Live Bird Markets in Nigeria: A Potential Reservoir for H9N2 Avian Influenza Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Genome Amplification and Sequencing
2.3. Illumina Sequencing Data Analysis
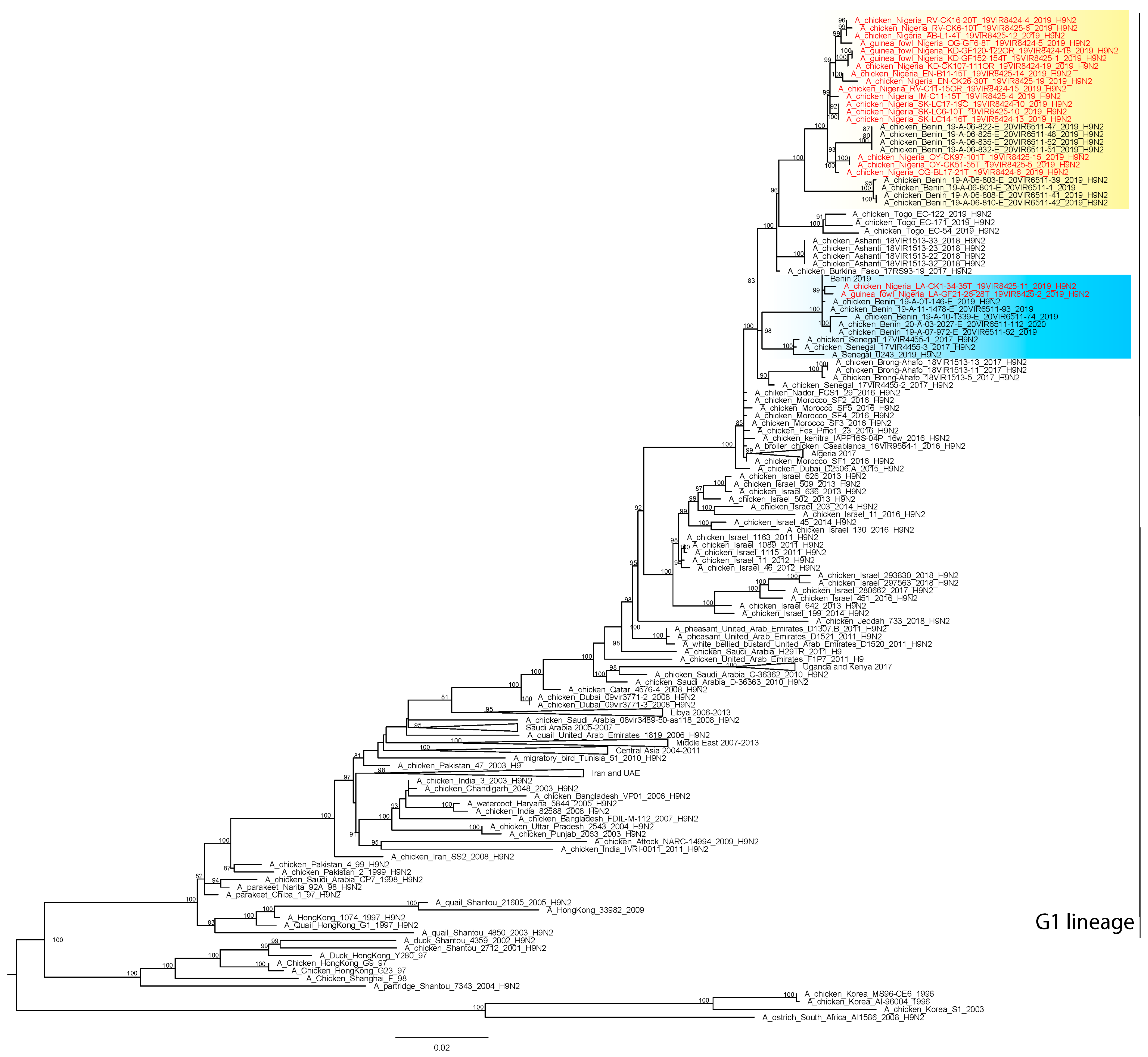
2.4. Phylogenetic Analyses
2.5. Bayesian Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joannis, T.; Lombin, L.H.; De Benedictis, P.; Cattoli, G.; Capua, I. Confirmation of H5N1 avian influenza in Africa. Vet. Rec. 2006, 158, 309–310. [Google Scholar] [CrossRef]
- Waziri, N.E.; Nguku, P.; Olayinka, A.; Ajayi, I.; Kabir, J.; Okolocha, E.; Tseggai, T.; Joannis, T.; Okewole, P.; Kumbish, P. Evaluating a surveillance system: Live-bird market surveillance for highly pathogenic avian influenza, a case study. Pan Afr. Med. J. 2014, 18 (Suppl. 1), 11. [Google Scholar] [CrossRef]
- Waziri, M.I.; Abdu, P.A.; Sa’idu, L.; Bello, M. Seroepidemiology and assessment of risk factors for the spread of avian influenza in birds in two Nigerian states. Vet. Med. Sci. 2017, 3, 227–238. [Google Scholar] [CrossRef]
- Samaan, G.; Gultom, A.; Indriani, R.; Lokuge, K.; Kelly, P.M. Critical control points for avian influenza A H5N1 in live bird markets in low resource settings. Prev. Vet. Med. 2011, 100, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.; Nelson, M.I.; Joannis, T.; Bertolotti, L.; Monne, I.; Salviato, A.; Olaleye, O.; Shittu, I.; Sulaiman, L.; Lombin, L.H.; et al. Evolutionary Dynamics of Multiple Sublineages of H5N1 Influenza Viruses in Nigeria from 2006 to 2008. J. Virol. 2010, 84, 3239–3247. [Google Scholar] [CrossRef] [Green Version]
- Coker, T.; Meseko, C.; Odaibo, G.; Olaleye, D. Circulation of the low pathogenic avian influenza subtype H5N2 virus in ducks at a live bird market in Ibadan, Nigeria. Infect. Dis. Poverty 2014, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Fusaro, A.; Zecchin, B.; Vrancken, B.; Abolnik, C.; Ademun, R.; Alassane, A.; Arafa, A.; Awuni, J.A.; Couacy-Hymann, E.; Coulibaly, M.B.; et al. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat. Commun. 2019, 10, 5310. [Google Scholar] [CrossRef] [Green Version]
- Laleye, A.T.; Bianco, A.; Shittu, I.; Sulaiman, L.; Fusaro, A.; Inuwa, B.; Oyetunde, J.; Zecchin, B.; Bakam, J.; Pastori, A.; et al. Genetic characterization of highly pathogenic avian Influenza H5Nx clade 2.3.4.4b reveals independent introductions in nigeria. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Shittu, I.; Bianco, A.; Gado, D.; Mkpuma, N.; Sulaiman, L.; Laleye, A.; Gobbo, F.; Bortolami, A.; Bonfante, F.; Vakuru, C.; et al. First detection of highly pathogenic H5N6 avian influenza virus on the African continent. Emerg. Microbes Infect. 2020, 9, 886–888. [Google Scholar] [CrossRef] [Green Version]
- Aiki-Raji, C.O.; Adebiyi, A.I.; Agbajelola, V.I.; Adetunji, S.A.; Lameed, Q.; Adesina, M.; Adekanye, G.; Omidokun, F.; Fagbohun, O.; Oluwayelu, D.O. Surveillance for low pathogenic avian influenza viruses in live-bird markets in Oyo and Ogun States, Nigeria. Asian Pac. J. Trop. Dis. 2015, 5, 369–373. [Google Scholar] [CrossRef]
- Oluwayelu, D.O.; Omolanwa, A.; Adebiyi, A.I.; Aiki-Raji, O.C. Flock-Based Surveillance for Low Pathogenic Avian Influenza Virus in Commercial Breeders and Layers, Southwest Nigeria. Afr. J. Infect. Dis. 2017, 11, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Peacock, T.P.; James, J.; Sealy, J.E.; Iqbal, M. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [Green Version]
- Zecchin, B.; Minoungou, G.; Fusaro, A.; Moctar, S.; Ouedraogo-Kaboré, A.; Schivo, A.; Salviato, A.; Marciano, S.; Monne, I. Influenza A(H9N2) Virus, Burkina Faso. Emerg. Infect. Dis. 2017, 23, 2118–2119. [Google Scholar] [CrossRef] [Green Version]
- Awuni, J.A.; Bianco, A.; Dogbey, O.J.; Fusaro, A.; Yingar, D.T.; Salviato, A.; Ababio, P.T.; Milani, A.; Bonfante, F.; Monne, I. Avian influenza H9N2 subtype in Ghana: Virus characterization and evidence of co-infection. Avian Pathol. 2019, 48, 470–476. [Google Scholar] [CrossRef]
- Fusade-Boyer, M.; Djegui, F.; Batawui, K.; Byuragaba, D.K.; Jones, J.C.; Wabwire-Mangeni, F.; Erima, B.; Atim, G.; Ukuli, Q.A.; Tugume, T.; et al. Antigenic and molecular characterization of low pathogenic avian influenza A(H9N2) viruses in sub-Saharan Africa from 2017 through 2019. Emerg. Microbes Infect. 2021, 10, 753–761. [Google Scholar] [CrossRef]
- Jallow, M.M.; Fall, A.; Barry, M.A.; Diop, B.; Sy, S.; Goudiaby, D.; Fall, M.; Enouf, V.; Niang, M.N.; Dia, N. Genetic characterization of the first detected human case of low pathogenic avian influenza A/H9N2 in sub-Saharan Africa, Senegal. Emerg. Microbes Infect. 2020, 9, 1092–1095. [Google Scholar] [CrossRef] [PubMed]
- Depristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Wilm, A.; Aw, P.P.K.; Bertrand, D.; Yeo, G.H.T.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S.; Thi Hoang, D.; Chernomor, O.; von Haeseler, A.; Quang Minh, B.; Sy Vinh, L.; et al. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, M.; Xu, L.; Wang, X.; Liu, X. Current situation of H9N2 subtype avian influenza in China. Vet. Res. 2017, 48, 49. [Google Scholar] [CrossRef] [Green Version]
- Ducatez, M.F.; Olinger, C.M.; Owoade, A.A.; De Landtsheer, S.; Ammerlaan, W.; Niesters, H.G.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M.; Muller, C.P. Avian Flu: Multiple introductions of H5N1 in Nigeria. Nature 2006, 442, 37. [Google Scholar] [CrossRef]
- Wan, H.; Perez, D.R. Amino Acid 226 in the Hemagglutinin of H9N2 Influenza Viruses Determines Cell Tropism and Replication in Human Airway Epithelial Cells. J. Virol. 2007, 81, 5181–5191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meseko, C.; Globig, A.; Ijomanta, J.; Joannis, T.; Nwosuh, C.; Shamaki, D.; Harder, T.; Hoffman, D.; Pohlmann, A.; Beer, M.; et al. Evidence of exposure of domestic pigs to Highly Pathogenic Avian Influenza H5N1 in Nigeria. Sci. Rep. 2018, 8, 5900. [Google Scholar] [CrossRef] [PubMed]

| S/N # | Isolates ID | Scientific Name | Location | Collection Date |
|---|---|---|---|---|
| 1 | A/chicken/Nigeria/RV-CK16-20T_19VIR8424-4/2019 | Gallus domesticus | Rivers | 7 May 2019 |
| 2 | A/chicken/Nigeria/RV-CK6-10T_19VIR8425-6/2019 | Gallus domesticus | Rivers | 19 July 2019 |
| 3 | A/chicken/Nigeria/RV-C11-15OR_19VIR8424-15/2019 | Gallus domesticus | Rivers | 7 December 2019 |
| 4 | A/chicken/Nigeria/SK-LC17-19C_19VIR8424-10/2019 | Gallus domesticus | Sokoto | 7 July 2019 |
| 5 | A/chicken/Nigeria/SK-LC14-16T_19VIR8424-13/2019 | Gallus domesticus | Sokoto | 7 July 2019 |
| 6 | A/chicken/Nigeria/SK-LC6-10T_19VIR8425-10/2019 | Gallus domesticus | Sokoto | 20 July 2019 |
| 7 | A/guinea_fowl/Nigeria/KD-GF152-154T_19VIR8425-1/2019 | Numida meleagris | Kaduna | 17 July 2019 |
| 8 | A/guinea_fowl/Nigeria/KD-GF120-122OR_19VIR8424-18/2019 | Numida meleagris | Kaduna | 7 November 2019 |
| 9 | A/chicken/Nigeria/KD-CK107-111OR_19VIR8424-19/2019 | Gallus domesticus | Kaduna | 7 November 2019 |
| 10 | A/guinea_fowl/Nigeria/LA-GF21-26-28T_19VIR8425-2/2019 | Numida meleagris | Lagos | 18 July 2019 |
| 11 | A/chicken/Nigeria/LA-CK1-34-35T_19VIR8425-11/2019 | Gallus domesticus | Lagos | 25 July 2019 |
| 12 | A/chicken/Nigeria/EN-CK26-30T_19VIR8425-19/2019 | Gallus domesticus | Enugu | 8 April 2019 |
| 13 | A/chicken/Nigeria/EN-B11-15T_19VIR8425-14/2019 | Gallus domesticus | Enugu | 28 July 2019 |
| 14 | A/guinea_fowl/Nigeria/OG-GF6-8T_19VIR8424-5/2019 | Numida meleagris | Ogun | 7 May 2019 |
| 15 | A/chicken/Nigeria/OG-BL17-21T_19VIR8424-6/2019 | Gallus domesticus | Ogun | 7 May 2019 |
| 16 | A/chicken/Nigeria/OY-CK97-101T_19VIR8425-15/2019 | Gallus domesticus | Oyo | 8 April 2019 |
| 17 | A/chicken/Nigeria/OY-CK51-55T_19VIR8425-5/2019 | Gallus domesticus | Oyo | 26 July 2019 |
| 18 | A/chicken/Nigeria/AB-L1-4T_19VIR8425-12/2019 | Gallus domesticus | Abia | 23 July 2019 |
| 19 | A/chicken/Nigeria/IM-C11-15T_19VIR8425-4/2019 | Gallus domesticus | Imo | 19 July 2019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulaiman, L.; Shittu, I.; Fusaro, A.; Inuwa, B.; Zecchin, B.; Gado, D.; Schivo, A.; Bianco, A.; Laleye, A.; Gobbo, F.; et al. Live Bird Markets in Nigeria: A Potential Reservoir for H9N2 Avian Influenza Viruses. Viruses 2021, 13, 1445. https://doi.org/10.3390/v13081445
Sulaiman L, Shittu I, Fusaro A, Inuwa B, Zecchin B, Gado D, Schivo A, Bianco A, Laleye A, Gobbo F, et al. Live Bird Markets in Nigeria: A Potential Reservoir for H9N2 Avian Influenza Viruses. Viruses. 2021; 13(8):1445. https://doi.org/10.3390/v13081445
Chicago/Turabian StyleSulaiman, Lanre, Ismaila Shittu, Alice Fusaro, Bitrus Inuwa, Bianca Zecchin, Dorcas Gado, Alessia Schivo, Alice Bianco, Agnes Laleye, Federica Gobbo, and et al. 2021. "Live Bird Markets in Nigeria: A Potential Reservoir for H9N2 Avian Influenza Viruses" Viruses 13, no. 8: 1445. https://doi.org/10.3390/v13081445
APA StyleSulaiman, L., Shittu, I., Fusaro, A., Inuwa, B., Zecchin, B., Gado, D., Schivo, A., Bianco, A., Laleye, A., Gobbo, F., Vakuru, C., Joannis, T., Monne, I., & Meseko, C. (2021). Live Bird Markets in Nigeria: A Potential Reservoir for H9N2 Avian Influenza Viruses. Viruses, 13(8), 1445. https://doi.org/10.3390/v13081445






