Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses
Abstract
1. Introduction
2. Human RNA Enteric Viruses
2.1. Rotavirus
2.2. Norovirus
2.3. Astrovirus
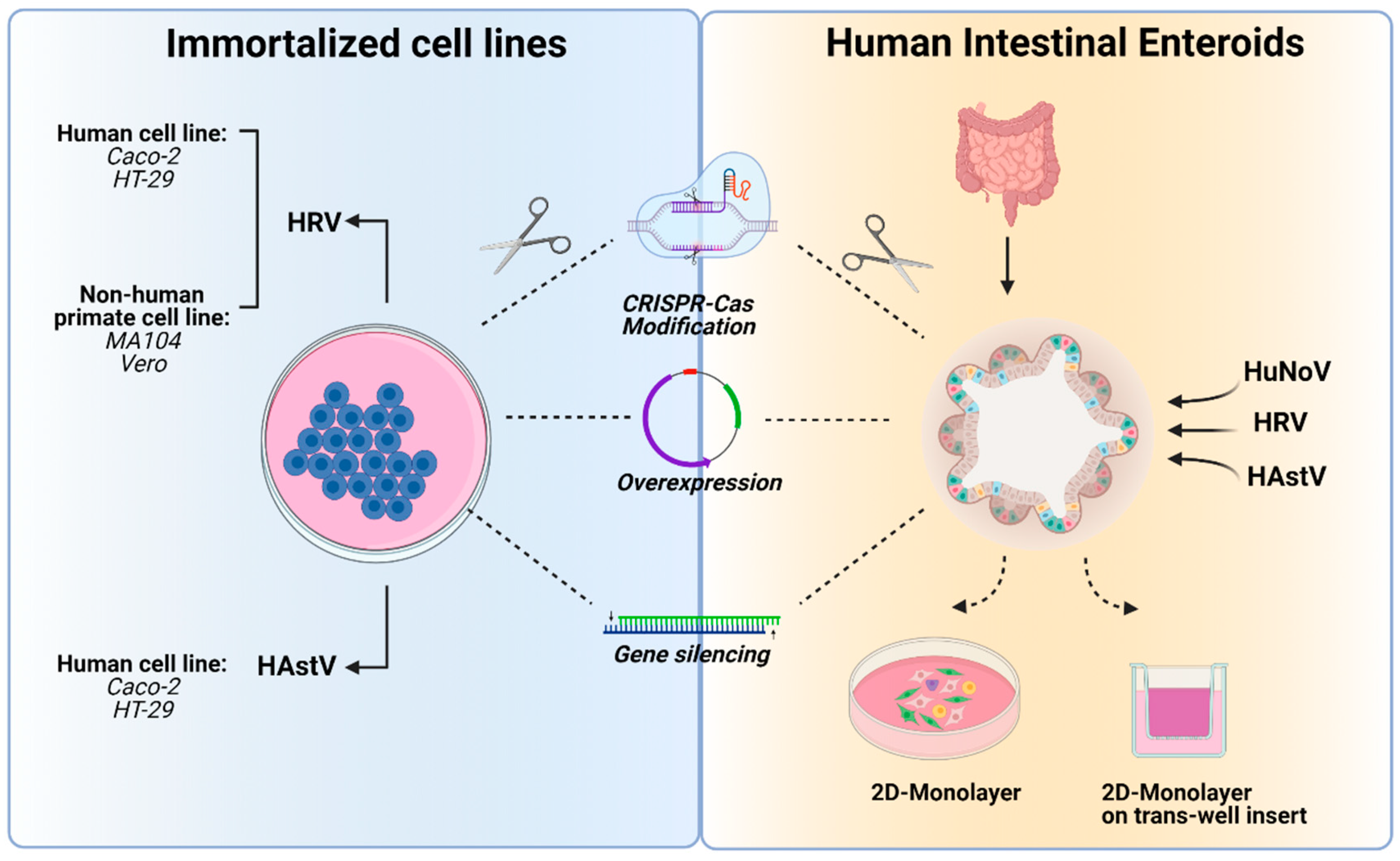
3. In Vitro Tools to Study Human Enteric RNA Viruses
3.1. Immortalized Cell Lines
3.1.1. Adenocarcinoma Cell Lines
3.1.2. B Cell Lines
3.1.3. Non-Human Primate Cell Lines
Advantages and Disadvantages of Using Immortalized Cell Lines
3.2. Primary Cells
Advantages and Disadvantages of Primary Cells
3.3. Intestinal Enteroids
Advantages and Disadvantages of HIEs
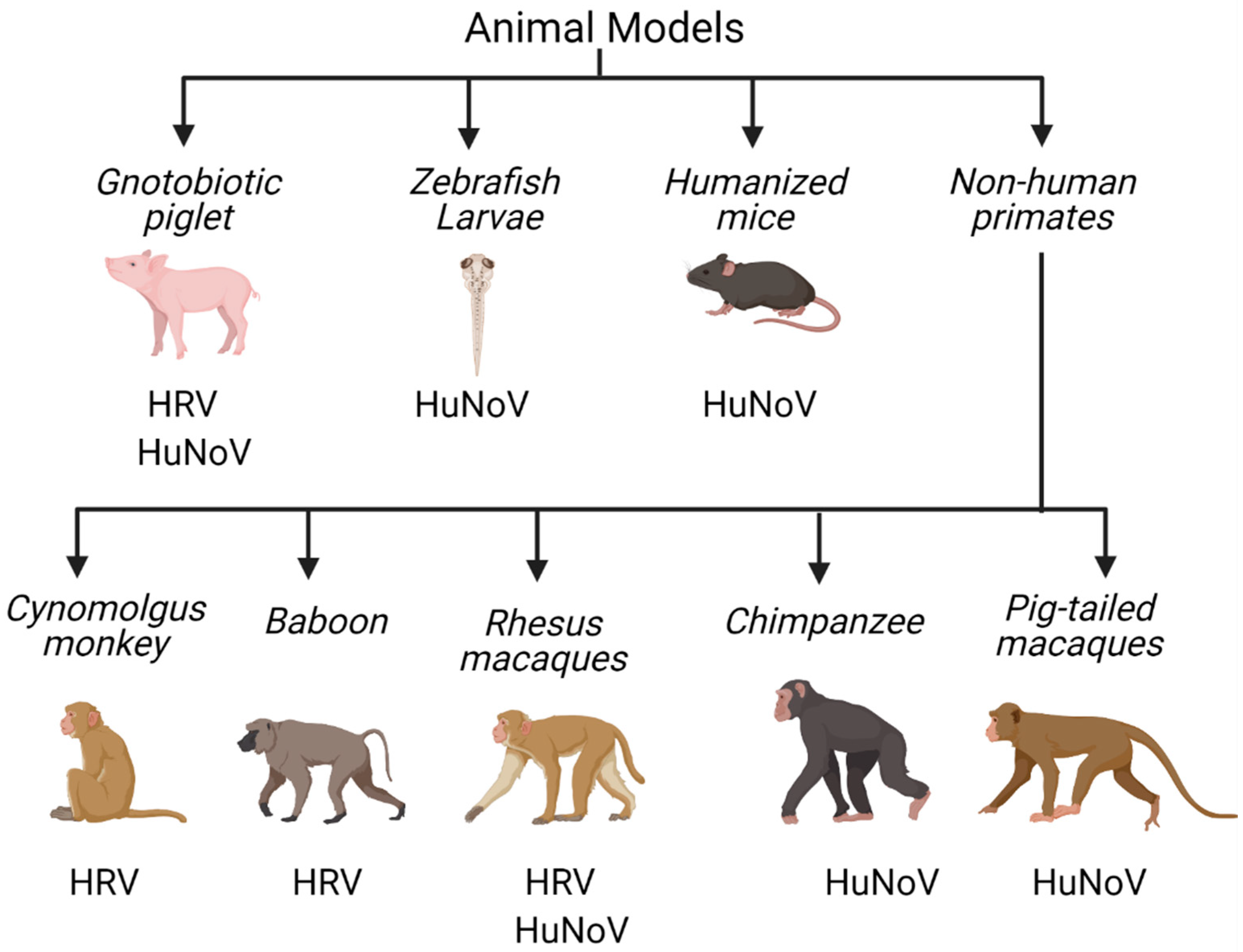
4. In Vivo Tools to Study Human Enteric RNA Viruses
4.1. Non-Human Primates
4.2. Gnotobiotic Pigs
4.3. Mouse Models
4.4. Zebrafish
4.5. Turkeys
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Bishop, R.F.; Kirkwood, C. Enteric viruses. Ref. Modul. Biomed. Sci. 2014. [Google Scholar] [CrossRef]
- Chiejina, M.; Samant, H. Viral Diarrhea. StatPearls [Internet]. 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK470525/ (accessed on 21 January 2021).
- Stuempfig, N.D.; Seroy, J. Viral gastroenteritis. StatPearls [Internet]. 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK518995/ (accessed on 21 January 2021).
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- McAtee, C.L.; Webman, R.; Gilman, R.H.; Mejia, C.; Bern, C.; Apaza, S.; Espetia, S.; Pajuelo, M.; Saito, M.; Challappa, R. Burden of norovirus and rotavirus in children after rotavirus vaccine introduction, Cochabamba, Bolivia. Am. J. Trop. Med. Hyg. 2016, 94, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.M.; Widdowson, M.-A.; Glass, R.I.; Akazawa, K.; Vinjé, J.; Parashar, U.D. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224. [Google Scholar] [CrossRef]
- Bosch, A.; Pintó, R.M.; Guix, S. Human astroviruses. Clin. Microbiol. Rev. 2014, 27, 1048–1074. [Google Scholar] [CrossRef]
- Ham, H.; Oh, S.; Jang, J.; Jo, S.; Choi, S.; Pak, S. Prevalence of human astrovirus in patients with acute gastroenteritis. Ann. Lab. Med. 2014, 34, 145. [Google Scholar] [CrossRef]
- Fischer, T.K.; Rasmussen, L.D.; Fonager, J. Taking gastro-surveillance into the 21st century. J. Clin. Virol. 2019, 117, 43–48. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Martínez-Puchol, S.; Silva-Sales, M.; Cornejo, T.; Bartolome, R.; Bofill-Mas, S.; Girones, R. Unveiling Viruses Associated with Gastroenteritis Using a Metagenomics Approach. Viruses 2020, 12, 1432. [Google Scholar] [CrossRef]
- Jourdan, N.; Laffitte, J.C.; Forestier, F.; Servin, A.; Quero, A. Infection of cultured human intestinal cells by monkey RRV and human Wa rotavirus as a function of intestinal epithelial cell differentiation. Res. Virol. 1995, 146, 325–331. [Google Scholar] [CrossRef]
- Casola, A.; Estes, M.K.; Crawford, S.E.; Ogra, P.L.; Ernst, P.B.; Garofalo, R.P.; Crowe, S.E. Rotavirus infection of cultured intestinal epithelial cells induces secretion of CXC and CC chemokines. Gastroenterology 1998, 114, 947–955. [Google Scholar] [CrossRef]
- Ding, S.; Diep, J.; Feng, N.; Ren, L.; Li, B.; Ooi, Y.S.; Wang, X.; Brulois, K.F.; Yasukawa, L.L.; Li, X. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Banks, L.D.; Engevik, K.A.; Chang-Graham, A.L.; Perry, J.L.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; Hyser, J.M. Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence. Gut Microbes 2020, 11, 1324–1347. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Patton, J.T.; McDonald, S.M. Culturing, storage, and quantification of rotaviruses. Curr. Protoc. Microbiol. 2009, 15, 15C.3.1–15C.3.24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tan, M.; Zhong, W.; Xia, M.; Huang, P.; Jiang, X. Human intestinal organoids express histo-blood group antigens, bind norovirus VLPs, and support limited norovirus replication. Sci. Rep. 2017, 7, 1–2. [Google Scholar] [CrossRef]
- Orr-Burks, N.; Murray, J.; Wu, W.; Kirkwood, C.D.; Todd, K.V.; Jones, L.; Bakre, A.; Wang, H.; Jiang, B.; Tripp, R.A. Gene-edited vero cells as rotavirus vaccine substrates. Vaccine X 2019, 3, 100045. [Google Scholar] [CrossRef]
- Finkbeiner, S.R.; Zeng, X.-L.; Utama, B.; Atmar, R.L.; Shroyer, N.F.; Estes, M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 2012, 3. [Google Scholar] [CrossRef]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.-L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R. Human intestinal enteroids: A new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef]
- Zou, W.Y.; Blutt, S.E.; Crawford, S.E.; Ettayebi, K.; Zeng, X.-L.; Saxena, K.; Ramani, S.; Karandikar, U.C.; Zachos, N.C.; Estes, M.K. Human intestinal enteroids: New models to study gastrointestinal virus infections. In Organoids; Springer: Berlin/Heidelberg, Germany, 2017; pp. 229–247. [Google Scholar]
- Chang-Graham, A.L.; Danhof, H.A.; Engevik, M.A.; Tomaro-Duchesneau, C.; Karandikar, U.C.; Estes, M.K.; Versalovic, J.; Britton, R.A.; Hyser, J.M. Human intestinal enteroids with inducible neurogenin-3 expression as a novel model of gut hormone secretion. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 209–229. [Google Scholar] [CrossRef]
- Wen, X.; Cao, D.; Jones, R.W.; Hoshino, Y.; Yuan, L. Tandem truncated rotavirus VP8* subunit protein with T cell epitope as non-replicating parenteral vaccine is highly immunogenic. Hum. Vaccines Immunother. 2015, 11, 2483–2489. [Google Scholar] [CrossRef]
- Wen, X.; Wen, K.; Cao, D.; Li, G.; Jones, R.W.; Li, J.; Szu, S.; Hoshino, Y.; Yuan, L. Inclusion of a universal tetanus toxoid CD4+ T cell epitope P2 significantly enhanced the immunogenicity of recombinant rotavirus ΔVP8* subunit parenteral vaccines. Vaccine 2014, 32, 4420–4427. [Google Scholar] [CrossRef]
- Torres, A.; Ji-Huang, L. Diarrheal response of gnotobiotic pigs after fetal infection and neonatal challenge with homologous and heterologous human rotavirus strains. J. Virol. 1986, 60, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ward, L.A.; Rosen, B.I.; To, T.L.; Saif, L.J. Systematic and intestinal antibody-secreting cell responses and correlates of protective immunity to human rotavirus in a gnotobiotic pig model of disease. J. Virol. 1996, 70, 3075–3083. [Google Scholar] [CrossRef] [PubMed]
- Bentes, G.A.; Guimarães, J.R.; Volotão, E.D.M.; Fialho, A.M.; Hooper, C.; Ganime, A.C.; Gardinali, N.R.; Lanzarini, N.M.; Da Silva, A.D.S.; Pitcovski, J. Cynomolgus monkeys (Macaca fascicularis) as an experimental infection model for human group a rotavirus. Viruses 2018, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- YAP, K.L.; ASRI, A. Experimental group A rotaviral infection in cynomolgus monkeys raised on formula diet. Microbiol. Immunol. 1990, 34, 153–162. [Google Scholar]
- Majer, M.; Behrens, F.; Weinmann, E.; Mauler, R.; Maass, G.; Baumeister, H.; Luthardt, T. Diarrhea in newborn cynomolgus monkeys infected with human rotavirus. Infection 1978, 6, 71–72. [Google Scholar] [CrossRef]
- Jones, M.K.; Grau, K.R.; Costantini, V.; Kolawole, A.O.; De Graaf, M.; Freiden, P.; Graves, C.L.; Koopmans, M.; Wallet, S.M.; Tibbetts, S.A. Human norovirus culture in B cells. Nat. Protoc. 2015, 10, 1939. [Google Scholar] [CrossRef]
- Todd, K.V.; Tripp, R.A. Vero cells as a mammalian cell substrate for human norovirus. Viruses 2020, 12, 439. [Google Scholar] [CrossRef]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.-L.; Qu, L. Replication of human noroviruses in stem cell–derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef]
- Lin, S.-C.; Qu, L.; Ettayebi, K.; Crawford, S.E.; Blutt, S.E.; Robertson, M.J.; Zeng, X.-L.; Tenge, V.R.; Ayyar, B.V.; Karandikar, U.C. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc. Natl. Acad. Sci. USA 2020, 117, 23782–23793. [Google Scholar] [CrossRef]
- Haga, K.; Ettayebi, K.; Tenge, V.R.; Karandikar, U.C.; Lewis, M.A.; Lin, S.-C.; Neill, F.H.; Ayyar, B.V.; Zeng, X.-L.; Larson, G. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. MBio 2020, 11. [Google Scholar] [CrossRef]
- Van Dycke, J.; Ny, A.; Conceição-Neto, N.; Maes, J.; Hosmillo, M.; Cuvry, A.; Goodfellow, I.; Nogueira, T.C.; Verbeken, E.; Matthijnssens, J. A robust human norovirus replication model in zebrafish larvae. PLoS Pathog. 2019, 15, e1008009. [Google Scholar] [CrossRef] [PubMed]
- Taube, S.; Kolawole, A.O.; Höhne, M.; Wilkinson, J.E.; Handley, S.A.; Perry, J.W.; Thackray, L.B.; Akkina, R.; Wobus, C.E. A mouse model for human norovirus. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; Meulia, T.; Grimes, S.; Han, M.G.; Saif, L.J. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 2006, 80, 10372–10381. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M. Median infectious dose of human norovirus GII. 4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J. Gen. Virol. 2013, 94 Pt 9, 2005. [Google Scholar] [CrossRef]
- Lei, S.; Ryu, J.; Wen, K.; Twitchell, E.; Bui, T.; Ramesh, A.; Weiss, M.; Li, G.; Samuel, H.; Clark-Deener, S. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci. Rep. 2016, 6, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Subekti, D.; Tjaniadi, P.; Lesmana, M.; McArdle, J.; Iskandriati, D.; Budiarsa, I.; Walujo, P.; Suparto, I.; Winoto, I.; Campbell, J. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 2002, 66, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Bok, K.; Parra, G.I.; Mitra, T.; Abente, E.; Shaver, C.K.; Boon, D.; Engle, R.; Yu, C.; Kapikian, A.Z.; Sosnovtsev, S.V. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. USA 2011, 108, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Brinker, J.; Blacklow, N.; Herrmann, J. Human astrovirus isolation and propagation in multiple cell lines. Arch. Virol. 2000, 145, 1847–1856. [Google Scholar] [CrossRef]
- Willcocks, M.; Carter, M.; Laidler, F.; Madeley, C. Growth and characterisation of human faecal astrovirus in a continuous cell line. Arch. Virol. 1990, 113, 73–81. [Google Scholar] [CrossRef]
- Kolawole, A.O.; Mirabelli, C.; Hill, D.R.; Svoboda, S.A.; Janowski, A.B.; Passalacqua, K.D.; Rodriguez, B.N.; Dame, M.K.; Freiden, P.; Berger, R.P. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog. 2019, 15, e1008057. [Google Scholar] [CrossRef]
- Méndez, E.; Muñoz-Yañez, C.; Sánchez-San Martín, C.; Aguirre-Crespo, G.; del Rocio Baños-Lara, M.; Gutierrez, M.; Espinosa, R.; Acevedo, Y.; Arias, C.F.; López, S. Characterization of human astrovirus cell entry. J. Virol. 2014, 88, 2452–2460. [Google Scholar] [CrossRef] [PubMed]
- Triana, S.; Stanifer, M.L.; Shahraz, M.; Mukenhirn, M.; Kee, C.; Rueda, D.O.; Paulsen, M.; Benes, V.; Boulant, S.; Alexandrov, T. Single-cell transcriptomics reveals immune response of intestinal cell types to viral infection. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ingle, H.; Hassan, E.; Gawron, J.; Mihi, B.; Li, Y.; Kennedy, E.A.; Kalugotla, G.; Makimaa, H.; Lee, S.; Desai, P. Murine astrovirus tropism for goblet cells and enterocytes facilitates an IFN-λ response in vivo and in enteroid cultures. Mucosal Immunol. 2021, 14, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Owusu, I.A.; Mirabelli, C.; Kolawole, A.O. Intestinal Enteroid Culture for Human Astroviruses. Bio Protoc. 2020, 10, e3687. [Google Scholar] [CrossRef]
- Flewett, T.; Bryden, A.; Davies, H. Virus particles in gastroenteritis. Lancet 1973, 2, 1497. [Google Scholar] [CrossRef]
- Bishop, R. Discovery of rotavirus: Implications for child health. J. Gastroenterol. Hepatol. 2009, 24, S81–S85. [Google Scholar] [CrossRef]
- Lundgren, O.; Svensson, L. Pathogenesis of rotavirus diarrhea. Microbes Infect. 2001, 3, 1145–1156. [Google Scholar] [CrossRef]
- Morris, A.P.; Estes, M.K. Microbes and microbial toxins: Paradigms for microbial-mucosal interactions VIII. Pathological consequences of rotavirus infection and its enterotoxin. Am. J. Physiol. 2001, 281, G303–G310. [Google Scholar]
- Chen, C.-C.; Huang, J.-L.; Chang, C.-J.; Kong, M.-S. Fecal calprotectin as a correlative marker in clinical severity of infectious diarrhea and usefulness in evaluating bacterial or viral pathogens in children. J. Pediatric Gastroenterol. Nutr. 2012, 55, 541–547. [Google Scholar] [CrossRef]
- Wiegering, V.; Kaiser, J.; Tappe, D.; Weißbrich, B.; Morbach, H.; Girschick, H.J. Gastroenteritis in childhood: A retrospective study of 650 hospitalized pediatric patients. Int. J. Infect. Dis. 2011, 15, e401–e407. [Google Scholar] [CrossRef]
- Gurwith, M.; Wenman, W.; Hinde, D.; Feltham, S.; Greenberg, H. A prospective study of rotavirus infection in infants and young children. J. Infect. Dis. 1981, 144, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Glass, R.I.; Kilgore, P.E.; Holman, R.C.; Jin, S.; Smith, J.C.; Woods, P.A.; Clarke, M.J.; Ho, M.S.; Gentsch, J.R. The epidemiology of rotavirus diarrhea in the United States: Surveillance and estimates of disease burden. J. Infect. Dis. 1996, 174 (Suppl. 1), S5–S11. [Google Scholar] [CrossRef]
- Desselberger, U.; Huppertz, H.-I. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J. Infect. Dis. 2011, 203, 188–195. [Google Scholar] [CrossRef]
- Luchs, A.; Timenetsky, M.d.C.S.T. Group A rotavirus gastroenteritis: Post-vaccine era, genotypes and zoonotic transmission. Einstein 2016, 14, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Arias, C. Early steps in rotavirus cell entry. Reoviruses Entry Assem. Morphog. 2006, 39–66. [Google Scholar] [CrossRef]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.-L.; Mutsafi, Y.; De Jésus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 2018, 24, 208–220.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiang, W.; Li, C.; Xu, J.; Zhou, D.; Shang, S. Molecular epidemiology of rotavirus A and adenovirus among children with acute diarrhea in Hangzhou, China. Gut Pathog. 2020, 12, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Aguirre, I.; Steyer, A.; Boben, J.; Gruden, K.; Poljšak-Prijatelj, M.; Ravnikar, M. Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J. Clin. Microbiol. 2008, 46, 2547–2554. [Google Scholar] [CrossRef]
- Vesikari, T.; Matson, D.O.; Dennehy, P.; Van Damme, P.; Santosham, M.; Rodriguez, Z.; Dallas, M.J.; Heyse, J.F.; Goveia, M.G.; Black, S.B. Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 2006, 354, 23–33. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Pérez-Schael, I.; Velázquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef]
- World Health Organization. Rotavirus vaccines = Vaccins antirotavirus. Wkly. Epidemiol. Rec. Relev. Épidémiol. Hebd. 2007, 82, 285–295. [Google Scholar]
- Linhares, A.C.; Velázquez, F.R.; Pérez-Schael, I.; Sáez-Llorens, X.; Abate, H.; Espinoza, F.; López, P.; Macías-Parra, M.; Ortega-Barría, E.; Rivera-Medina, D.M. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: A randomised, double-blind, placebo-controlled phase III study. Lancet 2008, 371, 1181–1189. [Google Scholar] [CrossRef]
- Vesikari, T.; Karvonen, A.; Prymula, R.; Schuster, V.; Tejedor, J.; Cohen, R.; Meurice, F.; Han, H.; Damaso, S.; Bouckenooghe, A. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: Randomised, double-blind controlled study. Lancet 2007, 370, 1757–1763. [Google Scholar] [CrossRef]
- Madhi, S.A.; Cunliffe, N.A.; Steele, D.; Witte, D.; Kirsten, M.; Louw, C.; Ngwira, B.; Victor, J.C.; Gillard, P.H.; Cheuvart, B.B. Research Article (New England Journal of Medicine) Effect of human rotavirus vaccine on severe diarrhea in African infants. Malawi Med. J. 2016, 28, 108–114. [Google Scholar] [PubMed]
- Kim, A.H.; Hogarty, M.P.; Harris, V.C.; Baldridge, M.T. The Complex Interactions Between Rotavirus and the Gut Microbiota. Front. Cell. Infect. Microbiol. 2021, 10, 820. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.C.; Armah, G.; Fuentes, S.; Korpela, K.E.; Parashar, U.; Victor, J.C.; Tate, J.; de Weerth, C.; Giaquinto, C.; Wiersinga, W.J. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J. Infect. Dis. 2017, 215, 34–41. [Google Scholar] [CrossRef]
- Dolin, R.; Blacklow, N.R.; DuPont, H.; Buscho, R.F.; Wyatt, R.G.; Kasel, J.A.; Hornick, R.; Chanock, R.M. Biological properties of Norwalk agent of acute infectious nonbacterial gastroenteritis. Proc. Soc. Exp. Biol. Med. 1972, 140, 578–583. [Google Scholar] [CrossRef]
- Kapikian, A.Z.; Wyatt, R.G.; Dolin, R.; Thornhill, T.S.; Kalica, A.R.; Chanock, R.M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 1972, 10, 1075–1081. [Google Scholar] [CrossRef]
- Atmar, R.L.; Opekun, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553. [Google Scholar] [CrossRef]
- Kirby, A.; Shi, J.; Montes, J.; Lichtenstein, M.; Moe, C. Disease course and viral shedding in experimental Norwalk virus and Snow Mountain virus infection. J. Med. Virol. 2014, 86, 2055–2064. [Google Scholar] [CrossRef]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.-W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.-P.; Widdowson, M.-A.; Glass, R.I.; Vinjé, J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J. Clin. Microbiol. 2010, 48, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Vinjé, J. Advances in laboratory methods for detection and typing of norovirus. J. Clin. Microbiol. 2015, 53, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinjé, J. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Karandikar, U.C.; Crawford, S.E.; Ajami, N.J.; Murakami, K.; Kou, B.; Ettayebi, K.; Papanicolaou, G.A.; Jongwutiwes, U.; Perales, M.-A.; Shia, J. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol. 2016, 97, 2291. [Google Scholar] [CrossRef] [PubMed]
- Green, K.Y.; Kaufman, S.S.; Nagata, B.M.; Chaimongkol, N.; Kim, D.Y.; Levenson, E.A.; Tin, C.M.; Yardley, A.B.; Johnson, J.A.; Barletta, A.B.F. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Zhang, H.; Morrison, S.; Tang, Y.-W. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin. Lab. Med. 2015, 35, 461–486. [Google Scholar] [CrossRef]
- Kocher, J.; Yuan, L. Norovirus vaccines and potential antinorovirus drugs: Recent advances and future perspectives. Future Virol. 2015, 10, 899–913. [Google Scholar] [CrossRef]
- Appleton, H. Viruses and gastroenteritis in infants. Lancet 1975, 1, 1297. [Google Scholar] [CrossRef]
- Madeley, C.; Cosgrove, B. 28 nm particles in faeces in infantile gastroenteritis. Lancet 1975, 306, 451–452. [Google Scholar] [CrossRef]
- González, G.G.; Pujol, F.H.; Liprandi, F.; Deibis, L.; Ludert, J.E. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J. Med. Virol. 1998, 55, 288–292. [Google Scholar] [CrossRef]
- Wunderli, W.; Meerbach, A.; Guengoer, T.; Berger, C.; Greiner, O.; Caduff, R.; Trkola, A.; Bossart, W.; Gerlach, D.; Schibler, M. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS ONE 2011, 6, e27483. [Google Scholar] [CrossRef]
- Méndez-Toss, M.; Griffin, D.D.; Calva, J.; Contreras, J.F.; Puerto, F.I.; Mota, F.; Guiscafré, H.; Cedillo, R.; Muñoz, O.; Herrera, I. Prevalence and genetic diversity of human astroviruses in Mexican children with symptomatic and asymptomatic infections. J. Clin. Microbiol. 2004, 42, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.; Lee, T.; Craig, J.; Reed, S.E. Astrovirus infection in volunteers. J. Med. Virol. 1979, 3, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Sebire, N.; Malone, M.; Shah, N.; Anderson, G.; Gaspar, H.; Cubitt, W. Pathology of astrovirus associated diarrhoea in a paediatric bone marrow transplant recipient. J. Clin. Pathol. 2004, 57, 1001–1003. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.; Angus, K.; Snodgrass, D. Ultrastructure of the small intestine in astrovirus-infected lambs. J. Gen. Virol. 1980, 49, 71–82. [Google Scholar] [CrossRef]
- Snodgrass, D.; Angus, K.; Gray, E.; Menzies, J.; Paul, G. Pathogenesis of diarrhoea caused by astrovirus infections in lambs. Arch. Virol. 1979, 60, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Le Cann, P.; Ranarijaona, S.; Monpoeho, S.; Le Guyader, F.; Ferré, V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 2004, 155, 11–15. [Google Scholar] [CrossRef]
- Finkbeiner, S.R.; Holtz, L.R.; Jiang, Y.; Rajendran, P.; Franz, C.J.; Zhao, G.; Kang, G.; Wang, D. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol. J. 2009, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.T.; Bauer, I.K.; Antonio, M.; Adeyemi, M.; Saha, D.; Oundo, J.O.; Ochieng, J.B.; Omore, R.; Stine, O.C.; Wang, D. Prevalence of classic, MLB-clade and VA-clade Astroviruses in Kenya and The Gambia. Virol. J. 2015, 12, 1–7. [Google Scholar] [CrossRef]
- Nollevaux, G.; Devillé, C.; El Moualij, B.; Zorzi, W.; Deloyer, P.; Schneider, Y.-J.; Peulen, O.; Dandrifosse, G. Development of a serum-free co-culture of human intestinal epithelium cell-lines (Caco-2/HT29-5M21). BMC Cell Biol. 2006, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.-Y.; Ko, H.-J.; Vallance, B.A. The intestinal epithelium: Central coordinator of mucosal immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Duizer, E.; Schwab, K.J.; Neill, F.H.; Atmar, R.L.; Koopmans, M.P.; Estes, M.K. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 2004, 85, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Karst, S.M.; Thackray, L.B.; Chang, K.-O.; Sosnovtsev, S.V.; Belliot, G.; Krug, A.; Mackenzie, J.M.; Green, K.Y.; Virgin IV, H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004, 2, e432. [Google Scholar] [CrossRef]
- Kitamoto, T.; Takai-Todaka, R.; Kato, A.; Kanamori, K.; Takagi, H.; Yoshida, K.; Katayama, K.; Nakanishi, A. Viral population changes during murine norovirus propagation in RAW 264.7 cells. Front. Microbiol. 2017, 8, 1091. [Google Scholar] [CrossRef] [PubMed]
- Kearney, K.; Chen, D.; Taraporewala, Z.F.; Vende, P.; Hoshino, Y.; Tortorici, M.A.; Barro, M.; Patton, J.T. Cell-line-induced mutation of the rotavirus genome alters expression of an IRF3-interacting protein. EMBO J. 2004, 23, 4072–4081. [Google Scholar] [CrossRef]
- Tsugawa, T.; Tatsumi, M.; Tsutsumi, H. Virulence-associated genome mutations of murine rotavirus identified by alternating serial passages in mice and cell cultures. J. Virol. 2014, 88, 5543–5558. [Google Scholar] [CrossRef] [PubMed]
- Govorkova, E.; Murti, G.; Meignier, B.; De Taisne, C.; Webster, R. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J. Virol. 1996, 70, 5519–5524. [Google Scholar] [CrossRef]
- Pinto, M. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 1983, 47, 323–330. [Google Scholar]
- Rousset, M. The human colon carcinoma cell lines HT-29 and Caco-2: Two in vitro models for the study of intestinal differentiation. Biochimie 1986, 68, 1035–1040. [Google Scholar] [CrossRef]
- Pintó, R.M.; Diez, J.M.; Bosch, A. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J. Med. Virol. 1994, 44, 310–315. [Google Scholar] [CrossRef]
- Peterson, M.D.; Mooseker, M.S. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J. Cell Sci. 1992, 102, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L.; Finlay, B.; Bass, D.; Von Bonsdorff, C.; Greenberg, H. Symmetric infection of rotavirus on polarized human intestinal epithelial (Caco-2) cells. J. Virol. 1991, 65, 4190–4197. [Google Scholar] [CrossRef]
- Jourdan, N.; Maurice, M.; Delautier, D.; Quero, A.M.; Servin, A.L.; Trugnan, G. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 1997, 71, 8268–8278. [Google Scholar] [CrossRef]
- Janowski, A.B.; Wang, D. Infection and propagation of astrovirus VA1 in cell culture. Curr. Protoc. Microbiol. 2019, 52, e73. [Google Scholar] [CrossRef]
- Harrington, P.R.; Vinjé, J.; Moe, C.L.; Baric, R.S. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 2004, 78, 3035–3045. [Google Scholar] [CrossRef]
- Harrington, P.R.; Lindesmith, L.; Yount, B.; Moe, C.L.; Baric, R.S. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 2002, 76, 12335–12343. [Google Scholar] [CrossRef]
- Hutson, A.M.; Atmar, R.L.; Marcus, D.M.; Estes, M.K. Norwalk virus-like particle hemagglutination by binding to H histo-blood group antigens. J. Virol. 2003, 77, 405–415. [Google Scholar] [CrossRef]
- Marionneau, S.; Ruvoën, N.; Le Moullac–Vaidye, B.; Clement, M.; Cailleau–Thomas, A.; Ruiz–Palacois, G.; Huang, P.; Jiang, X.; Le Pendu, J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002, 122, 1967–1977. [Google Scholar] [CrossRef]
- White, L.J.; Ball, J.M.; Hardy, M.E.; Tanaka, T.N.; Kitamoto, N.; Estes, M.K. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 1996, 70, 6589–6597. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Stoltzfus, G.T.; Zhu, C.; Jung, K.; Wang, Q.; Saif, L.J. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS ONE 2018, 13, e0178157. [Google Scholar] [CrossRef]
- Huet, G.; Kim, I.; De Bolos, C.; Lo-Guidice, J.; Moreau, O.; Hemon, B.; Richet, C.; Delannoy, P.; Real, F.; Degand, P. Characterization of mucins and proteoglycans synthesized by a mucin-secreting HT-29 cell subpopulation. J. Cell Sci. 1995, 108, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M. Enterocyte-like differentiation and polarization of the human colon cancer cells by replacement of glucose by galactose in the medium. Biol. Cell 1983, 44, 193–196. [Google Scholar]
- Fett, J.W.; Strydom, D.J.; Lobb, R.R.; Alderman, E.M.; Vallee, B.L. Lysozyme: A major secretory product of a human colon carcinoma cell line. Biochemistry 1985, 24, 965–975. [Google Scholar] [CrossRef]
- Superti, F.; Tinari, A.; Baldassarri, L.; Donelli, G. HT-29 cells: A new substrate for rotavirus growth. Arch. Virol. 1991, 116, 159–173. [Google Scholar] [CrossRef]
- Straub, T.M.; Zu Bentrup, K.H.; Coghlan, P.O.; Dohnalkova, A.; Mayer, B.K.; Bartholomew, R.A.; Valdez, C.O.; Bruckner-Lea, C.J.; Gerba, C.P.; Abbaszadegan, M.A. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 2007, 13, 396. [Google Scholar] [CrossRef]
- Straub, T.M.; Bartholomew, R.A.; Valdez, C.O.; Valentine, N.B.; Dohnalkova, A.; Ozanich, R.M.; Bruckner-Lea, C.J.; Call, D.R. Human norovirus infection of caco-2 cells grown as a three-dimensional tissue structure. J. Water Health 2011, 9, 225–240. [Google Scholar] [CrossRef]
- Papafragkou, E.; Hewitt, J.; Park, G.W.; Greening, G.; Vinje, J. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS ONE 2013, 8, e63485. [Google Scholar] [CrossRef] [PubMed]
- Herbst-Kralovetz, M.M.; Radtke, A.L.; Lay, M.K.; Hjelm, B.E.; Bolick, A.N.; Sarker, S.S.; Atmar, R.L.; Kingsley, D.H.; Arntzen, C.J.; Estes, M.K. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg. Infect. Dis. 2013, 19, 431. [Google Scholar] [CrossRef]
- Takanashi, S.; Saif, L.J.; Hughes, J.H.; Meulia, T.; Jung, K.; Scheuer, K.A.; Wang, Q. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Arch. Virol. 2014, 159, 257–266. [Google Scholar] [CrossRef]
- Wilen, C.B.; Lee, S.; Hsieh, L.L.; Orchard, R.C.; Desai, C.; Hykes, B.L.; McAllaster, M.R.; Balce, D.R.; Feehley, T.; Brestoff, J.R. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 2018, 360, 204–208. [Google Scholar] [CrossRef]
- Desmyter, J.; Melnick, J.L.; Rawls, W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 1968, 2, 955–961. [Google Scholar] [CrossRef]
- Osada, N.; Kohara, A.; Yamaji, T.; Hirayama, N.; Kasai, F.; Sekizuka, T.; Kuroda, M.; Hanada, K. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 2014, 21, 673–683. [Google Scholar] [CrossRef]
- Wyatt, R.G.; James, W.D.; Bohl, E.H.; Theil, K.W.; Saif, L.J.; Kalica, A.R.; Greenberg, H.B.; Kapikian, A.Z.; Chanock, R.M. Human rotavirus type 2: Cultivation in vitro. Science 1980, 207, 189–191. [Google Scholar] [CrossRef]
- Barrett, P.N.; Mundt, W.; Kistner, O.; Howard, M.K. Vero cell platform in vaccine production: Moving towards cell culture-based viral vaccines. Expert Rev. Vaccines 2009, 8, 607–618. [Google Scholar] [CrossRef]
- Wu, W.; Orr-Burks, N.; Karpilow, J.; Tripp, R.A. Development of improved vaccine cell lines against rotavirus. Sci. Data 2017, 4, 1–12. [Google Scholar] [CrossRef]
- Guix, S.; Pérez-Bosque, A.; Miró, L.; Moretó, M.; Bosch, A.; Pintó, R.M. Type I interferon response is delayed in human astrovirus infections. PLoS ONE 2015, 10, e0123087. [Google Scholar] [CrossRef]
- Hasegawa, A.; Matsuno, S.; Inouye, S.; Kono, R.; Tsurukubo, Y.; Mukoyama, A.; Saito, Y. Isolation of human rotaviruses in primary cultures of monkey kidney cells. J. Clin. Microbiol. 1982, 16, 387–390. [Google Scholar] [CrossRef]
- Whitaker, A.; Hayward, C. The characterization of three monkey kidney cell lines. Dev. Biol. Stand. 1985, 60, 125–131. [Google Scholar]
- Sato, K.; Inaba, Y.; Shinozaki, T.; Fujii, R.; Matumoto, M. Isolation of human rotavirus in cell cultures. Arch. Virol. 1981, 69, 155–160. [Google Scholar] [CrossRef]
- Urasawa, T.; Urasawa, S.; Taniguchi, K. Sequential passages of human rotavirus in MA-104 cells. Microbiol. Immunol. 1981, 25, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Sasaki, R.; Masuzaki, R.; Matsumoto, N.; Ogawa, M.; Moriyama, M. Cell culture systems and drug targets for hepatitis A virus infection. Viruses 2020, 12, 533. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.M.; Decker, C.C.; Dao Thi, V.L. Cell culture models for hepatitis E virus. Viruses 2019, 11, 608. [Google Scholar] [CrossRef]
- El-Mokhtar, M.A.; Sayed, I.M. Model systems for studying extrahepatic pathogenesis of hepatitis E virus. Current knowledge and future directions. Rev. Med. Virol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Dufour, J.M. Cell Lines: Valuable Tools or Useless Artifacts; Taylor & Francis: Abingdon, UK, 2012. [Google Scholar]
- Ward, R.L.; Knowlton, D.R.; Pierce, M.J. Efficiency of human rotavirus propagation in cell culture. J. Clin. Microbiol. 1984, 19, 748–753. [Google Scholar] [CrossRef]
- De La Rubia, L.; Herrera, M.; Cebrero, M.; De Jong, J. Acute pancreatitis associated with rotavirus infection. Pancreas 1996, 12, 98–99. [Google Scholar] [CrossRef]
- Honeyman, M.C.; Laine, D.; Zhan, Y.; Londrigan, S.; Kirkwood, C.; Harrison, L.C. Rotavirus infection induces transient pancreatic involution and hyperglycemia in weanling mice. PLoS ONE 2014, 9, e106560. [Google Scholar] [CrossRef]
- Coulson, B.S.; Witterick, P.D.; Tan, Y.; Hewish, M.J.; Mountford, J.N.; Harrison, L.C.; Honeyman, M.C. Growth of rotaviruses in primary pancreatic cells. J. Virol. 2002, 76, 9537–9544. [Google Scholar] [CrossRef]
- Marion, O.; Lhomme, S.; Nayrac, M.; Dubois, M.; Pucelle, M.; Requena, M.; Migueres, M.; Abravanel, F.; Peron, J.M.; Carrere, N. Hepatitis E virus replication in human intestinal cells. Gut 2020, 69, 901–910. [Google Scholar] [CrossRef]
- El-Mokhtar, M.A.; Othman, E.R.; Khashbah, M.Y.; Ismael, A.; Ghaliony, M.A.; Seddik, M.I.; Sayed, I.M. Evidence of the extrahepatic replication of Hepatitis E virus in human endometrial stromal cells. Pathogens 2020, 9, 295. [Google Scholar] [CrossRef]
- El-Mokhtar, M.A.; Seddik, M.I.; Osman, A.; Adel, S.; Abdel Aziz, E.M.; Mandour, S.A.; Mohammed, N.; Zarzour, M.A.; Abdel-Wahid, L.; Radwan, E. Hepatitis E virus mediates renal injury via the interaction between the immune cells and renal epithelium. Vaccines 2020, 8, 454. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Seddik, M.I.; Gaber, M.A.; Saber, S.H.; Mandour, S.A.; El-Mokhtar, M.A. Replication of Hepatitis E Virus (HEV) in primary human-derived monocytes and macrophages in vitro. Vaccines 2020, 8, 239. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Howell, J.C.; Wells, J.M.; Spence, J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011, 6, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Bijvelds, M.; Dang, W.; Xu, L.; van der Eijk, A.A.; Knipping, K.; Tuysuz, N.; Dekkers, J.F.; Wang, Y.; de Jonge, J. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res. 2015, 123, 120–131. [Google Scholar] [CrossRef]
- Saxena, K.; Simon, L.M.; Zeng, X.-L.; Blutt, S.E.; Crawford, S.E.; Sastri, N.P.; Karandikar, U.C.; Ajami, N.J.; Zachos, N.C.; Kovbasnjuk, O. A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc. Natl. Acad. Sci. USA 2017, 114, E570–E579. [Google Scholar] [CrossRef]
- Ettayebi, K.; Tenge, V.R.; Cortes-Penfield, N.W.; Crawford, S.E.; Neill, F.H.; Zeng, X.-L.; Yu, X.; Ayyar, B.V.; Burrin, D.; Ramani, S. New insights and enhanced human norovirus cultivation in human intestinal enteroids. Msphere 2021, 6, 1. [Google Scholar] [CrossRef]
- Costantini, V.; Morantz, E.K.; Browne, H.; Ettayebi, K.; Zeng, X.-L.; Atmar, R.L.; Estes, M.K.; Vinjé, J. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 2018, 24, 1453. [Google Scholar] [CrossRef]
- Li, L.; Xue, M.; Fu, F.; Yin, L.; Feng, L.; Liu, P. IFN-lambda 3 mediates antiviral protection against porcine epidemic diarrhea virus by inducing a distinct antiviral transcript profile in porcine intestinal epithelia. Front. Immunol. 2019, 10, 2394. [Google Scholar] [CrossRef]
- Murakami, K.; Tenge, V.R.; Karandikar, U.C.; Lin, S.-C.; Ramani, S.; Ettayebi, K.; Crawford, S.E.; Zeng, X.-L.; Neill, F.H.; Ayyar, B.V. Bile acids and ceramide overcome the entry restriction for GII. 3 human norovirus replication in human intestinal enteroids. Proc. Natl. Acad. Sci. USA 2020, 117, 1700–1710. [Google Scholar] [CrossRef]
- Wilson, R.L.; Swaminathan, G.; Ettayebi, K.; Bomidi, C.; Zeng, X.-L.; Blutt, S.E.; Estes, M.K.; Grande-Allen, K.J. Protein-functionalized poly (ethylene glycol) hydrogels as scaffolds for monolayer organoid culture. Tissue Eng. Part C Methods 2021, 27, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Atmar, R.L.; Ettayebi, K.; Ayyar, B.V.; Neill, F.H.; Braun, R.P.; Ramani, S.; Estes, M.K. Comparison of Microneutralization and Histo-Blood Group Antigen–Blocking Assays for Functional Norovirus Antibody Detection. J. Infect. Dis. 2020, 221, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Roodsant, T.; Navis, M.; Aknouch, I.; Renes, I.B.; Van Elburg, R.M.; Pajkrt, D.; Wolthers, K.C.; Schultsz, C.; Van Der Ark, K.C.; Sridhar, A. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front. Cell. Infect. Microbiol. 2020, 10, 272. [Google Scholar] [CrossRef]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- De Rudder, C.; Arroyo, M.C.; Lebeer, S.; Van de Wiele, T. Dual and Triple Epithelial Coculture Model Systems with Donor-Derived Microbiota and THP-1 Macrophages to Mimic Host-Microbe Interactions in the Human Sinonasal Cavities. Msphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Co, J.Y.; Margalef-Catala, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef]
- Zang, R.; Castro, M.F.G.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Giobbe, G.G.; Bonfante, F.; Zambaiti, E.; Gagliano, O.; Jones, B.C.; Luni, C.; Laterza, C.; Perin, S.; Stuart, H.T.; Pagliari, M. SARS-CoV-2 infection and replication in human fetal and pediatric gastric organoids. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gayer, C.P.; Basson, M.D. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signal. 2009, 21, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Lentle, R.; Janssen, P. Physical characteristics of digesta and their influence on flow and mixing in the mammalian intestine: A review. J. Comp. Physiol. B 2008, 178, 673–690. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab A Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Nasiri, R.; De Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseni, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 120196. [Google Scholar] [CrossRef]
- Wilson, R.L.; Hewes, S.A.; Rajan, A.; Lin, S.-C.; Bomidi, C.; Iida, T.; Estes, M.K.; Maresso, A.W.; Grande-Allen, K.J. A Millifluidic Perfusion Cassette for Studying the Pathogenesis of Enteric Infections Using Ex-Vivo Organoids. Ann. Biomed. Eng. 2021, 49, 1233–1244. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Crawford, S.E.; Blutt, S.E.; Estes, M.K. Human organoid cultures: Transformative new tools for human virus studies. Curr. Opin. Virol. 2018, 29, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Monedero, V.; Buesa, J.; Rodríguez-Díaz, J. The interactions between host glycobiology, bacterial microbiota, and viruses in the gut. Viruses 2018, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Walker, F.C.; Baldridge, M.T. Interactions between noroviruses, the host, and the microbiota. Curr. Opin. Virol. 2019, 37, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sestak, K. Non-human primate models of enteric viral infections. Viruses 2018, 10, 544. [Google Scholar] [CrossRef] [PubMed]
- Chege, G.K.; Steele, A.; Hart, C.; Snodgrass, D.; Omolo, E.O.; Mwenda, J.M. Experimental infection of non-human primates with a human rotavirus isolate. Vaccine 2005, 23, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.; Bogers, W.; Heeney, J.; van Amerongen, G.; Koopmans, M. Experimental norovirus infections in non-human primates. J. Med. Virol. 2005, 75, 313–320. [Google Scholar] [CrossRef]
- Wyatt, R.G.; Greenberg, H.B.; Dalgard, D.W.; Allen, W.P.; Sly, D.L.; Thornhill, T.S.; Chanock, R.M.; Kapikian, A.Z. Experimental infection of chimpanzees with the Norwalk agent of epidemic viral gastroenteritis. J. Med. Virol. 1978, 2, 89–96. [Google Scholar] [CrossRef]
- Malherbe, H.; Harwin, R. The cytopathic effects of vervet monkey viruses. S. Afr. Med. J. 1963, 37, 407–411. [Google Scholar] [PubMed]
- Yin, N.; Yang, F.-M.; Qiao, H.-T.; Zhou, Y.; Duan, S.-Q.; Lin, X.-C.; Wu, J.-Y.; Xie, Y.-P.; He, Z.-L.; Sun, M.-S. Neonatal rhesus monkeys as an animal model for rotavirus infection. World J. Gastroenterol. 2018, 24, 5109. [Google Scholar] [CrossRef] [PubMed]
- Ludert, J.E.; Feng, N.; Yu, J.H.; Broome, R.L.; Hoshino, Y.; Greenberg, H.B. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 1996, 70, 487–493. [Google Scholar] [CrossRef]
- McNeal, M.M.; Sestak, K.; Choi, A.H.-C.; Basu, M.; Cole, M.J.; Aye, P.P.; Bohm, R.P.; Ward, R.L. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 2005, 79, 944–954. [Google Scholar] [CrossRef]
- Hoshino, Y.; Honma, S.; Jones, R.W.; Santos, N.; Nakagomi, O.; Nakagomi, T.; Kapikian, A.Z.; Thouless, M.E. A rotavirus strain isolated from pig-tailed macaque (Macaca nemestrina) with diarrhea bears a P6 [1]: G8 specificity. Virology 2006, 345, 1–12. [Google Scholar] [CrossRef]
- Farkas, T.; Sestak, K.; Wei, C.; Jiang, X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008, 82, 5408–5416. [Google Scholar] [CrossRef]
- Sestak, K.; Feely, S.; Fey, B.; Dufour, J.; Hargitt, E.; Alvarez, X.; Pahar, B.; Gregoricus, N.; Vinjé, J.; Farkas, T. Experimental inoculation of juvenile rhesus macaques with primate enteric caliciviruses. PLoS ONE 2012, 7, e37973. [Google Scholar] [CrossRef]
- Xin, Y.-Y.; Li, L.-L.; Ao, Y.-Y.; Xie, Z.-P.; Li, J.-S.; Duan, Z.-J.; Yu, J.-M.; Zhang, B. A novel astrovirus identified in wild rhesus monkey feces in China. Arch. Virol. 2019, 164, 2385–2388. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.A.; Small, C.T.; Freiden, P.; Feeroz, M.; Matsen IV, F.A.; San, S.; Hasan, M.K.; Wang, D.; Jones-Engel, L.; Schultz-Cherry, S. Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog. 2015, 11, e1005225. [Google Scholar] [CrossRef] [PubMed]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Rajashekara, G.; Saif, L.J. Interactions between human microbiome, diet, enteric viruses and immune system: Novel insights from gnotobiotic pig research. Drug Discov. Today Dis. Models 2018, 28, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.; Ward, L.; Yuan, L.; Rosen, B.; To, T. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Viral Gastroenteritis 1996, 12, 153–161. [Google Scholar]
- Azevedo, M.P.; Vlasova, A.N.; Saif, L.J. Human rotavirus virus-like particle vaccines evaluated in a neonatal gnotobiotic pig model of human rotavirus disease. Expert Rev. Vaccines 2013, 12, 169–181. [Google Scholar] [CrossRef]
- Troeger, H.; Loddenkemper, C.; Schneider, T.; Schreier, E.; Epple, H.-J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Structural and functional changes of the duodenum in human norovirus infection. Gut 2009, 58, 1070–1077. [Google Scholar] [CrossRef]
- Wang, Q.-H.; Han, M.G.; Cheetham, S.; Souza, M.; Funk, J.A.; Saif, L.J. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 2005, 11, 1874. [Google Scholar] [CrossRef]
- Sugieda, M.; Nakajima, S. Viruses detected in the caecum contents of healthy pigs representing a new genetic cluster in genogroup II of the genus ‘Norwalk-like viruses’. Virus Res. 2002, 87, 165–172. [Google Scholar] [CrossRef]
- Indik, S.; Valíček, L.; Šmíd, B.; Dvořáková, H.; Rodák, L. Isolation and partial characterization of a novel porcine astrovirus. Vet. Microbiol. 2006, 117, 276–283. [Google Scholar] [CrossRef]
- Perlman, R.L. Mouse models of human diseaseAn evolutionary perspective. Evol. Med. Public Health 2016, 2016, 170–176. [Google Scholar] [PubMed]
- Gouvea, V.; Alencar, A.; Barth, O.; De Castro, L.; Fialho, A.; Araujo, H.; Majerowicz, S.; Pereira, H. Diarrhoea in mice infected with a human rotavirus. J. Gen. Virol. 1986, 67, 577–581. [Google Scholar] [CrossRef]
- Ebina, T.; Tsukada, K.; Umezu, K.; Nose, M.; Tsuda, K.; Hatta, H.; Kim, M.; Yamamoto, T. Gastroenteritis in suckling mice caused by human rotavirus can be prevented with egg yolk immunoglobulin (IgY) and treated with a protein-bound polysaccharide preparation (PSK). Microbiol. Immunol. 1990, 34, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Woode, G.; Bridger, J.C.; Jones, J.M.; Flewett, T.; Davies, H.; Davis, H.; White, G. Morphological and antigenic relationships between viruses (rotaviruses) from acute gastroenteritis of children, calves, piglets, mice, and foals. Infect. Immun. 1976, 14, 804–810. [Google Scholar] [CrossRef]
- Tajima, T.; Suzuki, E.; Ushijima, H.; Araki, K.; Kim, B.; Shinozaki, T.; Fujii, R. Isolation of murine rotavirus in cell cultures. Arch. Virol. 1984, 82, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Starkey, W.; Collins, J.; Wallis, T.; Clarke, G.; Spencer, A.; Haddon, S.; Osborne, M.; Candy, D.; Stephen, J. Kinetics, tissue specificity and pathological changes in murine rotavirus infection of mice. J. Gen. Virol. 1986, 67, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Much, D.H.; Zajac, I. Purification and characterization of epizootic diarrhea of infant mice virus. Infect. Immun. 1972, 6, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Pott, J.; Mahlakõiv, T.; Mordstein, M.; Duerr, C.U.; Michiels, T.; Stockinger, S.; Staeheli, P.; Hornef, M.W. IFN-λ determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA 2011, 108, 7944–7949. [Google Scholar] [CrossRef]
- Lin, J.-D.; Feng, N.; Sen, A.; Balan, M.; Tseng, H.-C.; McElrath, C.; Smirnov, S.V.; Peng, J.; Yasukawa, L.L.; Durbin, R.K. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 2016, 12, e1005600. [Google Scholar]
- Zhang, B.; Chassaing, B.; Shi, Z.; Uchiyama, R.; Zhang, Z.; Denning, T.L.; Crawford, S.E.; Pruijssers, A.J.; Iskarpatyoti, J.A.; Estes, M.K. Prevention and cure of rotavirus infection via TLR5/NLRC4–mediated production of IL-22 and IL-18. Science 2014, 346, 861–865. [Google Scholar] [CrossRef]
- Shi, Z.; Zou, J.; Zhang, Z.; Zhao, X.; Noriega, J.; Zhang, B.; Zhao, C.; Ingle, H.; Bittinger, K.; Mattei, L.M. Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 2019, 179, 644–658.e13. [Google Scholar] [CrossRef] [PubMed]
- Karst, S.M.; Wobus, C.E.; Lay, M.; Davidson, J.; Virgin, H.W. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2003, 299, 1575–1578. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, M.T.; Nice, T.J.; McCune, B.T.; Yokoyama, C.C.; Kambal, A.; Wheadon, M.; Diamond, M.S.; Ivanova, Y.; Artyomov, M.; Virgin, H.W. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 2015, 347, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Orchard, R.C.; Wilen, C.B.; Doench, J.G.; Baldridge, M.T.; McCune, B.T.; Lee, Y.-C.J.; Lee, S.; Pruett-Miller, S.M.; Nelson, C.A.; Fremont, D.H. Discovery of a proteinaceous cellular receptor for a norovirus. Science 2016, 353, 933–936. [Google Scholar] [CrossRef]
- Taube, S.; Perry, J.W.; McGreevy, E.; Yetming, K.; Perkins, C.; Henderson, K.; Wobus, C.E. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J. Virol. 2012, 86, 5584–5593. [Google Scholar] [CrossRef]
- Lee, S.; Wilen, C.B.; Orvedahl, A.; McCune, B.T.; Kim, K.-W.; Orchard, R.C.; Peterson, S.T.; Nice, T.J.; Baldridge, M.T.; Virgin, H.W. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe 2017, 22, 449–459.e4. [Google Scholar] [CrossRef]
- Grau, K.R.; Roth, A.N.; Zhu, S.; Hernandez, A.; Colliou, N.; DiVita, B.B.; Philip, D.T.; Riffe, C.; Giasson, B.; Wallet, S.M. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat. Microbiol. 2017, 2, 1586–1591. [Google Scholar] [CrossRef]
- Graziano, V.R.; Walker, F.C.; Kennedy, E.A.; Wei, J.; Ettayebi, K.; Strine, M.S.; Filler, R.B.; Hassan, E.; Hsieh, L.L.; Kim, A.S. CD300lf is the primary physiologic receptor of murine norovirus but not human norovirus. PLoS Pathog. 2020, 16, e1008242. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, C.C.; Loh, J.; Zhao, G.; Stappenbeck, T.S.; Wang, D.; Huang, H.V.; Virgin, H.W.; Thackray, L.B. Adaptive immunity restricts replication of novel murine astroviruses. J. Virol. 2012, 86, 12262–12270. [Google Scholar] [CrossRef]
- Cortez, V.; Sharp, B.; Yao, J.; Livingston, B.; Vogel, P.; Schultz-Cherry, S. Characterizing a murine model for astrovirus using viral isolates from persistently infected immunocompromised mice. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Ingle, H.; Lee, S.; Ai, T.; Orvedahl, A.; Rodgers, R.; Zhao, G.; Sullender, M.; Peterson, S.T.; Locke, M.; Liu, T.-C. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 2019, 4, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Cortez, V.; Boyd, D.F.; Crawford, J.C.; Sharp, B.; Livingston, B.; Rowe, H.M.; Davis, A.; Alsallaq, R.; Robinson, C.G.; Vogel, P. Astrovirus infects actively secreting goblet cells and alters the gut mucus barrier. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.F.; Nie, P. Zebrafish as a model for the study of host-virus interactions. In Innate Antiviral Immunity; Springer: Berlin/Heidelberg, Germany, 2017; pp. 57–78. [Google Scholar]
- Sullivan, C.; Soos, B.-L.; Millard, P.J.; Kim, C.H.; King, B.L. Modeling Virus-Induced Inflammation in Zebrafish: A Balance Between Infection Control and Excessive Inflammation. Front. Immunol. 2021, 12, 1635. [Google Scholar] [CrossRef]
- Tan, M.T.H.; Li, Y.; Eshaghi Gorji, M.; Gong, Z.; Li, D. Fucoidan But Not 2′-Fucosyllactose Inhibits Human Norovirus Replication in Zebrafish Larvae. Viruses 2021, 13, 461. [Google Scholar] [CrossRef]
- Van Dycke, J.; Cuvry, A.; Knickmann, J.; Ny, A.; Rakers, S.; Taube, S.; de Witte, P.; Neyts, J.; Rocha-Pereira, J. Infection of zebrafish larvae with human norovirus and evaluation of the in vivo efficacy of small-molecule inhibitors. Nat. Protoc. 2021, 16, 1830–1849. [Google Scholar] [CrossRef]
- Balla, K.M.; Rice, M.C.; Gagnon, J.A.; Elde, N.C. Linking virus discovery to immune responses visualized during zebrafish infections. Curr. Biol. 2020, 30, 2092–2103.e5. [Google Scholar] [CrossRef]
- Schultz-Cherry, S.; Kapczynski, D.R.; Simmons, V.M.; Koci, M.D.; Brown, C.; Barnes, H.J. Identifying agent (s) associated with poult enteritis mortality syndrome: Importance of the thymus. Avian Dis. 2000, 44, 256–265. [Google Scholar] [CrossRef]
- Koci, M.D.; Moser, L.A.; Kelley, L.A.; Larsen, D.; Brown, C.C.; Schultz-Cherry, S. Astrovirus induces diarrhea in the absence of inflammation and cell death. J. Virol. 2003, 77, 11798–11808. [Google Scholar] [CrossRef]
- Hargest, V.; Sharp, B.; Livingston, B.; Cortez, V.; Schultz-Cherry, S. Astrovirus replication is inhibited by nitazoxanide in vitro and in vivo. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Meliopoulos, V.A.; Marvin, S.A.; Freiden, P.; Moser, L.A.; Nighot, P.; Ali, R.; Blikslager, A.; Reddivari, M.; Heath, R.J.; Koci, M.D. Oral administration of astrovirus capsid protein is sufficient to induce acute diarrhea in vivo. MBio 2016, 7. [Google Scholar] [CrossRef]
- Staab, J.F.; Lemme-Dumit, J.M.; Latanich, R.; Pasetti, M.F.; Zachos, N.C. Co-Culture System of Human Enteroids/Colonoids with Innate Immune Cells. Curr. Protoc. Immunol. 2020, 131, e113. [Google Scholar] [CrossRef] [PubMed]
- In, J.G.; Foulke-Abel, J.; Clarke, E.; Kovbasnjuk, O. Human colonoid monolayers to study interactions between pathogens, commensals, and host intestinal epithelium. J. Vis. Exp. 2019, e59357. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Taya, C.; Aoki, J.; Matsuda, Y.; Ise, I.; Takeda, H.; Matsuzaki, T.; Amanuma, H.; Yonekawa, H.; Nomoto, A. Characterization of three different transgenic mouse lines that carry human poliovirus receptor gene—Influence of the transgene expression on pathogenesis. Arch. Virol. 1994, 139, 351–363. [Google Scholar] [CrossRef] [PubMed]
| Virus | Method | Origin | Experimental Model | Viral Strain(s) | References |
|---|---|---|---|---|---|
| Human Rotavirus | in vitro | Immortalized cells | |||
| Human colon adenocarcinoma | Caco-2 | Trypsin-activated HRV-Wa or SA114F | [11,12] | ||
| Human colorectal adenocarcinoma | HT-29 | Trypsin-activated SA114F | [12] | ||
| Human colorectal adenocarcinoma | Genetically-modified HT-29 (STAG2−/−, STAT1−/−, and STING−/−) HT-29 | Trypsin-activated HRV-Wa, G2P[4], G4P[6], G12P[4], G9P[8], G8P[11], G8P[10] | [13] | ||
| Human colorectal adenocarcinoma | HT29-MTX | Trypsin-activated SA114F | [14] | ||
| African Green Monkey Kidney | MA-104 and Vero | Trypsin-activated HRV | [15] | ||
| African Green Monkey Kidney | Genetically-modified (LRGUK−/−, WR62−/−, EMX2−/−) Vero cells | Trypsin-activated RV3, CDC-9. Rotarix and 116E | [16,17] | ||
| Human Intestinal Enteroids (HIEs) | |||||
| Human intestinal biopsies | 3D-HIEs | Trypsin activated G1P[8] and G9P[8] | [18,19] | ||
| Human intestinal biopsies | Differentiated 2D-monolayer and Transwell HIEs | Trypsin activated HRV | [20] | ||
| Human intestinal biopsies | 2D-Monolayer and transwell | Trypsin activated HRV G3P[8] | [21] | ||
| in vivo | Animal Model | ||||
| Rodent | Guinea pig | HRV-Wa | [22,23] | ||
| Pig | Gnotobiotic piglet | HRV-Wa | [24,25] | ||
| Non-human primate | Cynomolgus monkeys | HRV-Wa or infected stool filtrate | [26,27,28] | ||
| Human Norovirus | in vitro | Immortalized Cell lines | |||
| Human malignant B-cells | BJAB (with or without HT-29 co-culture) | GII.4 stool filtrate | [29] | ||
| African Green Monkey Kidney | Vero | Trypsin-activated GII.3 & GII.4 stool filtrate | [30] | ||
| HIEs | |||||
| Human intestinal biopsies | Differentiated 2D-monolayer & Transwell HIEs | Bile-treated GI.I, GII.3 & GII.4 stool filtrate | [20,31] | ||
| Human intestinal biopsies | Genetically-modified (IFNAR1−/−, IFNLR1−/−, STAT1−/−, MAVS−/−, and STAT1−/−STAT2−/−) HIEs | GII.3 & GII.4 stool filtrate | [32] | ||
| Human intestinal biopsies | Genetically-modified (FUT2−/−) HIEs | GII.3, GII.4 & GII.17 stool filtrate | [33] | ||
| in vivo | Animal Model | ||||
| Fish | Zebrafish | GI.7, GII.2, GII.3, GII.4, GII.6 from stool suspension | [34] | ||
| Rodent | Humanized BALB/c Rag−/−Il2rg−/− mice (engrafted with human CD34+) | GI.3a, GII.4, GII.6 from stool suspension | [35] | ||
| Pig | Gnotobiotic piglets | GII.4 stool filtrate | [36,37] | ||
| Pig | Rag2−/−Il2rg−/− gnotobiotic piglets | GII.4 stool filtrate | [38] | ||
| Non-human primate | Pig-tailed macaques | GII.3 from stool suspension | [39] | ||
| Non-human primate | Chimpanzee | GI.1 stool filtrate | [40] | ||
| Human Astrovirus | in vitro | Immortalized Cell line | |||
| Human colon adenocarcinoma | Caco-2 | HAstV-1, 2, 3, 4, 5, 6, 7 | [41,42,43] | ||
| Human colon adenocarcinoma | Genetically-modified Caco-2 (CLTC silencing) | Trypsin-activated HAstV-8 | [44] | ||
| Human colorectal adenocarcinoma | HT-29 | Trypsin-activated HAstV-1, 2, 3, 4, 5, 6, 7 | [41] | ||
| HIEs | |||||
| Human intestinal biopsies | 3D HIEs | HAstV-1 | [45,46] | ||
| Human intestinal biopsies | Differentiated 2D-monolayer in transwell | HAstV-VA1; HAstV-MLB1 & Trypsin- activated HAstV-1 | [43,47] |
| Experimental Considerations | Immortalized Cell Lines | Primary Cells | HIEs | Animal Model |
|---|---|---|---|---|
| Expense | Low | Moderate | High | High |
| Maintenance demands | Low | Moderate | Moderate to high | High |
| Biological relevance | Low | High | High | High |
| Reproducibility | High | Moderate | Moderate | Moderate (within institutions) |
| Genetic manipulation | Easy | Difficult | Moderate | Difficult |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aggarwal, S.; Hassan, E.; Baldridge, M.T. Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses. Viruses 2021, 13, 975. https://doi.org/10.3390/v13060975
Aggarwal S, Hassan E, Baldridge MT. Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses. Viruses. 2021; 13(6):975. https://doi.org/10.3390/v13060975
Chicago/Turabian StyleAggarwal, Somya, Ebrahim Hassan, and Megan T. Baldridge. 2021. "Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses" Viruses 13, no. 6: 975. https://doi.org/10.3390/v13060975
APA StyleAggarwal, S., Hassan, E., & Baldridge, M. T. (2021). Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses. Viruses, 13(6), 975. https://doi.org/10.3390/v13060975






