Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets
Abstract
1. Introduction
2. Materials and Methods
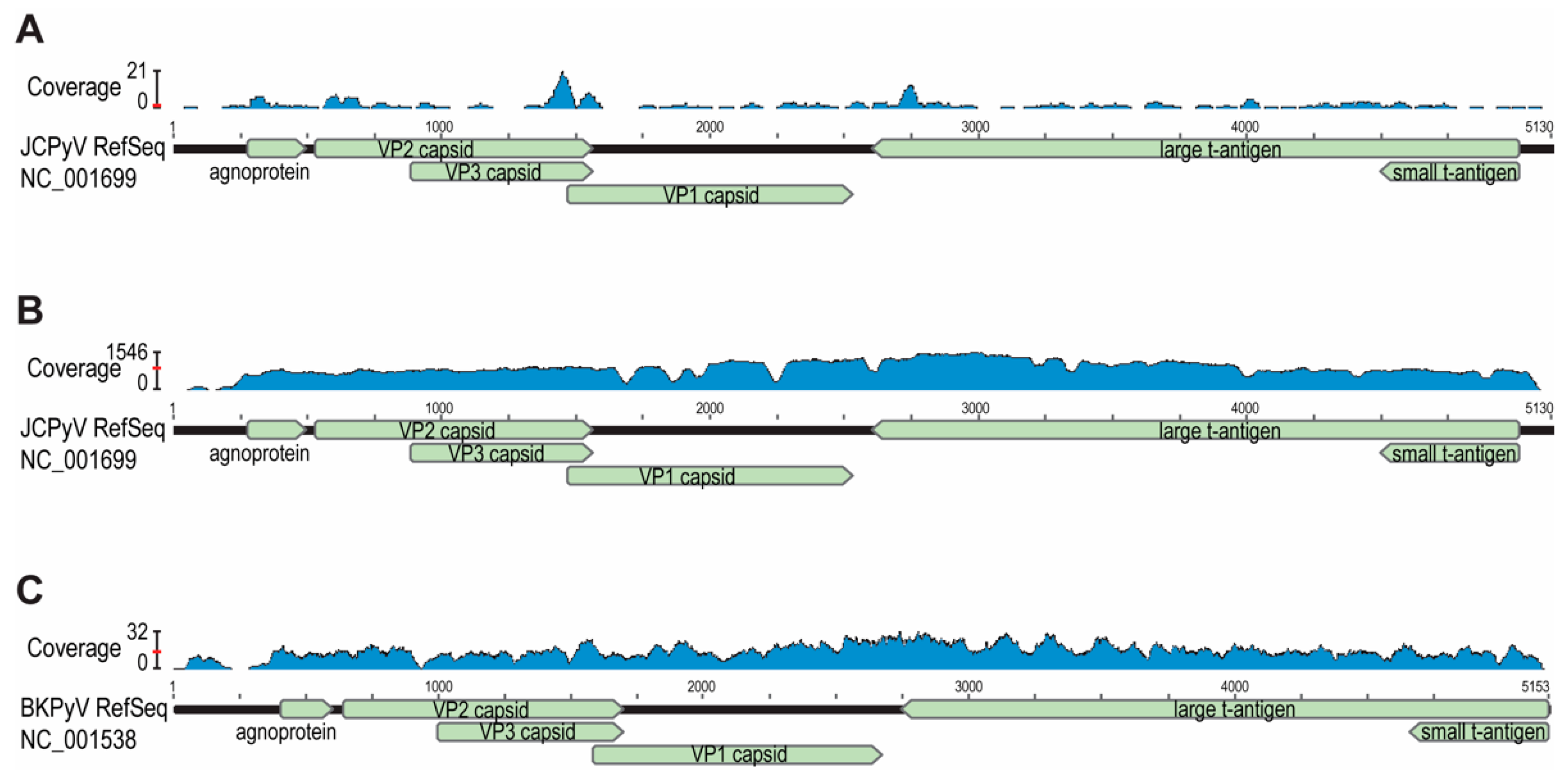
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neugent, M.L.; Hulyalkar, N.V.; Nguyen, V.H.; Zimmern, P.E.; De Nisco, N.J. Advances in Understanding the Human Urinary Microbiome and Its Potential Role in Urinary Tract Infection. mBio 2020, 11. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New Human Papovavirus (B.K.) Isolated from Urine after Renal Transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef]
- Wylie, K.M.; Mihindukulasuriya, K.A.; Zhou, Y.; Sodergren, E.; Storch, G.A.; Weinstock, G.M. Metagenomic Analysis of Double-Stranded DNA Viruses in Healthy Adults. BMC Biol. 2014, 12, 71. [Google Scholar] [CrossRef]
- Pinto, M.; Dobson, S. BK and JC Virus: A Review. J. Infect. 2014, 68 (Suppl. S1), S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Kunitake, T.; Guo, J.; Tominaga, T.; Kawabe, K.; Yogo, Y. Transmission of the Human Polyomavirus JC Virus Occurs Both within the Family and Outside the Family. J. Clin. Microbiol. 1994, 32, 2359–2363. [Google Scholar] [CrossRef] [PubMed]
- Agostini, H.T.; Yanagihara, R.; Davis, V.; Ryschkewitsch, C.F.; Stoner, G.L. Asian Genotypes of JC Virus in Native Americans and in a Pacific Island Population: Markers of Viral Evolution and Human Migration. Proc. Natl. Acad. Sci. USA 1997, 94, 14542–14546. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Girones, R. Excretion and Transmission of JCV in Human Populations. J. Neurovirol. 2001, 7, 345–349. [Google Scholar] [CrossRef][Green Version]
- Chang, H.; Wang, M.; Tsai, R.-T.; Lin, H.-S.; Huan, J.-S.; Wang, W.-C.; Chang, D. High Incidence of JC Viruria in JC-Seropositive Older Individuals. J. Neurovirol. 2002, 8, 447–451. [Google Scholar] [CrossRef]
- Agostini, H.T.; Deckhut, A.; Jobes, D.V.; Girones, R.; Schlunck, G.; Prost, M.G.; Frias, C.; Pérez-Trallero, E.; Ryschkewitsch, C.F.; Stoner, G.L. Genotypes of JC Virus in East, Central and Southwest Europe. J. Gen. Virol. 2001, 82, 1221–1331. [Google Scholar] [CrossRef]
- Baksh, F.K.; Finkelstein, S.D.; Swalsky, P.A.; Stoner, G.L.; Ryschkewitsch, C.F.; Randhawa, P. Molecular Genotyping of BK and JC Viruses in Human Polyomavirus-Associated Interstitial Nephritis after Renal Transplantation. Am. J. Kidney Dis. 2001, 38, 354–365. [Google Scholar] [CrossRef]
- Bogdanovic, G.; Brytting, M.; Cinque, P.; Grandien, M.; Fridell, E.; Ljungman, P.; Lönnqvist, B.; Hammarin, A.L. Nested PCR for Detection of BK Virus and JC Virus DNA. Clin. Diagn. Virol. 1994, 2, 211–220. [Google Scholar] [CrossRef]
- Cayres-Vallinoto, I.M.V.; Vallinoto, A.C.R.; Azevedo, V.N.; Machado, L.F.A.; Ishak, M.d.O.G.; Ishak, R. Human JCV Infections as a Bio-Anthropological Marker of the Formation of Brazilian Amazonian Populations. PLoS ONE 2012, 7, e46523. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.M.; Savassi-Ribas, F.; de Almeida, S.G.S.; da Silva, R.N.N.; Baez, C.F.; Zalis, M.G.; Guimarães, M.A.A.M.; Varella, R.B. A Globally Applicable PCR-Based Detection and Discrimination of BK and JC Polyomaviruses. Rev. Inst. Med. Trop. Sao Paulo 2018, 60, e47. [Google Scholar] [CrossRef]
- Del Valle, L.; Gordon, J.; Assimakopoulou, M.; Enam, S.; Geddes, J.F.; Varakis, J.N.; Katsetos, C.D.; Croul, S.; Khalili, K. Detection of JC Virus DNA Sequences and Expression of the Viral Regulatory Protein T-Antigen in Tumors of the Central Nervous System. Cancer Res. 2001, 61, 4287–4293. [Google Scholar] [PubMed]
- Dumonceaux, T.J.; Mesa, C.; Severini, A. Internally Controlled Triplex Quantitative PCR Assay for Human Polyomaviruses JC and BK. J. Clin. Microbiol. 2008, 46, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Dumoulin, A.; Hirsch, H.H. Reevaluating and Optimizing Polyomavirus BK and JC Real-Time PCR Assays to Detect Rare Sequence Polymorphisms. J. Clin. Microbiol. 2011, 49, 1382–1388. [Google Scholar] [CrossRef][Green Version]
- Funahashi, Y.; Iwata, S.; Ito, Y.; Kojima, S.; Yoshikawa, T.; Hattori, R.; Gotoh, M.; Nishiyama, Y.; Kimura, H. Multiplex Real-Time PCR Assay for Simultaneous Quantification of BK Polyomavirus, JC Polyomavirus, and Adenovirus DNA. J. Clin. Microbiol. 2010, 48, 825–830. [Google Scholar] [CrossRef]
- Giovannelli, I.; Ciccone, N.; Vaggelli, G.; Malva, N.D.; Torricelli, F.; Rossolini, G.M.; Giannecchini, S. Utility of Droplet Digital PCR for the Quantitative Detection of Polyomavirus JC in Clinical Samples. J. Clin. Virol. 2016, 82, 70–75. [Google Scholar] [CrossRef]
- Haghighi, M.F.; Seyyedi, N.; Farhadi, A.; Zare, F.; Kasraian, L.; Refiei Dehbidi, G.R.; Ranjbaran, R.; Behzad-Behbahani, A. Polyomaviruses BK and JC DNA Infection in Peripheral Blood Cells from Blood Donors. Braz. J. Infect. Dis. 2019, 23, 22–26. [Google Scholar] [CrossRef]
- Hori, R.; Murai, Y.; Tsuneyama, K.; Abdel-Aziz, H.O.; Nomoto, K.; Takahashi, H.; Cheng, C.; Kuchina, T.; Harman, B.V.; Takano, Y. Detection of JC Virus DNA Sequences in Colorectal Cancers in Japan. Virchows Archiv 2005, 447, 723–730. [Google Scholar] [CrossRef]
- Hussain, I.; Tasneem, F.; Umer, M.; Pervaiz, A.; Raza, M.; Arshad, M.I.; Shahzad, N. Specific and Quantitative Detection of Human Polyomaviruses BKPyV and JCPyV in the Healthy Pakistani Population. Virol. J. 2017, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Karalic, D.; Lazarevic, I.; Knezevic, A.; Cupic, M.; Jevtovic, D.; Jovanovic, T. Distribution of JC Virus Genotypes among Serbian Patients Infected with HIV and in Healthy Donors. J. Med. Virol. 2014, 86, 411–418. [Google Scholar] [CrossRef]
- Kruzel-Davila, E.; Divers, J.; Russell, G.B.; Kra-Oz, Z.; Cohen, M.S.; Langefeld, C.D.; Ma, L.; Lyles, D.S.; Hicks, P.J.; Skorecki, K.L.; et al. JC Viruria Is Associated with Reduced Risk of Diabetic Kidney Disease. J. Clin. Endocrinol. Metab. 2019, 104, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Makvandi, M.; Mombeini, H.; Haghighi, S.B.; Dastoorpoor, M.; Khodadad, N.; Babaahmadi, M.K.; Tabasi, M.; Pirmoradi, R. Molecular Epidemiology of JC Polyomavirus in HIV-Infected Patients and Healthy Individuals from Iran. Braz. J. Microbiol. 2020, 51, 37–43. [Google Scholar] [CrossRef] [PubMed]
- McNees, A.L.; White, Z.S.; Zanwar, P.; Vilchez, R.A.; Butel, J.S. Specific and Quantitative Detection of Human Polyomaviruses BKV, JCV, and SV40 by Real Time PCR. J. Clin. Virol. 2005, 34, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Merlino, C.; Bergallo, M.; Daniele, R.; Ponzi, A.N.; Cavallo, R. BKV-DNA and JCV-DNA Co-Quantification Assay to Evaluate Viral Load in Urine and Serum. Mol. Biotechnol. 2005, 30, 1–8. [Google Scholar] [CrossRef]
- Pal, A.; Sirota, L.; Maudru, T.; Peden, K.; Lewis, A.M. Real-Time, Quantitative PCR Assays for the Detection of Virus-Specific DNA in Samples with Mixed Populations of Polyomaviruses. J. Virol. Methods 2006, 135, 32–42. [Google Scholar] [CrossRef]
- Rafique, A.; Jiang, S.C. Genetic Diversity of Human Polyomavirus JCPyV in Southern California Wastewater. J. Water Health 2008, 6, 533–538. [Google Scholar] [CrossRef]
- Rencic, A.; Gordon, J.; Otte, J.; Curtis, M.; Kovatich, A.; Zoltick, P.; Khalili, K.; Andrews, D. Detection of JC Virus DNA Sequence and Expression of the Viral Oncoprotein, Tumor Antigen, in Brain of Immunocompetent Patient with Oligoastrocytoma. Proc. Natl. Acad. Sci. USA 1996, 93, 7352–7357. [Google Scholar] [CrossRef]
- Ryschkewitsch, C.; Jensen, P.; Hou, J.; Fahle, G.; Fischer, S.; Major, E.O. Comparison of PCR-Southern Hybridization and Quantitative Real-Time PCR for the Detection of JC and BK Viral Nucleotide Sequences in Urine and Cerebrospinal Fluid. J. Virol. Methods 2004, 121, 217–221. [Google Scholar] [CrossRef]
- Sehbani, L.; Kabamba-Mukadi, B.; Vandenbroucke, A.-T.; Bodéus, M.; Goubau, P. Specific and Quantitative Detection of Human Polyomaviruses BKV and JCV by LightCycler Real-Time PCR. J. Clin. Virol. 2006, 36, 159–162. [Google Scholar] [CrossRef]
- Tsai, R.T.; Wang, M.; Ou, W.C.; Lee, Y.L.; Li, S.Y.; Fung, C.Y.; Huang, Y.L.; Tzeng, T.Y.; Chen, Y.; Chang, D. Incidence of JC Viruria Is Higher than That of BK Viruria in Taiwan. J. Med. Virol. 1997, 52, 253–257. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Ly, M.; Bonilla, N.; Pride, D.T. The Human Urine Virome in Association with Urinary Tract Infections. Front. Microbiol. 2015, 6, 14. [Google Scholar] [CrossRef]
- Rani, A.; Ranjan, R.; McGee, H.S.; Metwally, A.; Hajjiri, Z.; Brennan, D.C.; Finn, P.W.; Perkins, D.L. A Diverse Virome in Kidney Transplant Patients Contains Multiple Viral Subtypes with Distinct Polymorphisms. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Garretto, A.; Thomas-White, K.; Wolfe, A.J.; Putonti, C. Detecting Viral Genomes in the Female Urinary Microbiome. J. Gen. Virol. 2018, 99, 1141–1146. [Google Scholar] [CrossRef]
- Moustafa, A.; Li, W.; Singh, H.; Moncera, K.J.; Torralba, M.G.; Yu, Y.; Manuel, O.; Biggs, W.; Venter, J.C.; Nelson, K.E.; et al. Microbial Metagenome of Urinary Tract Infection. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Carpenter, M.L.; Tan, S.K.; Watson, T.; Bacher, R.; Nagesh, V.; Watts, A.; Bentley, G.; Weber, J.; Huang, C.; Sahoo, M.K.; et al. Metagenomic Next-Generation Sequencing for Identification and Quantitation of Transplant-Related DNA Viruses. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Thomas-White, K.; Forster, S.C.; Kumar, N.; Van Kuiken, M.; Putonti, C.; Stares, M.D.; Hilt, E.E.; Price, T.K.; Wolfe, A.J.; Lawley, T.D. Culturing of Female Bladder Bacteria Reveals an Interconnected Urogenital Microbiota. Nat. Commun. 2018, 9, 1557. [Google Scholar] [CrossRef]
- Wommack, K.E.; Bhavsar, J.; Polson, S.W.; Chen, J.; Dumas, M.; Srinivasiah, S.; Furman, M.; Jamindar, S.; Nasko, D.J. VIROME: A Standard Operating Procedure for Analysis of Viral Metagenome Sequences. Stand. Genom. Sci. 2012, 6, 427–439. [Google Scholar] [CrossRef]
- Huson, D.H.; Weber, N. Microbial Community Analysis Using MEGAN. Methods Enzymol. 2013, 531, 465–485. [Google Scholar] [CrossRef]
- Roux, S.; Tournayre, J.; Mahul, A.; Debroas, D.; Enault, F. Metavir 2: New Tools for Viral Metagenome Comparison and Assembled Virome Analysis. BMC Bioinform. 2014, 15, 76. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function. Methods Mol. Biol. 2016, 1399, 207–233. [Google Scholar] [CrossRef]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A Multi-Classifier, Expert-Guided Approach to Detect Diverse DNA and RNA Viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.D.; Curty, G.; Xutao, D.; Hofer, C.B.; Machado, E.S.; Seuánez, H.N.; Soares, M.A.; Delwart, E.; Soares, E.A. Composite Analysis of the Virome and Bacteriome of HIV/HPV Co-Infected Women Reveals Proxies for Immunodeficiency. Viruses 2019, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-T.; Wang, H.; Wu, H.-S.; Zeng, J.; Yang, Y. Comparison of Viromes in Vaginal Secretion from Pregnant Women with and without Vaginitis. Virol. J. 2021, 18, 11. [Google Scholar] [CrossRef]
| Study | Accession No. | # Samples | # Reads Total |
|---|---|---|---|
| Kidney Transplant Virome [34] | PRJEB28510 1 | 27 | 157.5 M 3 |
| Pulmonary Tuberculosis Urinary Microbiome (unpublished) | PRJNA431965 1 | 3 | 9.5 M 4 |
| Microbial Metagenome of UTI [36] | PRJNA385350 1 | 49 | 532.0 M 4 |
| Virome in Healthy and BK Disease of Kidney Transplant (unpublished) | PRJNA587166 1 | 62 | 70.0 M 3 |
| Virome in Association with UTI [33] | cobian9680 2 | 20 | 14.5 M 3 |
| Portable Urinal Microbiome (unpublished) | PRJNA399057 1 | 4 | 228.3 M 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mormando, R.; Wolfe, A.J.; Putonti, C. Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets. Viruses 2021, 13, 1041. https://doi.org/10.3390/v13061041
Mormando R, Wolfe AJ, Putonti C. Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets. Viruses. 2021; 13(6):1041. https://doi.org/10.3390/v13061041
Chicago/Turabian StyleMormando, Rita, Alan J. Wolfe, and Catherine Putonti. 2021. "Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets" Viruses 13, no. 6: 1041. https://doi.org/10.3390/v13061041
APA StyleMormando, R., Wolfe, A. J., & Putonti, C. (2021). Discriminating between JCPyV and BKPyV in Urinary Virome Data Sets. Viruses, 13(6), 1041. https://doi.org/10.3390/v13061041






