Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Isolation, Propagation, and Purification
2.2. Phage Host Specificity Determination
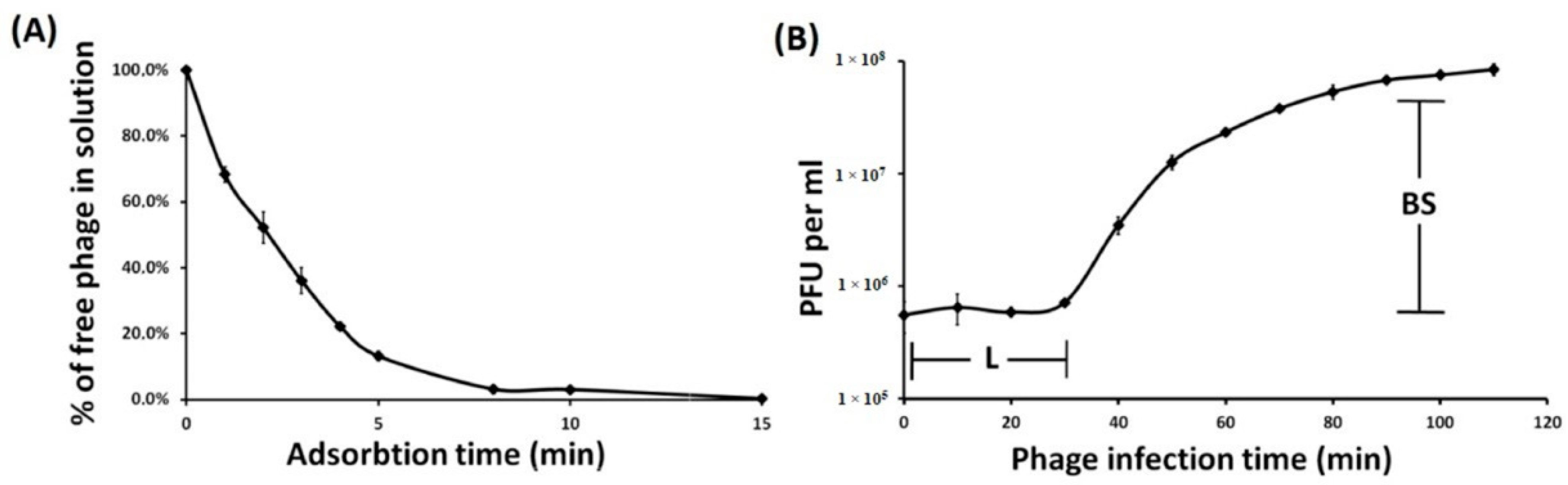
2.3. Phage Adsorption and One-Step Growth Experiments
2.4. Electron Microscopy
2.5. DNA Isolation and Sequencing
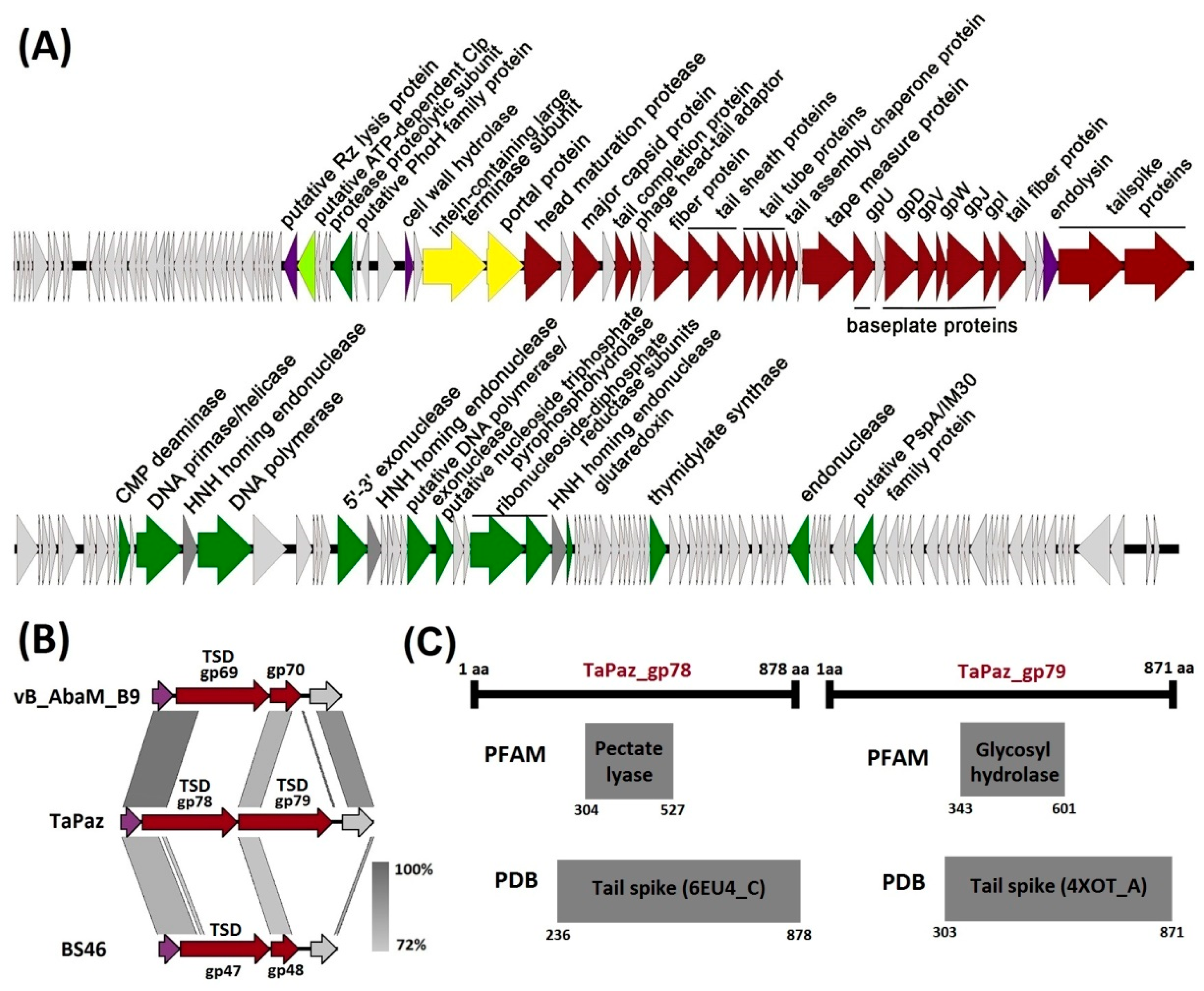
2.6. Phage Genome Analysis
2.7. Nucleotide Sequence Accession Number
2.8. Cloning, Expression, and Purification of the Recombinant Depolymerase
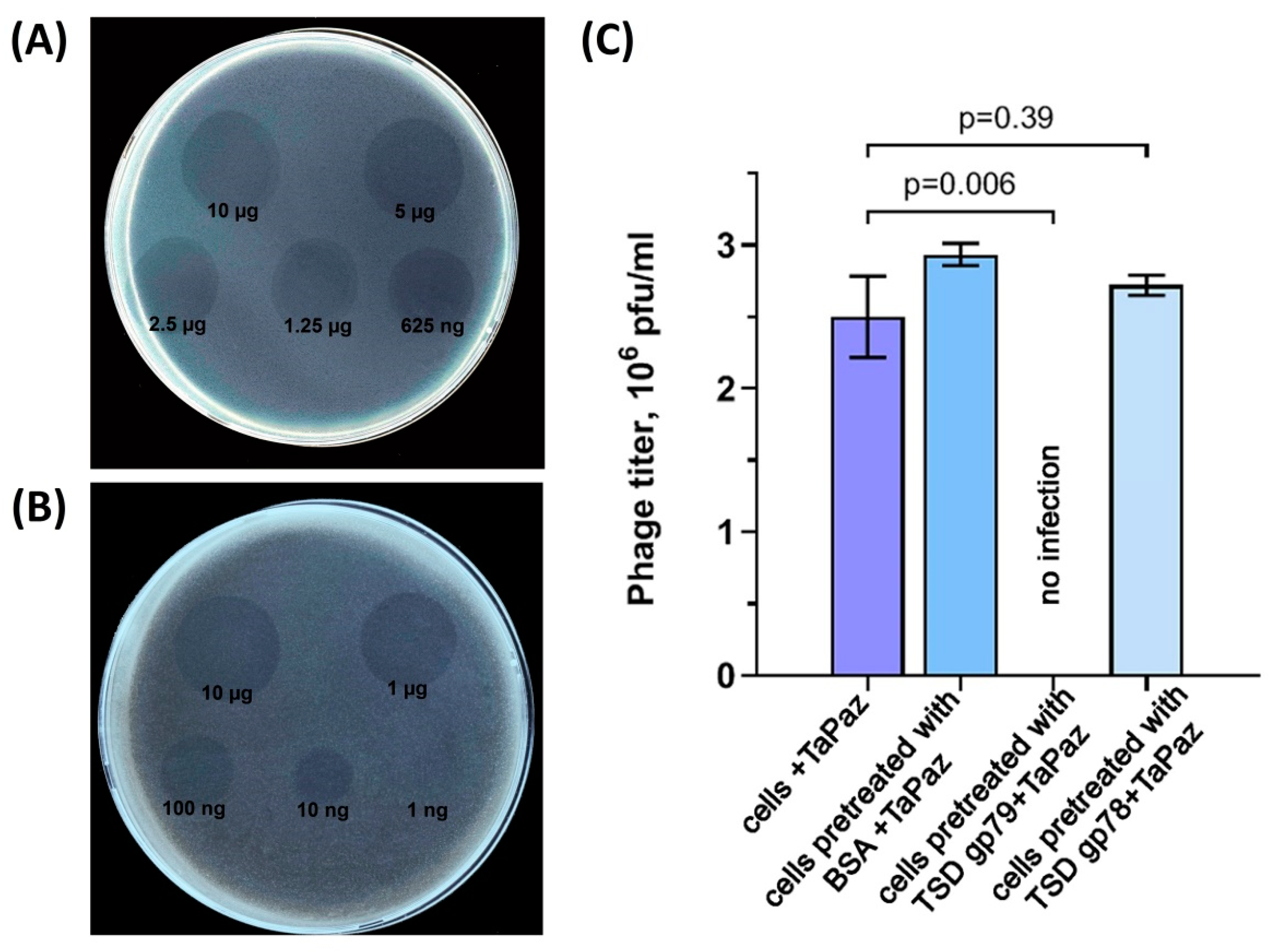
2.9. Phage Infection Inhibition Assay
2.10. Isolation and Purification of the CPS
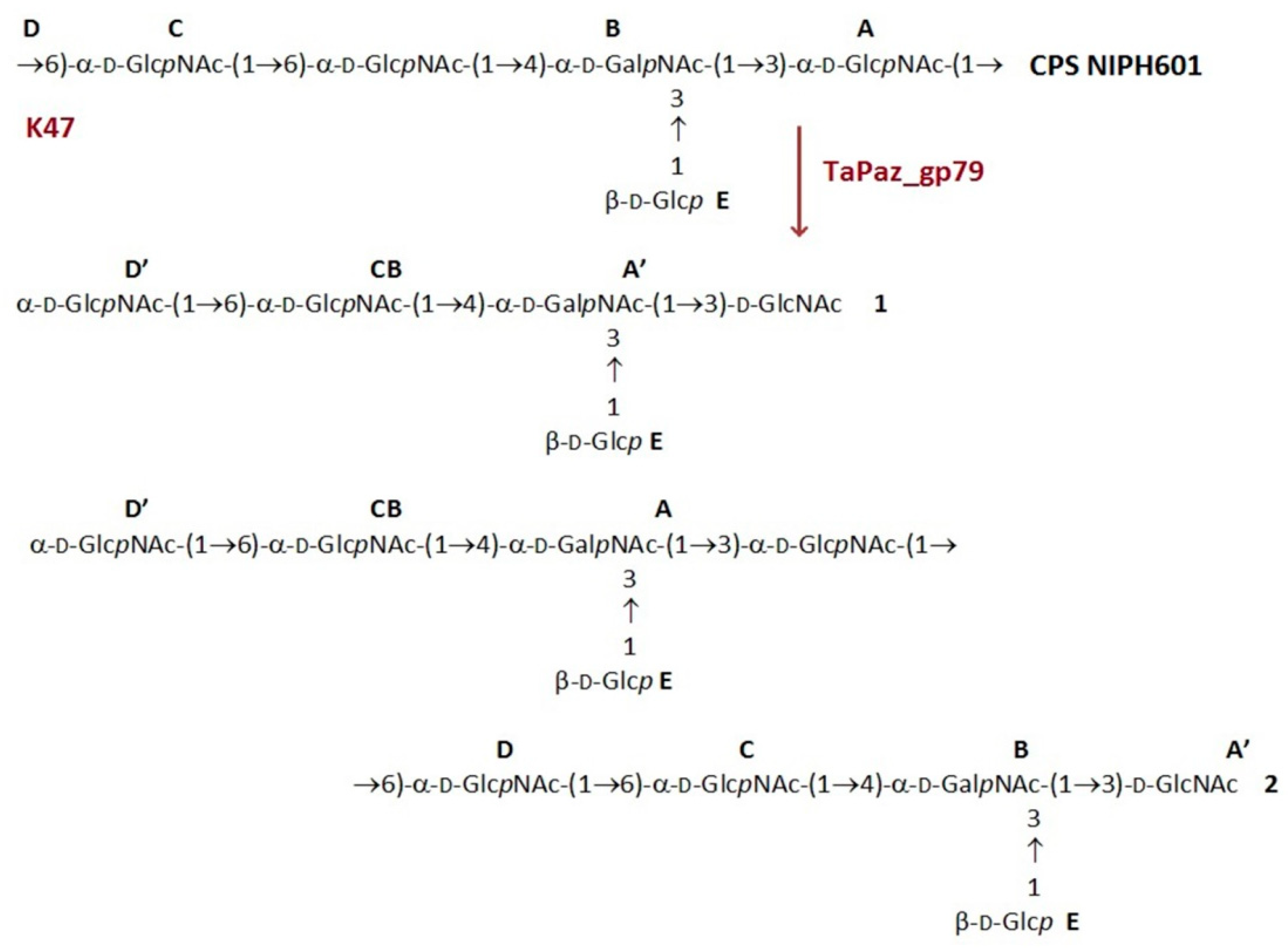
2.11. Cleavage of the CPS with TSD TaPaz_gp79
2.12. NMR Spectroscopy
2.13. Mass Spectrometry
3. Results and Discussion
3.1. Phage Isolation, Morphology, Host Specificity, and Infection Parameters
3.2. Phage Genome Organization and Comparison
3.3. Phage TaPaz Structural Tailspike Depolymerases
3.4. Cleavage of the A. baumannii NIPH 601 CPS by TSD TaPaz_gp79
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organisation. Prevention of Hospital-Acquired Infections; WHO Press: Geneva, Switzerland, 2002; Available online: https://www.who.int/csr/resources/publications/whocdscsreph200212.pdf (accessed on 23 April 2021).
- Vázquez-López, R.; Solano-Gálvez, S.G.; Juárez Vignon-Whaley, J.J.; Abello Vaamonde, J.A.; Padró Alonzo, L.A.; Rivera Reséndiz, A.; Muleiro Álvarez, M.; Vega López, E.N.; Franyuti-Kelly, G.; Álvarez-Hernández, D.A.; et al. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics; WHO Press: Geneva, Switzerland, 2017; Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 23 April 2021).
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.V.; Shneider, M.M.; Arbatsky, N.P.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Dmitrenok, A.S.; Chizhov, A.O.; Mikhailova, Y.V.; Shagin, D.A.; et al. Specific Interaction of Novel Friunavirus Phages Encoding Tailspike Depolymerases with Corresponding Acinetobacter baumannii Capsular Types. J. Virol. 2021, 95, e01714-20. [Google Scholar] [CrossRef]
- Lai, M.-J.; Chang, K.-C.; Huang, S.-W.; Luo, C.-H.; Chiou, P.-Y.; Wu, C.-C.; Lin, N.-T. The Tail Associated Protein of Acinetobacter baumannii Phage ΦAB6 Is the Host Specificity Determinant Possessing Exopolysaccharide Depolymerase Activity. PLoS ONE 2016, 11, e0153361. [Google Scholar] [CrossRef]
- Lee, I.-M.; Tu, I.-F.; Yang, F.-L.; Ko, T.-P.; Liao, J.-H.; Lin, N.-T.; Wu, C.-Y.; Ren, C.-T.; Wang, A.H.-J.; Chang, C.-M.; et al. Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage ΦAB6 tailspike protein. Sci. Rep. 2017, 7, 42711. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Costa, A.R.; Ferreira, A.; Konstantinides, N.; Santos, S.B.; Boon, M.; Noben, J.-P.; Lavigne, R.; Azeredo, J. Functional Analysis and Antivirulence Properties of a New Depolymerase from a Myovirus That Infects Acinetobacter baumannii Capsule K45. J. Virol. 2018, 93, e01163-18. [Google Scholar] [CrossRef]
- Oliveira, H.; Costa, A.R.; Konstantinides, N.; Ferreira, A.; Akturk, E.; Sillankorva, S.; Nemec, A.; Shneider, M.; Dötsch, A.; Azeredo, J. Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ. Microbiol. 2017, 19, 5060–5077. [Google Scholar] [CrossRef]
- Popova, A.V.; Shneider, M.M.; Myakinina, V.P.; Bannov, V.A.; Edelstein, M.V.; Rubalskii, E.O.; Aleshkin, A.V.; Fursova, N.K.; Volozhantsev, N.V. Characterization of myophage AM24 infecting Acinetobacter baumannii of the K9 capsular type. Arch. Virol. 2019, 164, 1493–1497. [Google Scholar] [CrossRef]
- Popova, A.V.; Shneider, M.M.; Mikhailova, Y.V.; Shelenkov, A.A.; Shagin, D.A.; Edelstein, M.V.; Kozlov, R.S. Complete Genome Sequence of Acinetobacter baumannii Phage BS46. Microbiol. Resour. Announc. 2020, 9, e00398-20. [Google Scholar] [CrossRef]
- Popova, A.; Lavysh, D.; Klimuk, E.; Edelstein, M.; Bogun, A.; Shneider, M.; Goncharov, A.; Leonov, S.; Severinov, K. Novel Fri1-like Viruses Infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12—Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses 2017, 9, 188. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Shneider, M.M.; Popova, A.V.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Chizhov, A.O. Mechanisms of Acinetobacter baumannii capsular polysaccharide cleavage by phage depolymerases. Biochemistry 2020, 85, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D. Bacteriophages; Interscience Publishers, Inc.: New York, NY, USA, 1959; OCLC 326505. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; ISBN 0-87969-309-6. [Google Scholar]
- Brenner, S.; Horne, R.W. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 1959, 34, 103–110. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009, 37, D205–D210. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, msab120. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.S.; Taylor, N.M.I.; Buth, S.A.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Kenyon, J.J.; Kasimova, A.A.; Shneider, M.M.; Shashkov, A.S.; Arbatsky, N.P.; Popova, A.V.; Miroshnikov, K.A.; Hall, R.M.; Knirel, Y.A. The KL24 gene cluster and a genomic island encoding a Wzy polymerase contribute genes needed for synthesis of the K24 capsular polysaccharide by the multiply antibiotic resistant Acinetobacter baumannii isolate RCH51. Microbiology 2017, 163, 355–363. [Google Scholar] [CrossRef]
- Tsedilin, A.M.; Fakhrutdinov, A.N.; Eremin, D.B.; Zalesskiy, S.S.; Chizhov, A.O.; Kolotyrkina, N.G.; Ananikov, V.P. How sensitive and accurate are routine NMR and MS measurements? Mendeleev Commun. 2015, 25, 454–456. [Google Scholar] [CrossRef]
- Duus, J.; Gotfredsen, C.H.; Bock, K. Carbohydrate Structural Determination by NMR Spectroscopy: Modern Methods and Limitations. Chem. Rev. 2000, 100, 4589–4614. [Google Scholar] [CrossRef]
- Farrugia, D.N.; Elbourne, L.D.H.; Hassan, K.A.; Eijkelkamp, B.A.; Tetu, S.G.; Brown, M.H.; Shah, B.S.; Peleg, A.; Mabbutt, B.C.; Paulsen, I.T. The Complete Genome and Phenome of a Community-Acquired Acinetobacter baumannii. PLoS ONE 2013, 8, e58628. [Google Scholar] [CrossRef]
- Novikova, O.; Jayachandran, P.; Kelley, D.S.; Morton, Z.; Merwin, S.; Topilina, N.I.; Belfort, M. Intein Clustering Suggests Functional Importance in Different Domains of Life. Mol. Biol. Evol. 2016, 33, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Karthikeyan, T.; Wadsworth, C.; Lewis, J.A.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Pannunzio, N.R.; et al. Origins of Highly Mosaic Mycobacteriophage Genomes. Cell 2003, 113, 171–182. [Google Scholar] [CrossRef]
- Dassa, B.; London, N.; Stoddard, B.L.; Schueler-Furman, O.; Pietrokovski, S. Fractured genes: A novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Res. 2009, 37, 2560–2573. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Kenyon, J.J.; Arbatsky, N.P.; Shneider, M.M.; Popova, A.V.; Miroshnikov, K.A.; Hall, R.M.; Knirel, Y.A. Related structures of neutral capsular polysaccharides of Acinetobacter baumannii isolates that carry related capsule gene clusters KL43, KL47, and KL88. Carbohydr. Res. 2016, 435, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Lipkind, G.M.; Shashkov, A.S.; Knirel, Y.A.; Vinogradov, E.V.; Kochetkov, N.K. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr. Res. 1988, 175, 59–75. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shchurova, A.S.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Skryabin, Y.P.; Mikhaylova, Y.V.; Sokolova, O.S.; Shelenkov, A.A.; Miroshnikov, K.A.; et al. Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy. Viruses 2021, 13, 978. https://doi.org/10.3390/v13060978
Shchurova AS, Shneider MM, Arbatsky NP, Shashkov AS, Chizhov AO, Skryabin YP, Mikhaylova YV, Sokolova OS, Shelenkov AA, Miroshnikov KA, et al. Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy. Viruses. 2021; 13(6):978. https://doi.org/10.3390/v13060978
Chicago/Turabian StyleShchurova, Anastasia S., Mikhail M. Shneider, Nikolay P. Arbatsky, Alexander S. Shashkov, Alexander O. Chizhov, Yuriy P. Skryabin, Yulia V. Mikhaylova, Olga S. Sokolova, Andrey A. Shelenkov, Konstantin A. Miroshnikov, and et al. 2021. "Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy" Viruses 13, no. 6: 978. https://doi.org/10.3390/v13060978
APA StyleShchurova, A. S., Shneider, M. M., Arbatsky, N. P., Shashkov, A. S., Chizhov, A. O., Skryabin, Y. P., Mikhaylova, Y. V., Sokolova, O. S., Shelenkov, A. A., Miroshnikov, K. A., Knirel, Y. A., & Popova, A. V. (2021). Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy. Viruses, 13(6), 978. https://doi.org/10.3390/v13060978







