Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry
Abstract
1. Introduction
1.1. AAV Biotechnology
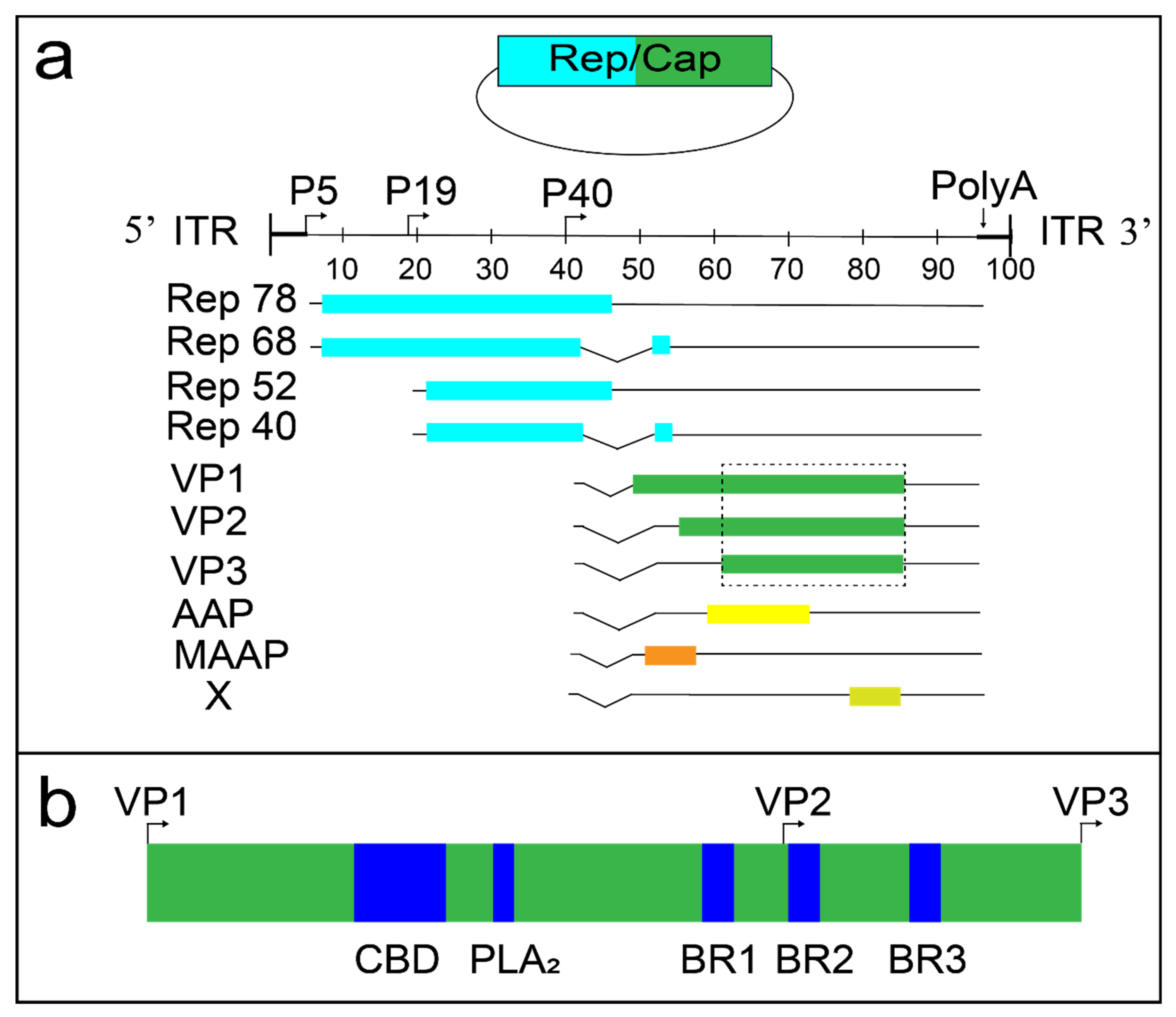
1.2. AAV Molecular Biology
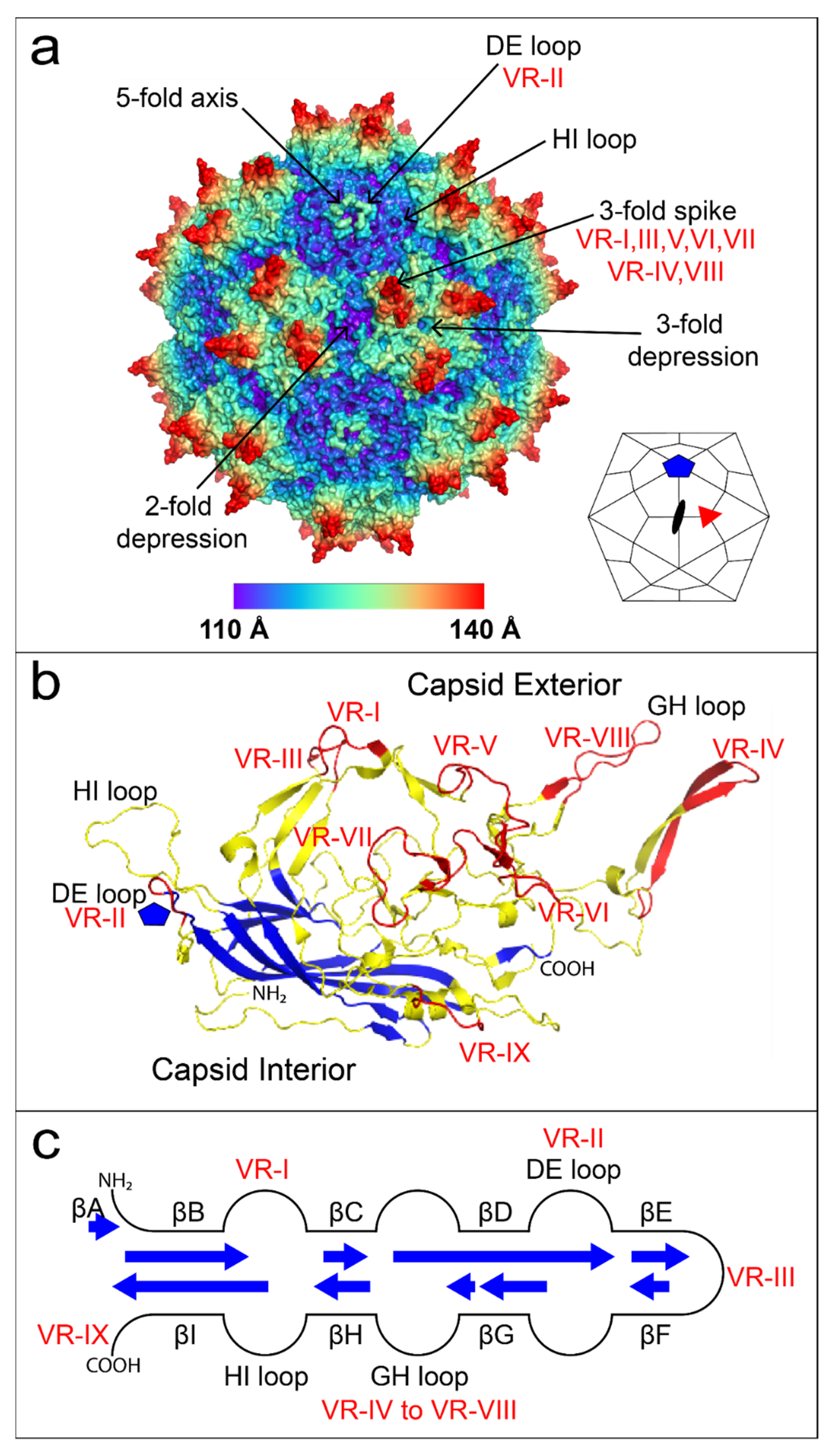
1.3. The First High-Resolution Dependovirus Structure
2. AAV Capsid Modification Strategies
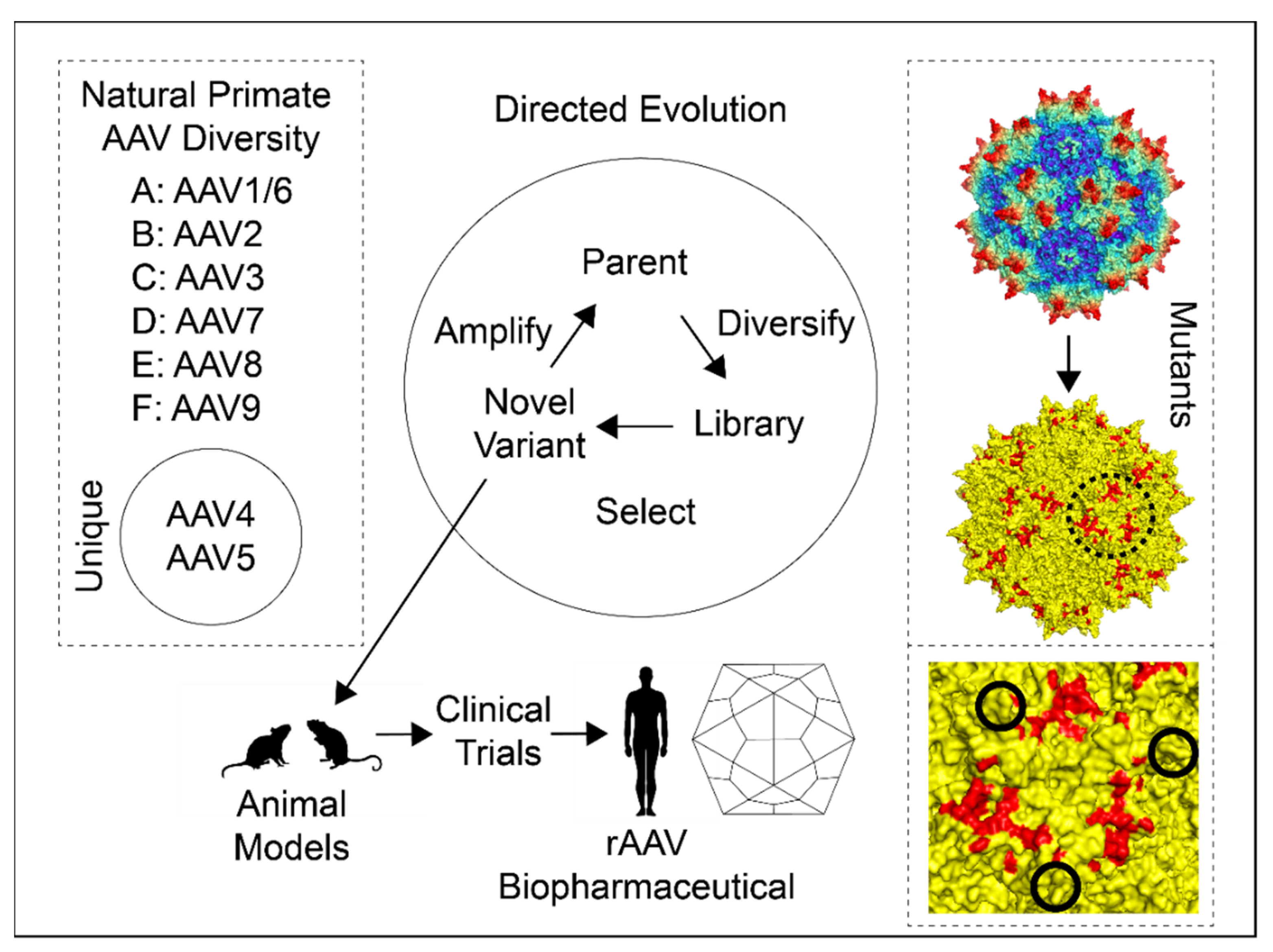
2.1. Natural AAV Genetic Diversity
2.2. AAV Directed Evolution
2.3. AAV Mutants
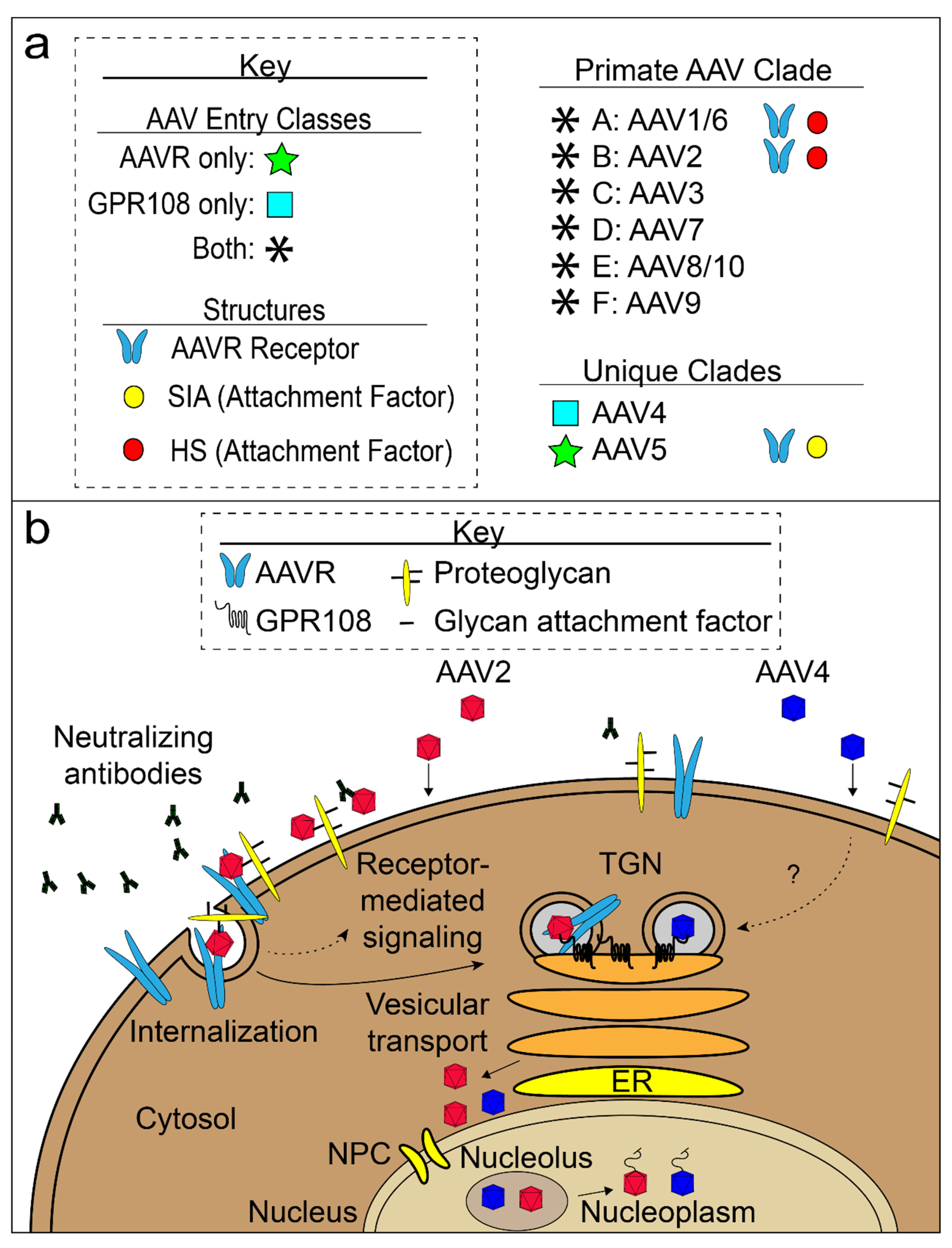
3. AAV Cell Attachment and Entry
3.1. AAV Glycan Attachment Factors
3.1.1. HSPG
3.1.2. SIA and GAL
3.2. AAV Proteinaceous Receptors
3.2.1. AAVR
3.2.2. GPR108 and VP1u
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Srivastava, A.; Weitzman, M.; Chatterjee, S.; Engelhardt, J.F.; Owens, R.A.; Muzyczka, N.; Ali, R. A Tribute to Barrie J. Carter. Hum. Gene Ther. 2020, 31, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Berns, K.I.; Muzyczka, N. AAV: An Overview of Unanswered Questions. Hum. Gene Ther. 2017, 28, 308–313. [Google Scholar] [CrossRef]
- Meier, A.F.; Fraefel, C.; Seyffert, M. The Interplay between Adeno-Associated Virus and its Helper Viruses. Viruses 2020, 12, 662. [Google Scholar] [CrossRef] [PubMed]
- Smalley, E. First AAV gene therapy poised for landmark approval. Nat. Biotechnol. 2017, 35, 998–999. [Google Scholar] [CrossRef] [PubMed]
- Al-Zaidy, S.; Pickard, A.S.; Kotha, K.; Alfano, L.N.; Lowes, L.; Paul, G.; Church, K.; Lehman, K.; Sproule, D.M.; Dabbous, O.; et al. Health outcomes in spinal muscular atrophy type 1 following AVXS-101 gene replacement therapy. Pediatr. Pulmonol. 2019, 54, 179–185. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.A.; Rodino-Klapac, L.R.; Goodspeed, K.; Gray, S.J.; Kay, C.N.; Boye, S.L.; Boye, S.E.; George, L.A.; Salabarria, S.; et al. Current Clinical Applications of In Vivo Gene Therapy with AAVs. Mol. Ther. 2021, 29, 464–488. [Google Scholar] [CrossRef]
- Greer, C.; Kozyak, B.; Stedman, H. Challenges at the Crossroads: Myopathy Trials in 2020 Hindsight. Mol. Ther. 2021, 29, 420–421. [Google Scholar] [CrossRef]
- Vandendriessche, T.; Thorrez, L.; Acosta-Sanchez, A.; Petrus, I.; Wang, L.; Ma, L.; Waele, L.D.E.; Iwasaki, Y.; Gillijns, V.; Wilson, J.M.; et al. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J. Thromb. Haemost. 2007, 5, 16–24. [Google Scholar] [CrossRef]
- Zaiss, A.K.; Liu, Q.; Bowen, G.P.; Wong, N.C.; Bartlett, J.S.; Muruve, D.A. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 2002, 76, 4580–4590. [Google Scholar] [CrossRef]
- Ferrari, F.K.; Xiao, X.; McCarty, D.; Samulski, R.J. New developments in the generation of Ad-free, high-titer rAAV gene therapy vectors. Nat. Med. 1997, 3, 1295–1297. [Google Scholar] [CrossRef]
- Matsushita, T.; Elliger, S.; Elliger, C.; Podsakoff, G.; Villarreal, L.; Kurtzman, G.J.; Iwaki, Y.; Colosi, P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998, 5, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998, 72, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Kern, A.; Rittner, K.; Kleinschmidt, J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998, 9, 2745–2760. [Google Scholar] [CrossRef]
- Clement, N.; Grieger, J.C. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol. Ther. Methods Clin. Dev. 2016, 3, 16002. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Li, M.; Zhong, L.; Su, Q.; Li, J.; Li, S.; He, R.; Zhang, Y.; Hendricks, G.; Wang, J. Empty virions in AAV8 vector preparations reduce transduction efficiency and may cause total viral particle dose-limiting side effects. Mol. Ther.-Methods Clin. Dev. 2014, 1, 9. [Google Scholar] [CrossRef]
- Gao, G.; Vandenberghe, L.H.; Alvira, M.R.; Lu, Y.; Calcedo, R.; Zhou, X.; Wilson, J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004, 78, 6381–6388. [Google Scholar] [CrossRef]
- Schmidt, M.; Govindasamy, L.; Afione, S.; Kaludov, N.; Agbandje-McKenna, M.; Chiorini, J.A. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942). J. Virol. 2008, 82, 8911–8916. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Voutetakis, A.; Afione, S.; Zheng, C.; Mandikian, D.; Chiorini, J.A. Adeno-associated virus type 12 (AAV12): A novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J. Virol. 2008, 82, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Takeuchi, T.; Enomoto, Y.; Kondo, K.; Sato, K.; Ono, F.; Sata, T.; Kanda, T. Tissue distribution of cynomolgus adeno-associated viruses AAV10, AAV11, and AAVcy.7 in naturally infected monkeys. Arch. Virol. 2008, 153, 375–380. [Google Scholar] [CrossRef]
- Mietzsch, M.; Jose, A.; Chipman, P.; Bhattacharya, N.; Daneshparvar, N.; McKenna, R.; Agbandje-McKenna, M. Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features. Viruses 2021, 13, 101. [Google Scholar] [CrossRef]
- Ng, R.; Govindasamy, L.; Gurda, B.L.; McKenna, R.; Kozyreva, O.G.; Samulski, R.J.; Parent, K.N.; Baker, T.S.; Agbandje-McKenna, M. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J. Virol. 2010, 84, 12945–12957. [Google Scholar] [CrossRef]
- Xie, Q.; Bu, W.; Bhatia, S.; Hare, J.; Somasundaram, T.; Azzi, A.; Chapman, M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 2002, 99, 10405–10410. [Google Scholar] [CrossRef] [PubMed]
- Lerch, T.F.; Xie, Q.; Chapman, M.S. The structure of adeno-associated virus serotype 3B (AAV-3B): Insights into receptor binding and immune evasion. Virology 2010, 403, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, L.; Padron, E.; McKenna, R.; Muzyczka, N.; Kaludov, N.; Chiorini, J.A.; Agbandje-McKenna, M. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 2006, 80, 11556–11570. [Google Scholar] [CrossRef]
- Govindasamy, L.; DiMattia, M.A.; Gurda, B.L.; Halder, S.; McKenna, R.; Chiorini, J.A.; Muzyczka, N.; Zolotukhin, S.; Agbandje-McKenna, M. Structural insights into adeno-associated virus serotype 5. J. Virol. 2013, 87, 11187–11199. [Google Scholar] [CrossRef]
- Xie, Q.; Lerch, T.F.; Meyer, N.L.; Chapman, M.S. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6). Virology 2011, 420, 10–19. [Google Scholar] [CrossRef]
- Nam, H.J.; Lane, M.D.; Padron, E.; Gurda, B.; McKenna, R.; Kohlbrenner, E.; Aslanidi, G.; Byrne, B.; Muzyczka, N.; Zolotukhin, S.; et al. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007, 81, 12260–12271. [Google Scholar] [CrossRef]
- DiMattia, M.A.; Nam, H.J.; Van Vliet, K.; Mitchell, M.; Bennett, A.; Gurda, B.L.; McKenna, R.; Olson, N.H.; Sinkovits, R.S.; Potter, M.; et al. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012, 86, 6947–6958. [Google Scholar] [CrossRef]
- Mietzsch, M.; Barnes, C.; Hull, J.A.; Chipman, P.; Xie, J.; Bhattacharya, N.; Sousa, D.; McKenna, R.; Gao, G.; Agbandje-McKenna, M. Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Lerch, T.F.; O’Donnell, J.K.; Meyer, N.L.; Xie, Q.; Taylor, K.A.; Stagg, S.M.; Chapman, M.S. Structure of AAV-DJ, a retargeted gene therapy vector: Cryo-electron microscopy at 4.5 A resolution. Structure 2012, 20, 1310–1320. [Google Scholar] [CrossRef]
- Xie, Q.; Yoshioka, C.K.; Chapman, M.S. Adeno-Associated Virus (AAV-DJ)-Cryo-EM Structure at 1.56 A Resolution. Viruses 2020, 12, 1194. [Google Scholar] [CrossRef]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.L.; Hirsch, M.L.; Barker, J.C.; Connelly, J.P.; Steininger, R.J., 3rd; Porteus, M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Lee, J.S.; Wang, L.; Desai, T.; Akache, B.; Storm, T.A.; Kay, M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008, 82, 5887–5911. [Google Scholar] [CrossRef]
- Ronzitti, G.; Gross, D.-A.; Mingozzi, F. Human Immune Responses to Adeno-Associated Virus (AAV) Vectors. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Summerford, C.; Samulski, R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998, 72, 1438–1445. [Google Scholar] [CrossRef]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An essential receptor for adeno-associated virus infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ogden, P.J.; Kelsic, E.D.; Sinai, S.; Church, G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 2019, 366, 1139–1143. [Google Scholar] [CrossRef]
- Im, D.S.; Muzyczka, N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J. Virol. 1992, 66, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- King, J.A.; Dubielzig, R.; Grimm, D.; Kleinschmidt, J.A. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001, 20, 3282–3291. [Google Scholar] [CrossRef]
- Girod, A.; Wobus, C.E.; Zadori, Z.; Ried, M.; Leike, K.; Tijssen, P.; Kleinschmidt, J.A.; Hallek, M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002, 83, 973–978. [Google Scholar] [CrossRef]
- Maurer, A.C.; Cepeda Diaz, A.K.; Vandenberghe, L.H. Residues on Adeno-associated Virus Capsid Lumen Dictate Interactions and Compatibility with the Assembly-Activating Protein. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, F.; Köther, K.; Schmidt, K.; Weghofer, M.; Raupp, C.; Nieto, K.; Kuck, A.; Gerlach, B.; Böttcher, B.; Müller, O.J.; et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011, 85, 12686–12697. [Google Scholar] [CrossRef] [PubMed]
- Naumer, M.; Sonntag, F.; Schmidt, K.; Nieto, K.; Panke, C.; Davey, N.E.; Popa-Wagner, R.; Kleinschmidt, J.A. Properties of the adeno-associated virus assembly-activating protein. J. Virol. 2012, 86, 13038–13048. [Google Scholar] [CrossRef] [PubMed]
- Earley, L.F.; Powers, J.M.; Adachi, K.; Baumgart, J.T.; Meyer, N.L.; Xie, Q.; Chapman, M.S.; Nakai, H. Adeno-associated Virus (AAV) Assembly-Activating Protein Is Not an Essential Requirement for Capsid Assembly of AAV Serotypes 4, 5, and 11. J. Virol. 2017, 91, e01980-16. [Google Scholar] [CrossRef]
- Tse, L.V.; Moller-Tank, S.; Meganck, R.M.; Asokan, A. Mapping and Engineering Functional Domains of the Assembly-Activating Protein of Adeno-associated Viruses. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Maurer, A.C.; Pacouret, S.; Cepeda Diaz, A.K.; Blake, J.; Andres-Mateos, E.; Vandenberghe, L.H. The Assembly-Activating Protein Promotes Stability and Interactions between AAV’s Viral Proteins to Nucleate Capsid Assembly. Cell Rep. 2018, 23, 1817–1830. [Google Scholar] [CrossRef]
- Stutika, C.; Gogol-Döring, A.; Botschen, L.; Mietzsch, M.; Weger, S.; Feldkamp, M.; Chen, W.; Heilbronn, R. A Comprehensive RNA Sequencing Analysis of the Adeno-Associated Virus (AAV) Type 2 Transcriptome Reveals Novel AAV Transcripts, Splice Variants, and Derived Proteins. J. Virol. 2016, 90, 1278–1289. [Google Scholar] [CrossRef]
- Cao, M.; Chiriva-Internati, M.; Hermonat, P.L. AAV2 X increases AAV6 rep/cap-driven rAAV production. Virology 2015, 482, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; You, H.; Hermonat, P.L. The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication. PLoS ONE 2014, 9, e104596. [Google Scholar] [CrossRef]
- Grieger, J.C.; Snowdy, S.; Samulski, R.J. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 2006, 80, 5199–5210. [Google Scholar] [CrossRef]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [PubMed]
- Zadori, Z.; Szelei, J.; Lacoste, M.C.; Li, Y.; Gariepy, S.; Raymond, P.; Allaire, M.; Nabi, I.R.; Tijssen, P. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 2001, 1, 291–302. [Google Scholar] [CrossRef]
- Zengel, J.; Carette, J.E. Structural and cellular biology of adeno-associated virus attachment and entry. Adv. Virus Res. 2020, 106, 39–84. [Google Scholar] [CrossRef]
- Rossmann, M.G. Structure of viruses: A short history. Q. Rev. Biophys. 2013, 46, 133–180. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.G.; Johnson, J.E. Icosahedral RNA Virus Structure. Ann. Rev. Biochem. 1989, 58, 533–573. [Google Scholar] [CrossRef]
- Chapman, M.S.; Liljas, L. Structural folds of viral proteins. Adv. Protein Chem. 2003, 64, 125–196. [Google Scholar] [CrossRef] [PubMed]
- Tsao, J.; Chapman, M.S.; Agbandje, M.; Keller, W.; Smith, K.; Wu, H.; Luo, M.; Smith, T.J.; Rossmann, M.G.; Compans, R.W.; et al. The three-dimensional structure of canine parvovirus and its functional implications. Science 1991, 251, 1456–1464. [Google Scholar] [CrossRef]
- Penzes, J.J.; Soderlund-Venermo, M.; Canuti, M.; Eis-Hubinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef]
- Agbandje, M.; McKenna, R.; Rossmann, M.G.; Strassheim, M.L.; Parrish, C.R. Structure Determination of Feline Panleukopenia Virus Empty Capsids. Proteins 1993, 16, 155–171. [Google Scholar] [CrossRef]
- Agbandje-McKenna, M.; Llamas-Saiz, A.L.; Wang, F.; Tattersall, P.; Rossmann, M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 1998, 6, 1369–1381. [Google Scholar] [CrossRef]
- Simpson, A.A.; Hebert, B.; Sullivan, G.M.; Parrish, C.R.; Zadori, Z.; Tijssen, P.; Rossmann, M.G. The structure of porcine parvovirus: Comparison with related viruses. J. Mol. Biol. 2002, 315, 1189–1198. [Google Scholar] [CrossRef]
- Simpson, A.A.; Chipman, P.R.; Baker, T.S.; Tijssen, P.; Rossmann, M.G. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 1998, 6, 1355–1367. [Google Scholar] [CrossRef]
- Chapman, M.S.; Agbandje-McKenna, M. Atomic structure of viral particles. In Parvoviruses; Kerr, J.R., Cotmore, S.F., Bloom, M.E., Linden, R.M., Parrish, C.R., Eds.; Hodder Arnold, Ltd.: London, UK, 2006; pp. 107–123. [Google Scholar]
- Chapman, M.S.; Rossmann, M.G. Structure, sequence, and function correlations among parvoviruses. Virology 1993, 194, 491–508. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Gao, G.; Alvira, M.R.; Somanathan, S.; Lu, Y.; Vandenberghe, L.H.; Rux, J.J.; Calcedo, R.; Sanmiguel, J.; Abbas, Z.; Wilson, J.M. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 2003, 100, 6081–6086. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wang, C.; Wu, J.; Ye, F.; Lv, R.; Hu, D.; Ai, L.; Yang, L.; Wu, T.; Li, B.; et al. Distribution and genetic diversity of adeno-associated viruses in bats from coastal areas of Southeast China. Sci. Rep. 2020, 10, 3725. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X.; Hon, C.C.; Zhang, H.; Zhou, P.; Zhang, Y.; Wu, Y.; Wang, L.F.; Shi, Z. Prevalence and genetic diversity of adeno-associated viruses in bats from China. J. Gen. Virol. 2010, 91, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Liu, Y.; Shi, Z.; Liu, H.; Wei, Y.; Yang, L. Bat adeno-associated viruses as gene therapy vectors with the potential to evade human neutralizing antibodies. Gene Ther. 2019, 26, 264–276. [Google Scholar] [CrossRef]
- Hoggan, M.D.; Blacklow, N.R.; Rowe, W.P. Studies of small DNA viruses found in various adenovirus preparations: Physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 1966, 55, 1467–1474. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, S.; Mizukami, H.; Young, N.S.; Brown, K.E. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology 1996, 221, 208–217. [Google Scholar] [CrossRef]
- Rutledge, E.; Halbert, C.; Russell, D. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998, 72, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Jensen, R.L.; Schnepp, B.C.; Connell, M.J.; Shell, R.; Sferra, T.J.; Bartlett, J.S.; Clark, K.R.; Johnson, P.R. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 2005, 79, 14781–14792. [Google Scholar] [CrossRef]
- Cabanes-Creus, M.; Hallwirth, C.V.; Westhaus, A.; Ng, B.H.; Liao, S.H.Y.; Zhu, E.; Navarro, R.G.; Baltazar, G.; Drouyer, M.; Scott, S.; et al. Restoring the natural tropism of AAV2 vectors for human liver. Sci. Transl. Med. 2020, 12, eaba3312. [Google Scholar] [CrossRef]
- Cabanes-Creus, M.; Westhaus, A.; Navarro, R.G.; Baltazar, G.; Zhu, E.; Amaya, A.K.; Liao, S.H.Y.; Scott, S.; Sallard, E.; Dilworth, K.L.; et al. Attenuation of Heparan Sulfate Proteoglycan Binding Enhances In Vivo Transduction of Human Primary Hepatocytes with AAV2. Mol. Ther. Methods Clin. Dev. 2020, 17, 1139–1154. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.L.; Brown, A.; Loveland, A.B.; Lotun, A.; Xu, M.; Luo, L.; Xu, G.; Li, J.; Ren, L.; Su, Q.; et al. Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties. Nat. Commun. 2020, 11, 3279. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, E.; Penzes, J.J.; Gifford, R.J.; Agbandje-Mckenna, M.; Kotin, R.M. Evolution of dependoparvoviruses across geological timescales-implications for design of AAV-based gene therapy vectors. Virus Evol. 2020, 6, veaa043. [Google Scholar] [CrossRef]
- Finnigan, G.C.; Hanson-Smith, V.; Stevens, T.H.; Thornton, J.W. Evolution of increased complexity in a molecular machine. Nature 2012, 481, 360–364. [Google Scholar] [CrossRef]
- Zinn, E.; Pacouret, S.; Khaychuk, V.; Turunen, H.T.; Carvalho, L.S.; Andres-Mateos, E.; Shah, S.; Shelke, R.; Maurer, A.C.; Plovie, E.; et al. In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector. Cell Rep. 2015, 12, 1056–1068. [Google Scholar] [CrossRef]
- Santiago-Ortiz, J.; Ojala, D.S.; Westesson, O.; Weinstein, J.R.; Wong, S.Y.; Steinsapir, A.; Kumar, S.; Holmes, I.; Schaffer, D.V. AAV ancestral reconstruction library enables selection of broadly infectious viral variants. Gene Ther. 2015, 22, 934–946. [Google Scholar] [CrossRef]
- Landegger, L.D.; Pan, B.; Askew, C.; Wassmer, S.J.; Gluck, S.D.; Galvin, A.; Taylor, R.; Forge, A.; Stankovic, K.M.; Holt, J.R.; et al. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017, 35, 280–284. [Google Scholar] [CrossRef]
- Hu, C.J.; Lu, Y.C.; Tsai, Y.H.; Cheng, H.Y.; Takeda, H.; Huang, C.Y.; Xiao, R.; Hsu, C.J.; Tsai, J.W.; Vandenberghe, L.H.; et al. Efficient in Utero Gene Transfer to the Mammalian Inner Ears by the Synthetic Adeno-Associated Viral Vector Anc80L65. Mol. Ther. Methods Clin. Dev. 2020, 18, 493–500. [Google Scholar] [CrossRef]
- Gurda, B.L.; DiMattia, M.A.; Miller, E.B.; Bennett, A.; McKenna, R.; Weichert, W.S.; Nelson, C.D.; Chen, W.J.; Muzyczka, N.; Olson, N.H.; et al. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J. Virol. 2013, 87, 9111–9124. [Google Scholar] [CrossRef] [PubMed]
- Mingozzi, F.; Maus, M.V.; Hui, D.J.; Sabatino, D.E.; Murphy, S.L.; Rasko, J.E.; Ragni, M.V.; Manno, C.S.; Sommer, J.; Jiang, H.; et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007, 13, 419–422. [Google Scholar] [CrossRef]
- Popa-Wagner, R.; Porwal, M.; Kann, M.; Reuss, M.; Weimer, M.; Florin, L.; Kleinschmidt, J.A. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J. Virol. 2012, 86, 9163–9174. [Google Scholar] [CrossRef]
- Johnson, J.S.; Li, C.; DiPrimio, N.; Weinberg, M.S.; McCown, T.J.; Samulski, R.J. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 2010, 84, 8888–8902. [Google Scholar] [CrossRef] [PubMed]
- Wörner, T.P.; Bennett, A.; Habka, S.; Snijder, J.; Friese, O.; Powers, T.; Agbandje-McKenna, M.; Heck, A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021, 12, 1642. [Google Scholar] [CrossRef]
- Becerra, S.P.; Koczot, F.; Fabisch, P.; Rose, J.A. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 1988, 62, 2745–2754. [Google Scholar] [CrossRef]
- Excoffon, K.J.; Koerber, J.T.; Dickey, D.D.; Murtha, M.; Keshavjee, S.; Kaspar, B.K.; Zabner, J.; Schaffer, D.V. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. USA 2009, 106, 3865–3870. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, Z.; Zhang, J.; Firrman, J.; Wei, H.; Zhuang, Z.; Liu, L.; Miao, L.; Hu, Y.; Li, D.; et al. A Robust System for Production of Superabundant VP1 Recombinant AAV Vectors. Mol. Ther.-Methods Clin. Dev. 2017, 7, 146–156. [Google Scholar] [CrossRef]
- Salganik, M.; Venkatakrishnan, B.; Bennett, A.; Lins, B.; Yarbrough, J.; Muzyczka, N.; Agbandje-McKenna, M.; McKenna, R. Evidence for pH-dependent protease activity in the adeno-associated virus capsid. J. Virol. 2012, 86, 11877–11885. [Google Scholar] [CrossRef] [PubMed]
- Mietzsch, M.; Broecker, F.; Reinhardt, A.; Seeberger, P.H.; Heilbronn, R. Differential adeno-associated virus serotype-specific interaction patterns with synthetic heparins and other glycans. J. Virol. 2014, 88, 2991–3003. [Google Scholar] [CrossRef]
- Mietzsch, M.; Casteleyn, V.; Weger, S.; Zolotukhin, S.; Heilbronn, R. OneBac 2.0: Sf9 Cell Lines for Production of AAV5 Vectors with Enhanced Infectivity and Minimal Encapsidation of Foreign DNA. Hum. Gene Ther. 2015, 26, 688–697. [Google Scholar] [CrossRef]
- Fakhiri, J.; Grimm, D. Best of most possible worlds: Hybrid gene therapy vectors based on parvoviruses and heterologous viruses. Mol. Ther. 2021. [Google Scholar] [CrossRef] [PubMed]
- Schmit, P.F.; Pacouret, S.; Zinn, E.; Telford, E.; Nicolaou, F.; Broucque, F.; Andres-Mateos, E.; Xiao, R.; Penaud-Budloo, M.; Bouzelha, M.; et al. Cross-Packaging and Capsid Mosaic Formation in Multiplexed AAV Libraries. Mol. Ther. Methods Clin. Dev. 2020, 17, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.E.; Rolling, F.; Li, C.; Conrath, H.; Xiao, W.; Xiao, X.; Samulski, R.J. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 2002, 76, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Hauck, B.; Xiao, W. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 2003, 77, 2768–2774. [Google Scholar] [CrossRef]
- Rabinowitz, J.E.; Bowles, D.E.; Faust, S.M.; Ledford, J.G.; Cunningham, S.E.; Samulski, R.J. Cross-dressing the virion: The transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J. Virol. 2004, 78, 4421–4432. [Google Scholar] [CrossRef]
- Chai, Z.; Sun, J.; Rigsbee, K.M.; Wang, M.; Samulski, R.J.; Li, C. Application of polyploid adeno-associated virus vectors for transduction enhancement and neutralizing antibody evasion. J. Control. Release 2017, 262, 348–356. [Google Scholar] [CrossRef]
- Chai, Z.; Zhang, X.; Dobbins, A.L.; Frost, E.A.; Samulski, R.J.; Li, C. Chimeric Capsid Proteins Impact Transduction Efficiency of Haploid Adeno-Associated Virus Vectors. Viruses 2019, 11, 1138. [Google Scholar] [CrossRef]
- Mills, D.R.; Peterson, R.L.; Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl. Acad. Sci. USA 1967, 58, 217–224. [Google Scholar] [CrossRef]
- Clackson, T.; Hoogenboom, H.R.; Griffiths, A.D.; Winter, G. Making antibody fragments using phage display libraries. Nature 1991, 352, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Arnold, F.H. Tuning the activity of an enzyme for unusual environments: Sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl. Acad. Sci. USA 1993, 90, 5618–5622. [Google Scholar] [CrossRef] [PubMed]
- Gibney, E.; Van Noorden, R.; Ledford, H.; Castelvecchi, D.; Warren, M. ‘Test-tube’ evolution wins Chemistry Nobel Prize. Nature 2018, 562, 176. [Google Scholar] [CrossRef]
- Stemmer, W.P. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 10747–10751. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.P. Rapid evolution of a protein in vitro by DNA shuffling. Nature 1994, 370, 389–391. [Google Scholar] [CrossRef]
- Lukashov, V.V.; Goudsmit, J. Evolutionary relationships among parvoviruses: Virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 2001, 75, 2729–2740. [Google Scholar] [CrossRef]
- Senapathy, P.; Carter, B.J. Molecular cloning of adeno-associated virus variant genomes and generation of infectious virus by recombination in mammalian cells. J. Biol. Chem. 1984, 259, 4661–4666. [Google Scholar] [CrossRef]
- Bowles, D.E.; Rabinowitz, J.E.; Samulski, R.J. Marker rescue of adeno-associated virus (AAV) capsid mutants: A novel approach for chimeric AAV production. J. Virol. 2003, 77, 423–432. [Google Scholar] [CrossRef][Green Version]
- Perabo, L.; Endell, J.; King, S.; Lux, K.; Goldnau, D.; Hallek, M.; Buning, H. Combinatorial engineering of a gene therapy vector: Directed evolution of adeno-associated virus. J. Gene Med. 2006, 8, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Maheshri, N.; Koerber, J.T.; Kaspar, B.K.; Schaffer, D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006, 24, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Giver, L.; Shao, Z.; Affholter, J.A.; Arnold, F.H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998, 16, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Cabanes-Creus, M.; Ginn, S.L.; Amaya, A.K.; Liao, S.H.Y.; Westhaus, A.; Hallwirth, C.V.; Wilmott, P.; Ward, J.; Dilworth, K.L.; Santilli, G.; et al. Codon-Optimization of Wild-Type Adeno-Associated Virus Capsid Sequences Enhances DNA Family Shuffling while Conserving Functionality. Mol. Ther. Methods Clin. Dev. 2019, 12, 71–84. [Google Scholar] [CrossRef]
- Soong, N.W.; Nomura, L.; Pekrun, K.; Reed, M.; Sheppard, L.; Dawes, G.; Stemmer, W.P. Molecular breeding of viruses. Nat. Genet. 2000, 25, 436–439. [Google Scholar] [CrossRef]
- Lochrie, M.A.; Tatsuno, G.P.; Christie, B.; McDonnell, J.W.; Zhou, S.; Surosky, R.; Pierce, G.F.; Colosi, P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006, 80, 821–834. [Google Scholar] [CrossRef]
- Meyer, N.L.; Hu, G.; Davulcu, O.; Xie, Q.; Noble, A.J.; Yoshioka, C.; Gingerich, D.S.; Trzynka, A.; David, L.; Stagg, S.M.; et al. Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR. Elife 2019, 8, e44707. [Google Scholar] [CrossRef]
- Zhang, R.; Cao, L.; Cui, M.; Sun, Z.; Hu, M.; Zhang, R.; Stuart, W.; Zhao, X.; Yang, Z.; Li, X.; et al. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat. Microbiol. 2019, 4, 675–682. [Google Scholar] [CrossRef]
- Adachi, K.; Enoki, T.; Kawano, Y.; Veraz, M.; Nakai, H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 2014, 5, 3075. [Google Scholar] [CrossRef]
- Levy, S.F.; Blundell, J.R.; Venkataram, S.; Petrov, D.A.; Fisher, D.S.; Sherlock, G. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 2015, 519, 181–186. [Google Scholar] [CrossRef]
- Pekrun, K.; De Alencastro, G.; Luo, Q.J.; Liu, J.; Kim, Y.; Nygaard, S.; Galivo, F.; Zhang, F.; Song, R.; Tiffany, M.R.; et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- de Alencastro, G.; Pekrun, K.; Valdmanis, P.; Tiffany, M.; Xu, J.; Kay, M.A. Tracking Adeno-Associated Virus Capsid Evolution by High-Throughput Sequencing. Hum. Gene Ther. 2020, 31, 553–564. [Google Scholar] [CrossRef]
- Marsic, D.; Mendez-Gomez, H.R.; Zolotukhin, S. High-accuracy biodistribution analysis of adeno-associated virus variants by double barcode sequencing. Mol. Ther. Methods Clin. Dev. 2015, 2, 15041. [Google Scholar] [CrossRef]
- Herrmann, A.K.; Bender, C.; Kienle, E.; Grosse, S.; El Andari, J.; Botta, J.; Schurmann, N.; Wiedtke, E.; Niopek, D.; Grimm, D. A Robust and All-Inclusive Pipeline for Shuffling of Adeno-Associated Viruses. ACS Synth. Biol. 2019, 8, 194–206. [Google Scholar] [CrossRef]
- Davidsson, M.; Diaz-Fernandez, P.; Schwich, O.D.; Torroba, M.; Wang, G.; Bjorklund, T. A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing. Sci. Rep. 2016, 6, 37563. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.H.; Bashir, A.; Sinai, S.; Jain, N.K.; Ogden, P.J.; Riley, P.F.; Church, G.M.; Colwell, L.J.; Kelsic, E.D. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 2021, 39, 691–696. [Google Scholar] [CrossRef]
- Spector, L.P.; Tiffany, M.; Ferraro, N.M.; Abell, N.S.; Montgomery, S.B.; Kay, M.A. Evaluating the Genomic Parameters Governing rAAV-Mediated Homologous Recombination. Mol. Ther. 2021, 29, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, J.; Drouin, L.M.; Agbandje-McKenna, M.; Chen, C.; Qiao, C.; Pu, D.; Hu, X.; Wang, D.Z.; Li, J.; et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. USA 2009, 106, 3946–3951. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Asokan, A.; Wu, Z.; Van Dyke, T.; DiPrimio, N.; Johnson, J.S.; Govindaswamy, L.; Agbandje-McKenna, M.; Leichtle, S.; Redmond, D.E., Jr.; et al. Engineering and selection of shuffled AAV genomes: A new strategy for producing targeted biological nanoparticles. Mol. Ther. 2008, 16, 1252–1260. [Google Scholar] [CrossRef]
- Koerber, J.T.; Jang, J.H.; Schaffer, D.V. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 2008, 16, 1703–1709. [Google Scholar] [CrossRef]
- O’Donnell, J.; Taylor, K.A.; Chapman, M.S. Adeno-associated virus-2 and its primary cellular receptor--Cryo-EM structure of a heparin complex. Virology 2009, 385, 434–443. [Google Scholar] [CrossRef]
- Wobus, C.E.; Hugle-Dorr, B.; Girod, A.; Petersen, G.; Hallek, M.; Kleinschmidt, J.A. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: Epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 2000, 74, 9281–9293. [Google Scholar] [CrossRef]
- McCraw, D.M.; O’Donnell, J.K.; Taylor, K.A.; Stagg, S.M.; Chapman, M.S. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 2012, 431, 40–49. [Google Scholar] [CrossRef]
- Foust, K.D.; Nurre, E.; Montgomery, C.L.; Hernandez, A.; Chan, C.M.; Kaspar, B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009, 27, 59–65. [Google Scholar] [CrossRef]
- Duque, S.; Joussemet, B.; Riviere, C.; Marais, T.; Dubreil, L.; Douar, A.M.; Fyfe, J.; Moullier, P.; Colle, M.A.; Barkats, M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 2009, 17, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Bevan, A.K.; Duque, S.; Foust, K.D.; Morales, P.R.; Braun, L.; Schmelzer, L.; Chan, C.M.; McCrate, M.; Chicoine, L.G.; Coley, B.D.; et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther. 2011, 19, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Deverman, B.E.; Pravdo, P.L.; Simpson, B.P.; Kumar, S.R.; Chan, K.Y.; Banerjee, A.; Wu, W.L.; Yang, B.; Huber, N.; Pasca, S.P.; et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016, 34, 204–209. [Google Scholar] [CrossRef]
- Hordeaux, J.; Wang, Q.; Katz, N.; Buza, E.L.; Bell, P.; Wilson, J.M. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther. 2018, 26, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Hordeaux, J.; Yuan, Y.; Clark, P.M.; Wang, Q.; Martino, R.A.; Sims, J.J.; Bell, P.; Raymond, A.; Stanford, W.L.; Wilson, J.M. The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier. Mol. Ther. 2019, 27, 912–921. [Google Scholar] [CrossRef]
- Nonnenmacher, M.; Weber, T. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe 2011, 10, 563–576. [Google Scholar] [CrossRef]
- Di Pasquale, G.; Chiorini, J.A. AAV transcytosis through barrier epithelia and endothelium. Mol. Ther. 2006, 13, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.S.; Meltzer, J.C.; Buzhdygan, T.; Cheng, M.J.; Sena-Esteves, M.; Bennett, R.E.; Sullivan, T.P.; Razmpour, R.; Gong, Y.; Ng, C.; et al. Selection of an Efficient AAV Vector for Robust CNS Transgene Expression. Mol. Ther. Methods Clin. Dev. 2019, 15, 320–332. [Google Scholar] [CrossRef]
- Perabo, L.; Goldnau, D.; White, K.; Endell, J.; Boucas, J.; Humme, S.; Work, L.M.; Janicki, H.; Hallek, M.; Baker, A.H.; et al. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J. Virol. 2006, 80, 7265–7269. [Google Scholar] [CrossRef]
- Girod, A.; Ried, M.; Wobus, C.; Lahm, H.; Leike, K.; Kleinschmidt, J.; Deleage, G.; Hallek, M. Genetic capsid modifications allow efficient re-targeting of adeno- associated virus type 2. Nat. Med. 1999, 5, 1052–1056. [Google Scholar] [CrossRef]
- Moskalenko, M.; Chen, L.; van Roey, M.; Donahue, B.A.; Snyder, R.O.; McArthur, J.G.; Patel, S.D. Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: Implications for gene therapy and virus structure. J. Virol. 2000, 74, 1761–1766. [Google Scholar] [CrossRef]
- Wu, P.; Xiao, W.; Conlon, T.; Hughes, J.; Agbandje-McKenna, M.; Ferkol, T.; Flotte, T.; Muzyczka, N. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 2000, 74, 8635–8647. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Mahajan, S.P.; Sulam, J.; Gray, J.J. Deep Learning in Protein Structural Modeling and Design. Patterns 2020, 1, 100142. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.D.; Kummer, M.; Kondratov, O.; Banerjee, A.; Moskalenko, O.; Zolotukhin, S. Applying machine learning to predict viral assembly for adeno-associated virus capsid libraries. Mol. Ther. Methods Clin. Dev. 2021, 20, 276–286. [Google Scholar] [CrossRef]
- Havlik, L.P.; Simon, K.E.; Smith, J.K.; Klinc, K.A.; Tse, L.V.; Oh, D.K.; Fanous, M.M.; Meganck, R.M.; Mietzsch, M.; Kleinschmidt, J.; et al. Coevolution of Adeno-associated Virus Capsid Antigenicity and Tropism through a Structure-Guided Approach. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Fromm, J.R.; Hileman, R.E.; Caldwell, E.E.; Weiler, J.M.; Linhardt, R.J. Differences in the interaction of heparin with arginine and lysine and the importance of these basic amino acids in the binding of heparin to acidic fibroblast growth factor. Arch. Biochem. Biophys. 1995, 323, 279–287. [Google Scholar] [CrossRef]
- Conrad, H.E. Heparin-Binding Proteins, 1st ed.; Academic Press: San Diego, CA, USA, 1998; p. 527. [Google Scholar]
- Capila, I.; Linhardt, R.J. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 2002, 41, 391–412. [Google Scholar] [CrossRef]
- Kern, A.; Schmidt, K.; Leder, C.; Muller, O.J.; Wobus, C.E.; Bettinger, K.; Von der Lieth, C.W.; King, J.A.; Kleinschmidt, J.A. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003, 77, 11072–11081. [Google Scholar] [CrossRef]
- Opie, S.R.; Warrington, K.H., Jr.; Agbandje-McKenna, M.; Zolotukhin, S.; Muzyczka, N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003, 77, 6995–7006. [Google Scholar] [CrossRef]
- Lerch, T.F.; Chapman, M.S. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B). Virology 2012, 423, 6–13. [Google Scholar] [CrossRef]
- Afione, S.; DiMattia, M.A.; Halder, S.; Di Pasquale, G.; Agbandje-McKenna, M.; Chiorini, J.A. Identification and mutagenesis of the adeno-associated virus 5 sialic acid binding region. J. Virol. 2015, 89, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Y.; Patel, A.; Ng, R.; Miller, E.B.; Halder, S.; McKenna, R.; Asokan, A.; Agbandje-McKenna, M. Characterization of the Adeno-associated virus 1 and 6 sialic acid binding site. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Spear, J.M.; Noble, A.J.; Sousa, D.R.; Meyer, N.L.; Davulcu, O.; Zhang, F.; Linhardt, R.J.; Stagg, S.M.; Chapman, M.S. The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide. Mol. Ther. Methods Clin. Dev. 2017, 5, 1–12. [Google Scholar] [CrossRef][Green Version]
- Handa, A.; Muramatsu, S.; Qiu, J.; Mizukami, H.; Brown, K.E. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 2000, 81, 2077–2084. [Google Scholar] [CrossRef] [PubMed]
- Levy, H.C.; Bowman, V.D.; Govindasamy, L.; McKenna, R.; Nash, K.; Warrington, K.; Chen, W.; Muzyczka, N.; Yan, X.; Baker, T.S.; et al. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J. Struct. Biol. 2009, 165, 146–156. [Google Scholar] [CrossRef]
- Connell, B.J.; Lortat-Jacob, H. Human Immunodeficiency Virus and Heparan Sulfate: From Attachment to Entry Inhibition. Front. Immunol. 2013, 4, 385. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Spilman, M.; Meyer, N.L.; Lerch, T.F.; Stagg, S.M.; Chapman, M.S. Electron microscopy analysis of a disaccharide analog complex reveals receptor interactions of adeno-associated virus. J. Struct. Biol. 2013, 184, 129–135. [Google Scholar] [CrossRef]
- Zhang, F.; Aguilera, J.; Beaudet, J.M.; Xie, Q.; Lerch, T.F.; Davulcu, O.; Colon, W.; Chapman, M.S.; Linhardt, R.J. Characterization of interactions between heparin/glycosaminoglycan and adeno-associated virus. Biochemistry 2013, 52, 6275–6285. [Google Scholar] [CrossRef] [PubMed]
- Hirst, G.K. The Agglutination of Red Cells by Allantoic Fluid of Chick Embryos Infected with Influenza Virus. Science 1941, 94, 22–23. [Google Scholar] [CrossRef] [PubMed]
- McClelland, L.; Hare, R. The adsorption of influenza virus by red cells and a new in vitro method of measuring antibodies for influenza virus. Can. Public Health J. 1941, 32, 530–538. [Google Scholar]
- Kuchipudi, S.V.; Nelli, R.K.; Gontu, A.; Satyakumar, R.; Surendran Nair, M.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bueno, A.; Rubio, M.P.; Bryant, N.; McKenna, R.; Agbandje-McKenna, M.; Almendral, J.M. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 2006, 80, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Palermo, L.M.; Hafenstein, S.L.; Parrish, C.R. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 2006, 80, 8482–8492. [Google Scholar] [CrossRef] [PubMed]
- Kaludov, N.; Brown, K.E.; Walters, R.W.; Zabner, J.; Chiorini, J.A. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 2001, 75, 6884–6893. [Google Scholar] [CrossRef]
- Wu, Z.; Miller, E.; Agbandje-McKenna, M.; Samulski, R.J. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J. Virol. 2006, 80, 9093–9103. [Google Scholar] [CrossRef]
- Shen, S.; Bryant, K.D.; Brown, S.M.; Randell, S.H.; Asokan, A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J. Biol. Chem. 2011, 286, 13532–13540. [Google Scholar] [CrossRef]
- Bell, C.L.; Gurda, B.L.; Van Vliet, K.; Agbandje-McKenna, M.; Wilson, J.M. Identification of the galactose binding domain of the AAV9 capsid. J. Virol. 2012, 86, 7326–7333. [Google Scholar] [CrossRef]
- Qing, K.; Mah, C.; Hansen, J.; Zhou, S.; Dwarki, V.; Srivastava, A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 1999, 5, 71–77. [Google Scholar] [CrossRef]
- Kashiwakura, Y.; Tamayose, K.; Iwabuchi, K.; Hirai, Y.; Shimada, T.; Matsumoto, K.; Nakamura, T.; Watanabe, M.; Oshimi, K.; Daida, H. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J. Virol. 2005, 79, 609–614. [Google Scholar] [CrossRef]
- Di Pasquale, G.; Davidson, B.L.; Stein, C.S.; Martins, I.; Scudiero, D.; Monks, A.; Chiorini, J.A. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 2003, 9, 1306–1312. [Google Scholar] [CrossRef]
- Weller, M.L.; Amornphimoltham, P.; Schmidt, M.; Wilson, P.A.; Gutkind, J.S.; Chiorini, J.A. Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat. Med. 2010, 16, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Summerford, C.; Bartlett, J.S.; Samulski, R.J. AlphaVbeta5 integrin: A co-receptor for adeno-associated virus type 2 infection. Nat. Med. 1999, 5, 78–82. [Google Scholar] [CrossRef]
- Qiu, J.; Brown, K.E. Integrin alphaVbeta5 is not involved in adeno-associated virus type 2 (AAV2) infection. Virology 1999, 264, 436–440. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asokan, A.; Hamra, J.B.; Govindasamy, L.; Agbandje-McKenna, M.; Samulski, R.J. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J. Virol. 2006, 80, 8961–8969. [Google Scholar] [CrossRef] [PubMed]
- Akache, B.; Grimm, D.; Pandey, K.; Yant, S.R.; Xu, H.; Kay, M.A. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 2006, 80, 9831–9836. [Google Scholar] [CrossRef]
- Mizukami, H.; Young, N.; Brown, K. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glyco protein. Virology 1996, 217, 124–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carette, J.E.; Guimaraes, C.P.; Wuethrich, I.; Blomen, V.A.; Varadarajan, M.; Sun, C.; Bell, G.; Yuan, B.; Muellner, M.K.; Nijman, S.M.; et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 2011, 29, 542–546. [Google Scholar] [CrossRef]
- Hamilton, B.A.; Li, X.; Pezzulo, A.A.; Abou Alaiwa, M.H.; Zabner, J. Polarized AAVR expression determines infectivity by AAV gene therapy vectors. Gene Ther. 2019, 26, 240–249. [Google Scholar] [CrossRef]
- Pillay, S.; Zou, W.; Cheng, F.; Puschnik, A.S.; Meyer, N.L.; Ganaie, S.S.; Deng, X.; Wosen, J.E.; Davulcu, O.; Yan, Z.; et al. AAV serotypes have distinctive interactions with domains of the cellular receptor AAVR. J. Virol. 2017, 91, e00391-17. [Google Scholar] [CrossRef]
- Bhella, D. The role of cellular adhesion molecules in virus attachment and entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140035. [Google Scholar] [CrossRef] [PubMed]
- Silveria, M.A.; Large, E.E.; Zane, G.M.; White, T.A.; Chapman, M.S. The Structure of an AAV5-AAVR Complex at 2.5 A Resolution: Implications for Cellular Entry and Immune Neutralization of AAV Gene Therapy Vectors. Viruses 2020, 12, 1326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, G.; Cao, L.; Sun, Z.; He, Y.; Cui, M.; Sun, Y.; Li, S.; Li, H.; Qin, L.; et al. Divergent engagements between adeno-associated viruses with their cellular receptor AAVR. Nat. Commun. 2019, 10, 3760. [Google Scholar] [CrossRef] [PubMed]
- Dudek, A.M.; Pillay, S.; Puschnik, A.S.; Nagamine, C.M.; Cheng, F.; Qiu, J.; Carette, J.E.; Vandenberghe, L.H. An Alternate Route for Adeno-associated Virus (AAV) Entry Independent of AAV Receptor. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Dudek, A.M.; Zabaleta, N.; Zinn, E.; Pillay, S.; Zengel, J.; Porter, C.; Franceschini, J.S.; Estelien, R.; Carette, J.E.; Zhou, G.L.; et al. GPR108 Is a Highly Conserved AAV Entry Factor. Mol. Ther. 2020, 28, 367–381. [Google Scholar] [CrossRef]
- Meisen, W.H.; Nejad, Z.B.; Hardy, M.; Zhao, H.; Oliverio, O.; Wang, S.; Hale, C.; Ollmann, M.M.; Collins, P.J. Pooled Screens Identify GPR108 and TM9SF2 as Host Cell Factors Critical for AAV Transduction. Mol. Ther. Methods Clin. Dev. 2020, 17, 601–611. [Google Scholar] [CrossRef]
- Micaroni, M.; Perinetti, G.; Berrie, C.P.; Mironov, A.A. The SPCA1 Ca2+ pump and intracellular membrane trafficking. Traffic 2010, 11, 1315–1333. [Google Scholar] [CrossRef]
- Madigan, V.J.; Tyson, T.O.; Yuziuk, J.A.; Pillai, M.; Moller-Tank, S.; Asokan, A. A CRISPR screen identifies the cell polarity determinant Crumbs 3 as an AAV restriction factor in hepatocytes. J. Virol. 2019. [Google Scholar] [CrossRef]
- Dorsch, S.; Liebisch, G.; Kaufmann, B.; von Landenberg, P.; Hoffmann, J.H.; Drobnik, W.; Modrow, S. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 2002, 76, 2014–2018. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, S.; Bottcher, B.; von der Lieth, C.W.; Bleker, S.; Kleinschmidt, J.A. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 2005, 79, 5296–5303. [Google Scholar] [CrossRef]
- Kurian, J.J.; Lakshmanan, R.; Chmely, W.M.; Hull, J.A.; Yu, J.C.; Bennett, A.; McKenna, R.; Agbandje-McKenna, M. Adeno-Associated Virus VP1u Exhibits Protease Activity. Viruses 2019, 11, 399. [Google Scholar] [CrossRef] [PubMed]
- Ertl, H.C.J. T Cell-Mediated Immune Responses to AAV and AAV Vectors. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Woodard, K.T.; Liang, K.J.; Bennett, W.C.; Samulski, R.J. Heparan Sulfate Binding Promotes Accumulation of Intravitreally Delivered Adeno-associated Viral Vectors at the Retina for Enhanced Transduction but Weakly Influences Tropism. J. Virol. 2016, 90, 9878–9888. [Google Scholar] [CrossRef] [PubMed]
- Stroh, L.J.; Stehle, T. Glycan Engagement by Viruses: Receptor Switches and Specificity. Annu. Rev. Virol. 2014, 1, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; de Vries, R.P.; Paulson, J.C. Virus recognition of glycan receptors. Curr. Opin. Virol. 2019, 34, 117–129. [Google Scholar] [CrossRef] [PubMed]

| Serotype | Clade | Resolution | Year | Tropism 1 | Tropism 2 | Tropism 3 | Reference | PDBid |
|---|---|---|---|---|---|---|---|---|
| AAV1 | A | X-ray 2.5 Å | 2010 | Muscle, CNS, heart | Skin, lung, kidney, cervix, bone | Kidney, skin | Ng et al. [21] | 3NG9 |
| AAV2 | B | X-ray 3.0 Å | 2002 | Liver, CNS, muscle | Skin, lung, kidney, cervix, liver, bone | Liver, kidney, cervix, retina, skin | Xie et al. [22] | 1LP3 |
| AAV3 | C | X-ray 2.6 Å | 2010 | Muscle, stem cells | Skin, lung, kidney, cervix, liver, bone | Skin | Lerch et al. [23] | 3KIC |
| AAV4 | Unique | X-ray 3.2 Å | 2006 | Eye, CNS | Bone | Not detected | Govindasamy et al. [24] | 2G8G |
| AAV5 | Unique | X-ray 3.5 Å | 2013 | CNS, lung, eye | Not detected | Not detected | Govindasamy et al. [25] | 3NTT |
| AAV6 | A | X-ray 3.0 Å | 2011 | Muscle, CNS, heart, lung | Skin, lung, kidney, cervix, bone | Skin | Xie et al. [26] | 4V86 |
| AAV7 | D | Cryo-EM 3.0 Å | 2021 | Muscle, CNS | Not detected | Not detected | Mietzsch et al. [20] | 7L5Q |
| AAV8 | E | X-ray 2.6 Å | 2007 | Liver, muscle, pancreas, CNS | Not detected | Not detected | Nam et al. [27] | 2QA0 |
| AAV9 | F | X-ray 2.8 Å | 2012 | Broad distribution | Not detected | Not detected | Dimattia et al. [28] | 3UX1 |
| AAVrh.39 (AAV10-like) | Cryo-EM 3.4 Å | 2020 | Muscle (AAV10) | Not tested | Not tested | Mietzsch et al. [29] | 6V1T | |
| AAV11 | Cryo-EM 2.9 Å | 2021 | Unknown | Not tested | Not tested | Mietzsch et al. [20] | 7L6F | |
| AAV12 | Cryo-EM 2.5 Å | 2021 | Nasal | Not tested | Not tested | Mietzsch et al. [20] | 7L6B | |
| AAV13 | Cryo-EM 3.0 Å | 2021 | Not shown | Not tested | Not tested | Mietzsch et al. [20] | 7L6I | |
| AAVDJ | Cryo-EM 4.5 Å | 2012 | Not shown | Not tested | Liver, kidney, cervix, retina, skin, lung | Lerch et al. [30] | 3J1Q | |
| AAVDJ | Cryo-EM 1.6 Å | 2020 | Not shown | Not tested | Liver, kidney, cervix, retina, skin, lung | Xie et al. [31] | 7KFR |
| Serotype | Resolution | Year | Reference |
|---|---|---|---|
| AAV2 | Cryo-EM 8.3 Å | 2009 | O’Donnell et al. [133] |
| AAV3 | X-ray 6.5 Å | 2012 | Lerch et al. [157] |
| AAV5 | X-ray 3.5 Å | 2015 | Afione et al. [158] |
| AAV1 | X-ray 3.0 Å | 2016 | Huang et al. [159] |
| AAVDJ | Cryo-EM 3.0 Å | 2017 | Xie et al. [160] |
| Serotype | Resolution | Year | Reference | PDBid |
|---|---|---|---|---|
| AAV1 | Cryo-EM 3.3 Å | 2019 | Zhang et al. [189] | 6JCQ |
| AAV2 | Cryo-EM 2.8 Å | 2019 | Zhang et al. [120] | 6IHB |
| AAV2 | Cryo-EM 2.4 Å | 2019 | Meyer et al. [118] | 6NZO |
| AAV5 | Cryo-EM 3.2 Å | 2019 | Zhang et al. [189] | 6JCS |
| AAV5 | Cryo-EM 2.5 Å | 2020 | Silveria et al. [188] | 7KPN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Large, E.E.; Silveria, M.A.; Zane, G.M.; Weerakoon, O.; Chapman, M.S. Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry. Viruses 2021, 13, 1336. https://doi.org/10.3390/v13071336
Large EE, Silveria MA, Zane GM, Weerakoon O, Chapman MS. Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry. Viruses. 2021; 13(7):1336. https://doi.org/10.3390/v13071336
Chicago/Turabian StyleLarge, Edward E., Mark A. Silveria, Grant M. Zane, Onellah Weerakoon, and Michael S. Chapman. 2021. "Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry" Viruses 13, no. 7: 1336. https://doi.org/10.3390/v13071336
APA StyleLarge, E. E., Silveria, M. A., Zane, G. M., Weerakoon, O., & Chapman, M. S. (2021). Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry. Viruses, 13(7), 1336. https://doi.org/10.3390/v13071336






