.; Wong, R.L.-Y.; Liu, D.; Huang, J.; He, J.; Zhu, A.; Zhao, J.; et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020, 182, 734–743.e5. [Google Scholar] [CrossRef]
- Rathnasinghe, R.; Strohmeier, S.; Amanat, F.; Gillespie, V.L.; Krammer, F.; García-Sastre, A.; Coughlan, L.; Schotsaert, M.; Uccellini, M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2433–2445. [Google Scholar] [CrossRef]
- Jaume, M.; Yip, M.S.; Cheung, C.Y.; Leung, H.L.; Li, P.H.; Kien, F.; Dutry, I.; Callendret, B.; Escriou, N.; Altmeyer, R.; et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011, 85, 10582–10597. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chu, H.; Li, C.; Wong, B.H.-Y.; Cheng, Z.-S.; Poon, V.K.-M.; Sun, T.; Lau, C.C.-Y.; Wong, K.K.-Y.; Chan, J.Y.-W.; et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: Implications for pathogenesis. J. Infect. Dis. 2014, 209, 1331–1342. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Chan, K.-H.; Choi, G.K.-Y.; To, K.K.-W.; Tse, H.; Cai, J.-P.; Yeung, M.L.; Cheng, V.C.-C.; Chen, H.; Che, X.-Y.; et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: Implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013, 207, 1743–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, K.P.Y.; Cheung, M.-C.; Perera, R.A.P.M.; Ng, K.-C.; Bui, C.H.T.; Ho, J.C.W.; Ng, M.M.T.; Kuok, D.I.T.; Shih, K.C.; Tsao, S.-W.; et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: An analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020, 8, 687–695. [Google Scholar] [CrossRef]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Boumaza, A.; Gay, L.; Mezouar, S.; Bestion, E.; Diallo, A.B.; Michel, M.; Desnues, B.; Raoult, D.; La Scola, B.; Halfon, P.; et al. Monocytes and macrophages, targets of SARS-CoV-2: The clue for Covid-19 immunoparalysis. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Finkelstein, M.T.; Mermelstein, A.G.; Parker Miller, E.; Seth, P.C.; Stancofski, E.-S.D.; Fera, D. Structural Analysis of Neutralizing Epitopes of the SARS-CoV-2 Spike to Guide Therapy and Vaccine Design Strategies. Viruses 2021, 13, 134. [Google Scholar] [CrossRef]
- Huo, J.; Zhao, Y.; Ren, J.; Zhou, D.; Duyvesteyn, H.M.E.; Ginn, H.M.; Carrique, L.; Malinauskas, T.; Ruza, R.R.; Shah, P.N.M.; et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe 2020, 28, 445–454.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Duyvesteyn, H.M.E.; Chen, C.-P.; Huang, C.-G.; Chen, T.-H.; Shih, S.-R.; Lin, Y.-C.; Cheng, C.-Y.; Cheng, S.-H.; Huang, Y.-C.; et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020, 27, 950–958. [Google Scholar] [CrossRef]
- Wrapp, D.; de Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; van Breedam, W.; Roose, K.; van Schie, L.; Hoffmann, M.; Pöhlmann, S.; et al. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015.e15. [Google Scholar] [CrossRef]
- Lv, Z.; Deng, Y.-Q.; Ye, Q.; Cao, L.; Sun, C.-Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L.; et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020, 369, 1505–1509. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Dagotto, G.; Mercado, N.B.; Martinez, D.R.; Hou, Y.J.; Nkolola, J.P.; Carnahan, R.H.; Crowe, J.E.; Baric, R.S.; Barouch, D.H. Comparison of Subgenomic and Total RNA in SARS-CoV-2 Challenged Rhesus Macaques. J. Virol. 2021. [Google Scholar] [CrossRef]
- Cleary, S.J.; Pitchford, S.C.; Amison, R.T.; Carrington, R.; Robaina Cabrera, C.L.; Magnen, M.; Looney, M.R.; Gray, E.; Page, C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020, 177, 4851–4865. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Grado, V.H.; Tan, G.S.; Leon, P.E.; Yondola, M.; Palese, P. Direct administration in the respiratory tract improves efficacy of broadly neutralizing anti-influenza virus monoclonal antibodies. Antimicrob. Agents Chemother. 2015, 59, 4162–4172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, G.A.; Hemming, V.G.; Horswood, R.L.; Baron, P.A.; Chanock, R.M. Effectiveness of topically administered neutralizing antibodies in experimental immunotherapy of respiratory syncytial virus infection in cotton rats. J. Virol. 1987, 61, 1851–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weltzin, R.; Traina-Dorge, V.; Soike, K.; Zhang, J.Y.; Mack, P.; Soman, G.; Drabik, G.; Monath, T.P. Intranasal monoclonal IgA antibody to respiratory syncytial virus protects rhesus monkeys against upper and lower respiratory tract infection. J. Infect. Dis. 1996, 174, 256–261. [Google Scholar] [CrossRef] [Green Version]
- Piepenbrink, M.S.; Park, J.-G.; Oladunni, F.S.; Deshpande, A.; Basu, M.; Sarkar, S.; Loos, A.; Woo, J.; Lovalenti, P.; Sloan, D.; et al. Therapeutic activity of an inhaled potent SARS-CoV-2 neutralizing human monoclonal antibody in hamsters. Cell Rep. Med. 2021, 2, 100218. [Google Scholar] [CrossRef]
- Visweswaraiah, A. Tracking the tissue distribution of marker dye following intranasal delivery in mice and chinchillas: A multifactorial analysis of parameters affecting nasal retention. Vaccine 2002, 20, 3209–3220. [Google Scholar] [CrossRef]
- Ku, Z.; Xie, X.; Hinton, P.R.; Liu, X.; Ye, X.; Muruato, A.E.; Ng, D.C.; Biswas, S.; Zou, J.; Liu, Y.; et al. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature 2021, 1–6. [Google Scholar] [CrossRef]
- Ladel, S.; Flamm, J.; Zadeh, A.S.; Filzwieser, D.; Walter, J.-C.; Schlossbauer, P.; Kinscherf, R.; Lischka, K.; Luksch, H.; Schindowski, K. Allogenic Fc Domain-Facilitated Uptake of IgG in Nasal Lamina Propria: Friend or Foe for Intranasal CNS Delivery? Pharmaceutics 2018, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Ladel, S.; Maigler, F.; Flamm, J.; Schlossbauer, P.; Handl, A.; Hermann, R.; Herzog, H.; Hummel, T.; Mizaikoff, B.; Schindowski, K. Impact of Glycosylation and Species Origin on the Uptake and Permeation of IgGs through the Nasal Airway Mucosa. Pharmaceutics 2020, 12, 1014. [Google Scholar] [CrossRef]
- Yoshida, M.; Claypool, S.M.; Wagner, J.S.; Mizoguchi, E.; Mizoguchi, A.; Roopenian, D.C.; Lencer, W.I.; Blumberg, R.S. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity 2004, 20, 769–783. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020, 9. [Google Scholar] [CrossRef]
- Arvin, A.M.; Fink, K.; Schmid, M.A.; Cathcart, A.; Spreafico, R.; Havenar-Daughton, C.; Lanzavecchia, A.; Corti, D.; Virgin, H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 2020, 584, 353–363. [Google Scholar] [CrossRef]
- Quinlan, B.D.; He, W.; Mou, H.; Zhang, L.; Guo, Y.; Chang, J.; Peng, S.; Ojha, A.; Tavora, R.; Parcells, M.S.; et al. An engineered receptor-binding domain improves the immunogenicity of multivalent SARS-CoV-2 vaccines. bioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Du Bois, I.; Marsico, A.; Bertrams, W.; Schweiger, M.R.; Caffrey, B.E.; Sittka-Stark, A.; Eberhardt, M.; Vera, J.; Vingron, M.; Schmeck, B.T. Genome-wide Chromatin Profiling of Legionella pneumophila-Infected Human Macrophages Reveals Activation of the Probacterial Host Factor TNFAIP2. J. Infect. Dis. 2016, 214, 454–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herkt, C.E.; Caffrey, B.E.; Surmann, K.; Blankenburg, S.; Gesell Salazar, M.; Jung, A.L.; Herbel, S.M.; Hoffmann, K.; Schulte, L.N.; Chen, W.; et al. A MicroRNA Network Controls Legionella pneumophila Replication in Human Macrophages via LGALS8 and MX1. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chand, M.; Hopkins, S.; Dabrera, G.; Achison, C.; Barclay, W.; Ferguson, N.; Volz, E.; Loman, N.; Rambaut, A.; Barrett, J. Investigation of novel SARS-COV-2 variant, Variant of Concern 202012/01. In PHE Gateway Number: GW-1824; Public Health England: London, UK, 2020. [Google Scholar]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Krähling, V.; Halwe, S.; Rohde, C.; Becker, D.; Berghöfer, S.; Dahlke, C.; Eickmann, M.; Ercanoglu, M.S.; Gieselmann, L.; Herwig, A.; et al. Development and characterization of an indirect ELISA to detect SARS-CoV-2 spike protein-specific antibodies. J. Immunol. Methods 2021, 490, 112958. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Killington, R.A. Virus isolation and quantitation. Virol. Methods Man. 1996, 25–46. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corman, V.M.; Eckerle, I.; Bleicker, T.; Zaki, A.; Landt, O.; Eschbach-Bludau, M.; van Boheemen, S.; Gopal, R.; Ballhause, M.; Bestebroer, T.M.; et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 2012, 17, 20285. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halwe, S.; Kupke, A.; Vanshylla, K.; Liberta, F.; Gruell, H.; Zehner, M.; Rohde, C.; Krähling, V.; Gellhorn Serra, M.; Kreer, C.; et al. Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses 2021, 13, 1498. https://doi.org/10.3390/v13081498
Halwe S, Kupke A, Vanshylla K, Liberta F, Gruell H, Zehner M, Rohde C, Krähling V, Gellhorn Serra M, Kreer C, et al. Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses. 2021; 13(8):1498. https://doi.org/10.3390/v13081498
Chicago/Turabian StyleHalwe, Sandro, Alexandra Kupke, Kanika Vanshylla, Falk Liberta, Henning Gruell, Matthias Zehner, Cornelius Rohde, Verena Krähling, Michelle Gellhorn Serra, Christoph Kreer, and et al. 2021. "Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection" Viruses 13, no. 8: 1498. https://doi.org/10.3390/v13081498
APA StyleHalwe, S., Kupke, A., Vanshylla, K., Liberta, F., Gruell, H., Zehner, M., Rohde, C., Krähling, V., Gellhorn Serra, M., Kreer, C., Klüver, M., Sauerhering, L., Schmidt, J., Cai, Z., Han, F., Young, D., Yang, G., Widera, M., Koch, M., ... Becker, S. (2021). Intranasal Administration of a Monoclonal Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. Viruses, 13(8), 1498. https://doi.org/10.3390/v13081498





