The Portal Vertex of KSHV Promotes Docking of Capsids at the Nuclear Pores
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Viruses
2.2. Lytic Reactivation of KSHV in iSLK Infected Cells and Virion Purification
2.3. Virus Quantification
2.4. De Novo Infection
2.5. Immunofluorescence
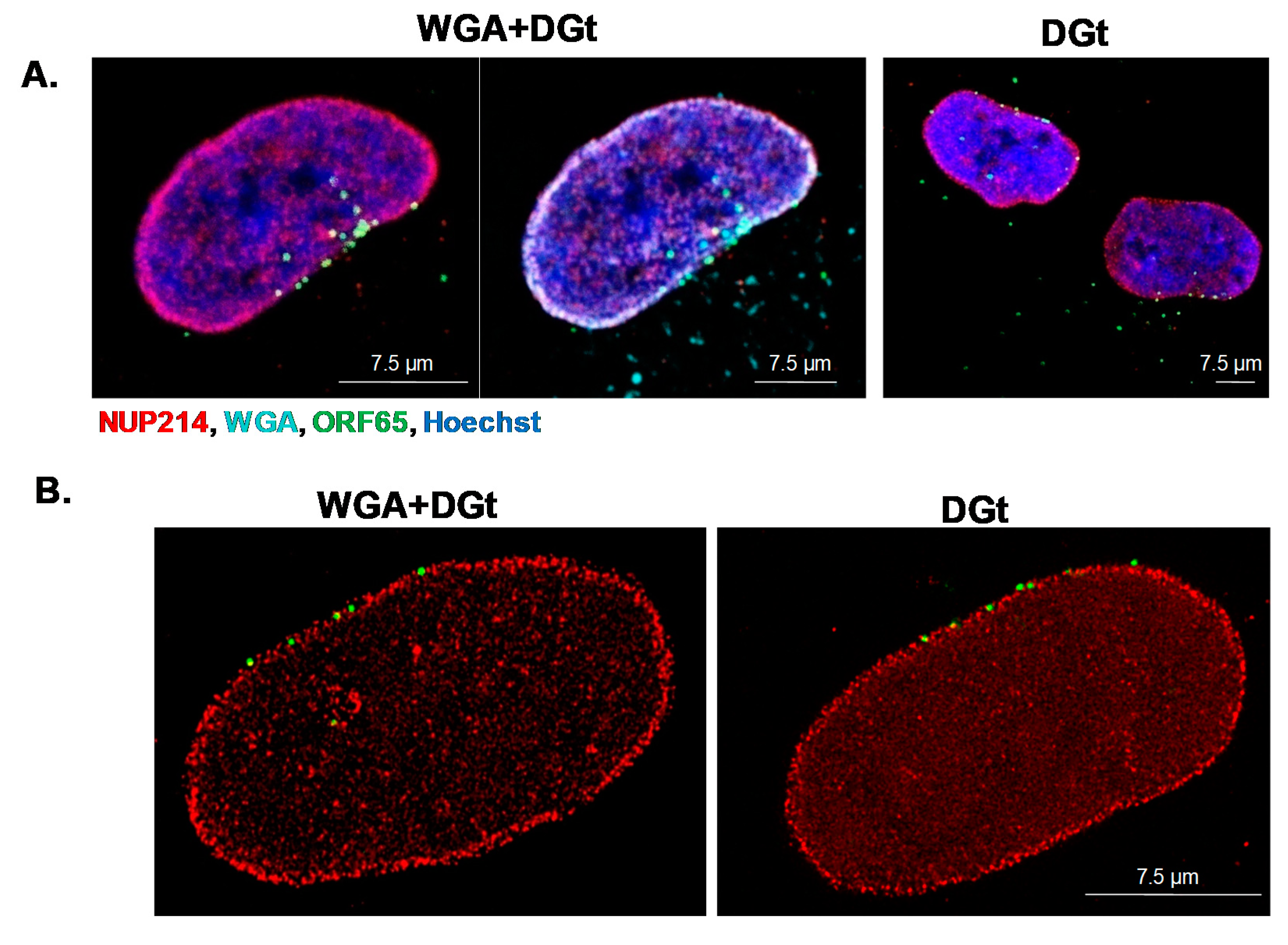
2.6. Inhibition of Nuclear Export by WGA
3. Results
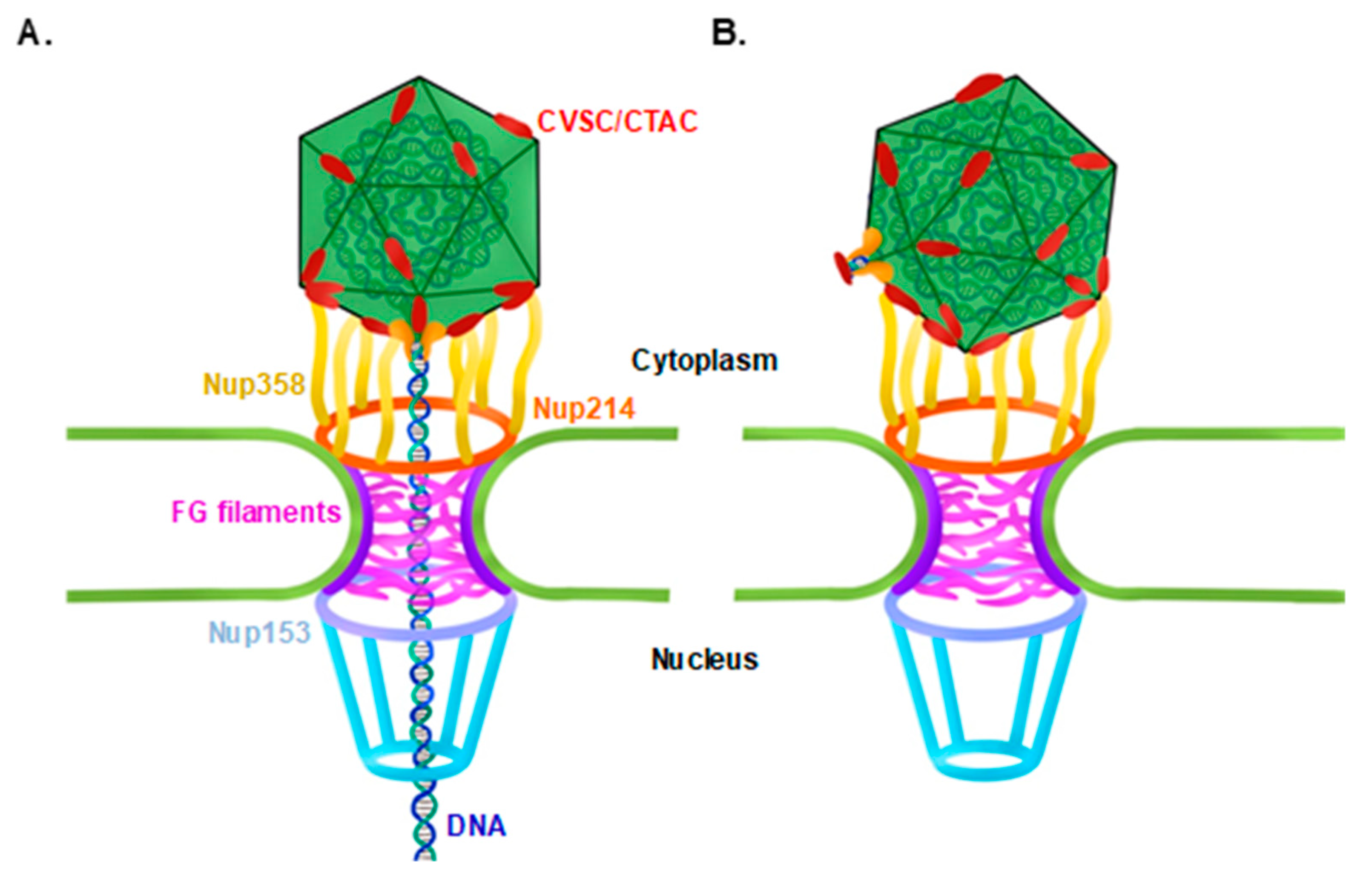
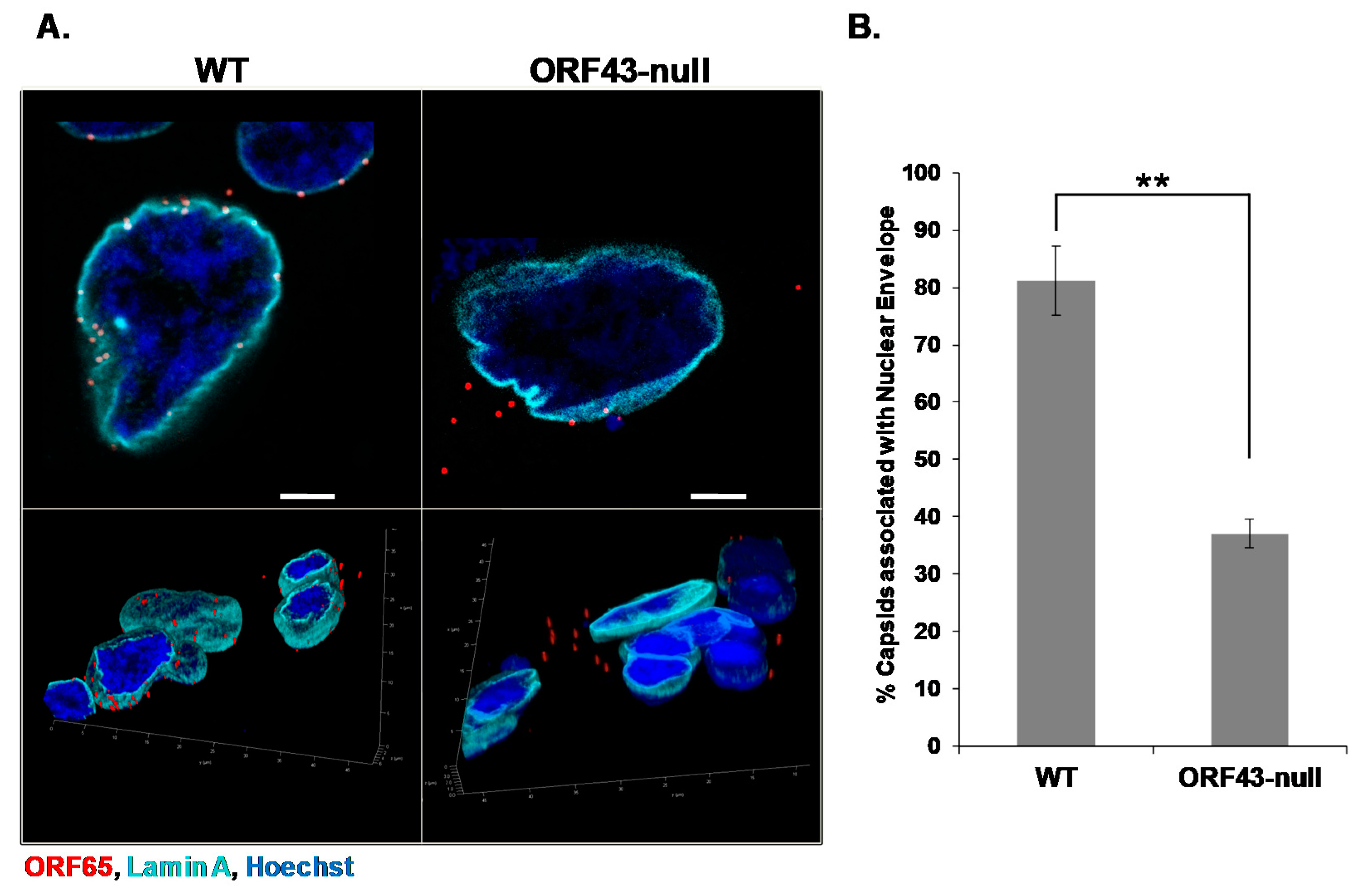
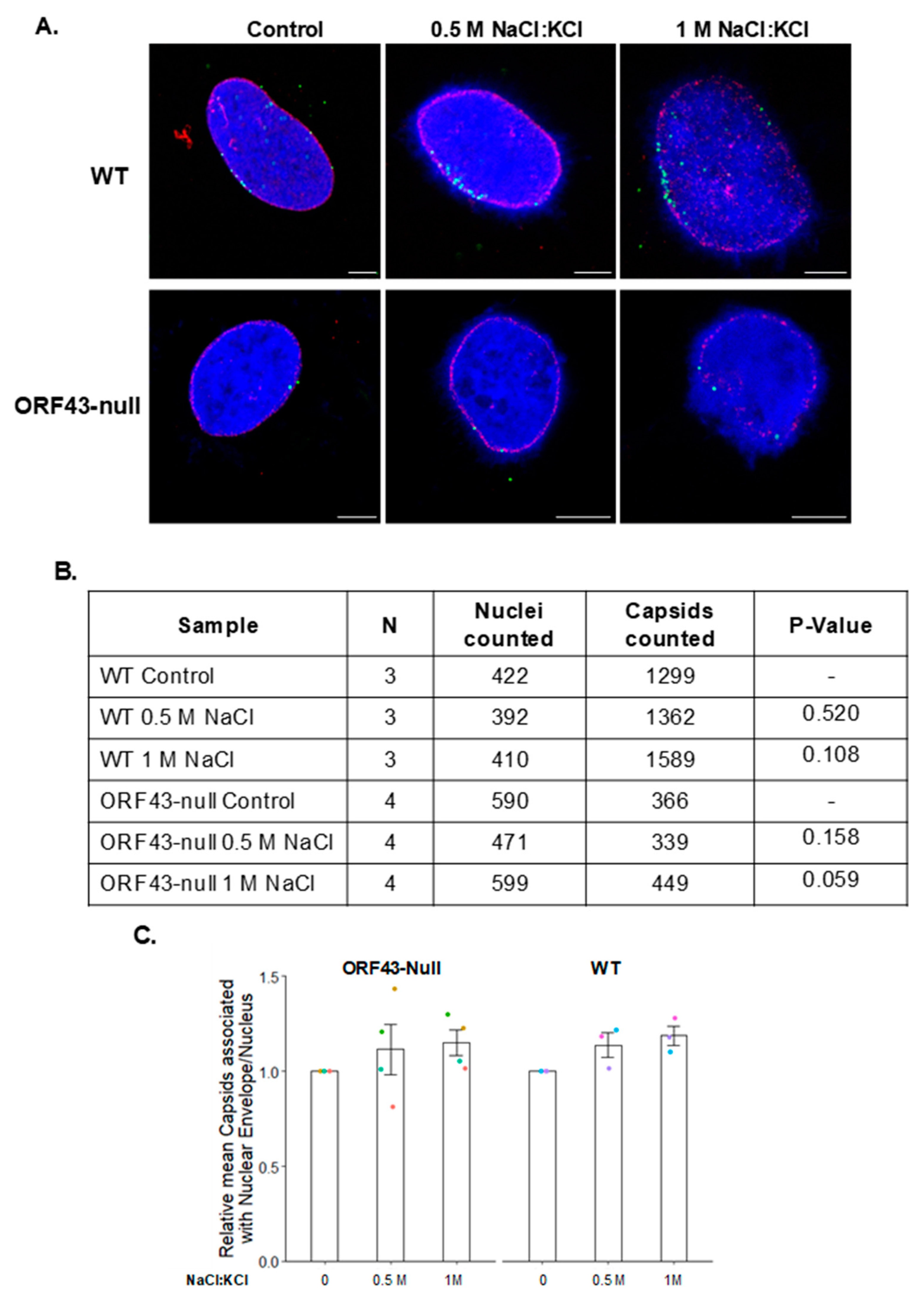
3.1. KSHV Capsids That Lack the Portal Structure Accumulate at the Nuclear Envelope at a Lower Extent than Wild-Type Capsids
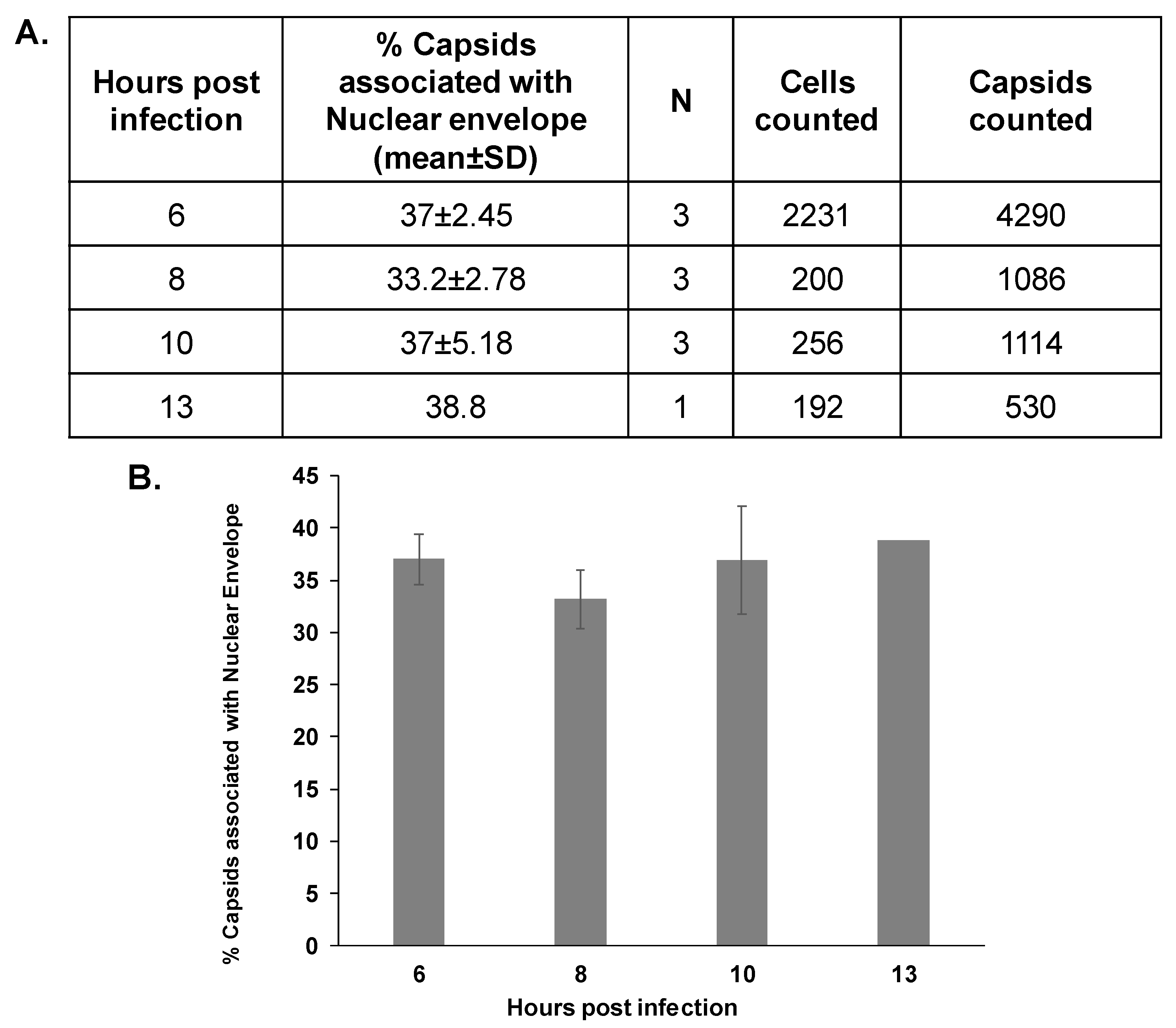
3.2. The Association Extent of ORF43-Null Capsids with the Nuclear Envelope Do Not Increase at Later Time Points Post-Infection
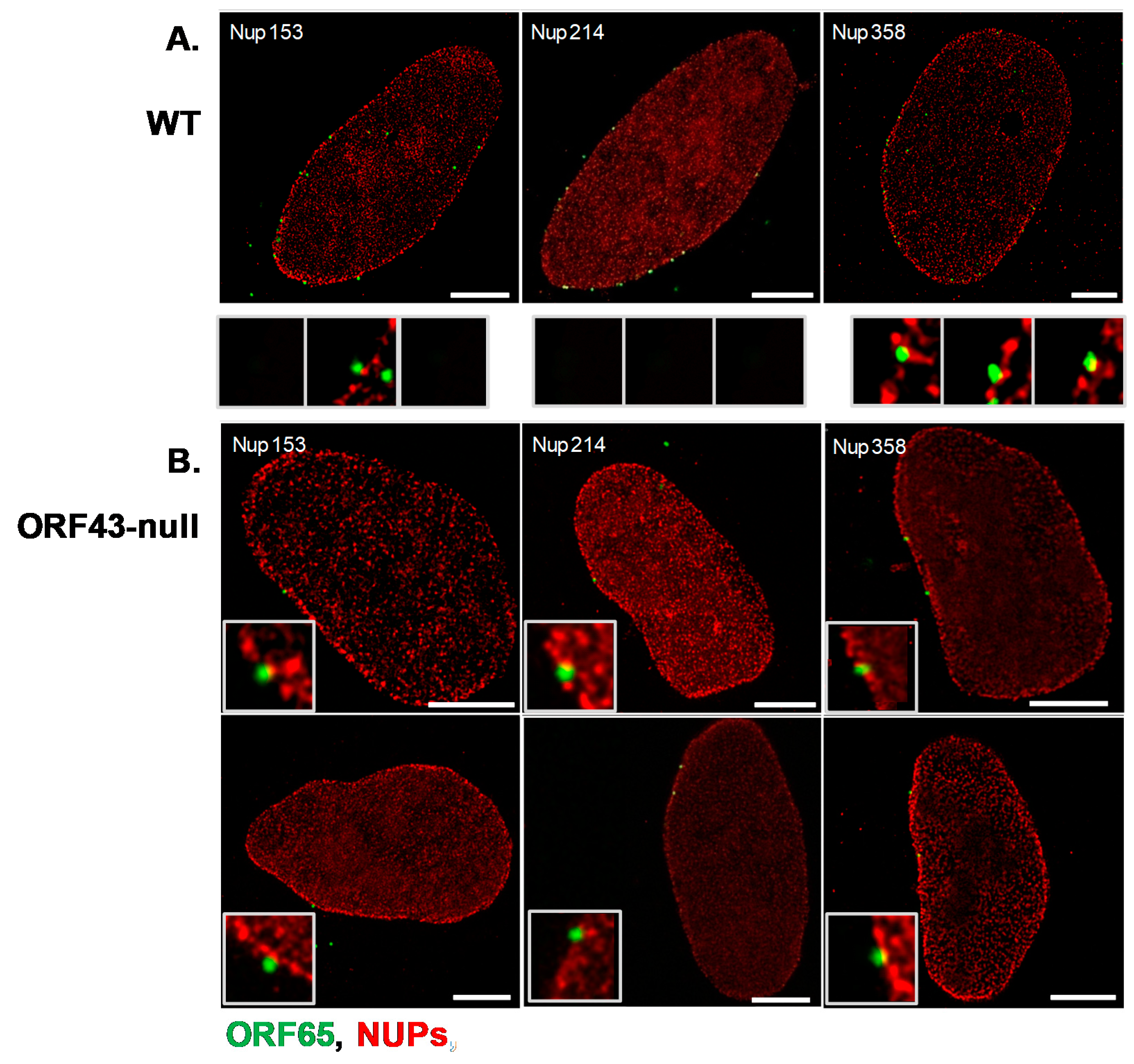
3.3. Co-Localization of WT and ORF43-Null Capsids with Nucleoporins
3.4. High Salt Treatment Does Not Alter the Proportion of Bound Capsids to the Nuclear Envelope
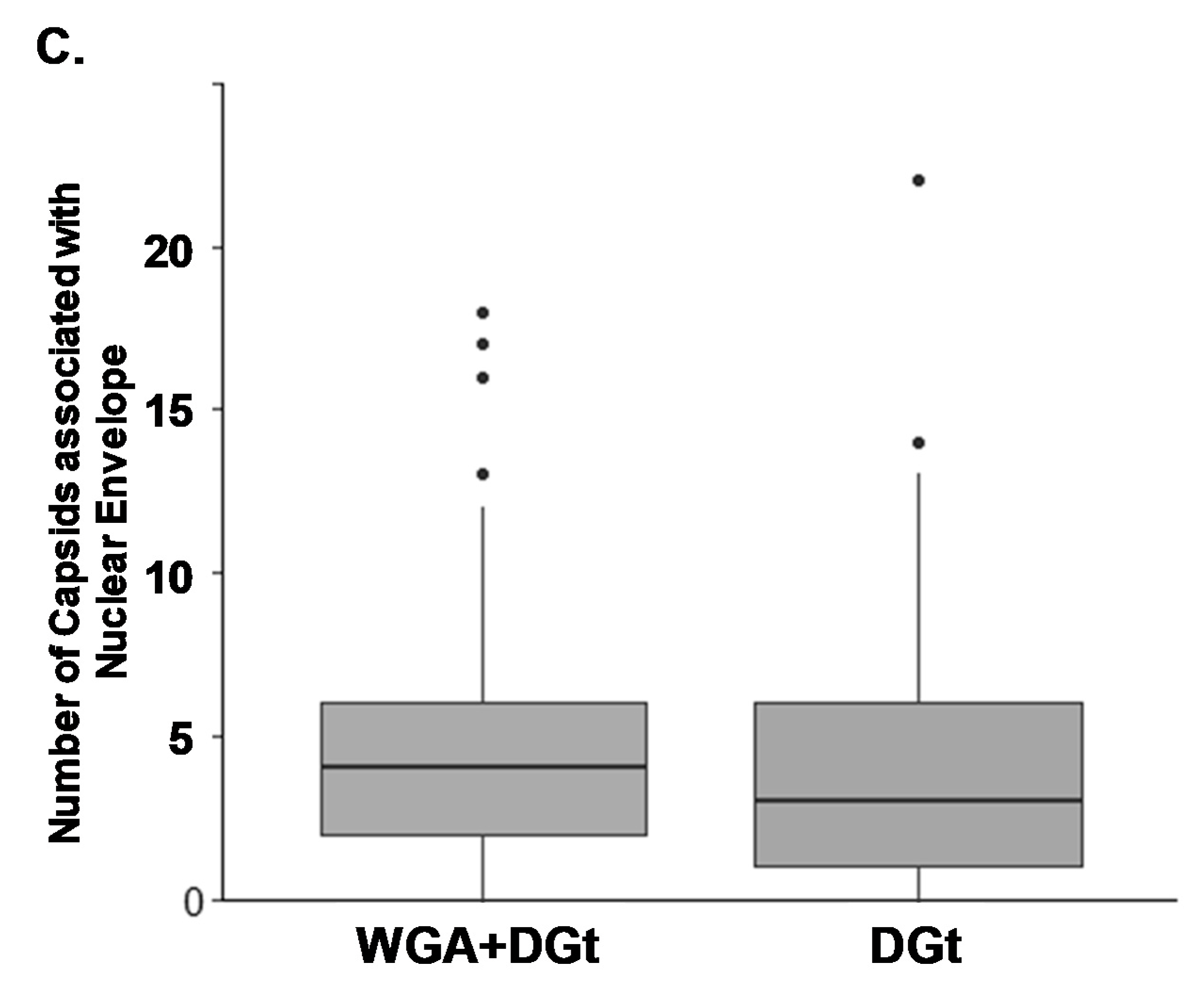
3.5. Export Inhibition Does Not Affect the Proportion of Nuclear Envelope-Associated Capsids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Kalt, I.; Masa, S.R.; Sarid, R. Linking the Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) to human malignancies. Methods Mol. Biol. 2009, 471, 387–407. [Google Scholar] [CrossRef]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef]
- Goncalves, P.H.; Ziegelbauer, J.; Uldrick, T.S.; Yarchoan, R. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr. Opin. HIV AIDS 2017, 12, 47–56. [Google Scholar] [CrossRef]
- Gramolelli, S.; Schulz, T.F. The role of Kaposi sarcoma-associated herpesvirus in the pathogenesis of Kaposi sarcoma. J. Pathol. 2015, 235, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Calabro, M.L.; Sarid, R. Human Herpesvirus 8 and Lymphoproliferative Disorders. Mediterr. J. Hematol. Infect. Dis. 2018, 10, 2018061. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; O’Connor, C.M.; Kedes, D.H.; Zhou, Z.H. Cryo-electron tomography of Kaposi’s sarcoma-associated herpesvirus capsids reveals dynamic scaffolding structures essential to capsid assembly and maturation. J. Struct. Biol. 2008, 161, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Dai, X.; Jih, J.; Liu, Y.T.; Bi, G.Q.; Sun, R. DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV. Cell 2019, 178, 1329–4312. [Google Scholar] [CrossRef]
- Liu, Y.T.; Jih, J.; Dai, X.; Bi, G.Q.; Zhou, Z.H. Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 2019, 570, 257–261. [Google Scholar] [CrossRef] [PubMed]
- McElwee, M.; Vijayakrishnan, S.; Rixon, F.; Bhella, D. Structure of the herpes simplex virus portal-vertex. PLoS Biol. 2018, 16, 2006191. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T.; Schmid, M.F.; Rixon, F.J.; Chiu, W. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 2007, 81, 2065–2068. [Google Scholar] [CrossRef] [PubMed]
- Dedeo, C.L.; Cingolani, G.; Teschke, C.M. Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses. Annu. Rev. Virol. 2019, 6, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Dunn-Kittenplon, D.D.; Kalt, I.; Lellouche, J.M.; Sarid, R. The KSHV portal protein ORF43 is essential for the production of infectious viral particles. Virology 2019, 529, 205–215. [Google Scholar] [CrossRef]
- Trus, B.L.; Cheng, N.; Newcomb, W.W.; Homa, F.L.; Brown, J.C.; Steven, A.C. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 2004, 78, 12668–12671. [Google Scholar] [CrossRef]
- Nabiee, R.; Syed, B.; Ramirez Castano, J.; Lalani, R.; Totonchy, J.E. An Update of the Virion Proteome of Kaposi Sarcoma-Associated Herpesvirus. Viruses 2020, 12, 1382. [Google Scholar] [CrossRef]
- Dai, X.; Gong, D.; Wu, T.T.; Sun, R.; Zhou, Z.H. Organization of capsid-associated tegument components in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2014, 88, 12694–12702. [Google Scholar] [CrossRef] [PubMed]
- Huet, A.; Huffman, J.B.; Conway, J.F.; Homa, F.L. Role of the Herpes Simplex Virus CVSC Proteins at the Capsid Portal Vertex. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Trus, B.L.; Heymann, J.B.; Nealon, K.; Cheng, N.; Newcomb, W.W.; Brown, J.C. Capsid structure of Kaposi’s sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 2001, 75, 2879–2890. [Google Scholar] [CrossRef] [PubMed]
- Nealon, K.; Newcomb, W.W.; Pray, T.R.; Craik, C.S.; Brown, J.C.; Kedes, D.H. Lytic replication of Kaposi’s sarcoma-associated herpesvirus results in the formation of multiple capsid species: Isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 2001, 75, 2866–2878. [Google Scholar] [CrossRef][Green Version]
- Tandon, R.; Mocarski, E.S.; Conway, J.F. The A, B, Cs of herpesvirus capsids. Viruses 2015, 7, 899–914. [Google Scholar] [CrossRef]
- Dai, X.; Gong, D.; Lim, H.; Jih, J.; Wu, T.T.; Sun, R. Structure and mutagenesis reveal essential capsid protein interactions for KSHV replication. Nature 2018, 553, 521–525. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, Z.H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 360. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, W.; Zhu, L.; Zhu, D.; Feng, R.; Wang, J. Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation. Protein Cell 2020, 11, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yuan, S.; Zhu, D.; Tang, H.; Wang, N.; Chen, W. Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component. Nat. Commun. 2018, 9, 3668. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, J.; Zhu, D.; Wang, N.; Gao, Q.; Chen, W. Cryo-EM structure of a herpesvirus capsid at 3.1 A. Science 2018, 360. [Google Scholar] [CrossRef] [PubMed]
- Machon, C.; Fabrega-Ferrer, M.; Zhou, D.; Cuervo, A.; Carrascosa, J.L.; Stuart, D.I. Atomic structure of the Epstein-Barr virus portal. Nat. Commun. 2019, 10, 3891. [Google Scholar] [CrossRef]
- Visalli, R.J.; Schwartz, A.M.; Patel, S.; Visalli, M.A. Identification of the Epstein Barr Virus portal. Virology 2019, 529, 152–159. [Google Scholar] [CrossRef]
- Sun, J.; Liu, C.; Peng, R.; Zhang, F.K.; Tong, Z.; Liu, S. Cryo-EM structure of the varicella-zoster virus A-capsid. Nat. Commun. 2020, 11, 4795. [Google Scholar] [CrossRef]
- Huffman, J.B.; Daniel, G.R.; Falck-Pedersen, E.; Huet, A.; Smith, G.A.; Conway, J.F. The C Terminus of the Herpes Simplex Virus UL25 Protein Is Required for Release of Viral Genomes from Capsids Bound to Nuclear Pores. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Pasdeloup, D.; Blondel, D.; Isidro, A.L.; Rixon, F.J. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 2009, 83, 6610–6623. [Google Scholar] [CrossRef]
- Jovasevic, V.; Liang, L.; Roizman, B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 2008, 82, 3311–3319. [Google Scholar] [CrossRef] [PubMed]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes simplex virus replication: Roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef]
- Flatt, J.W.; Greber, U.F. Misdelivery at the Nuclear Pore Complex-Stopping a Virus Dead in Its Tracks. Cells 2015, 4, 277–296. [Google Scholar] [CrossRef]
- Bauer, D.W.; Huffman, J.B.; Homa, F.L.; Evilevitch, A. Herpes virus genome, the pressure is on. J. Am. Chem. Soc. 2013, 135, 11216–11221. [Google Scholar] [CrossRef] [PubMed]
- Brandariz-Nunez, A.; Liu, T.; Du, T.; Evilevitch, A. Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection. elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Brandariz-Nunez, A.; Robinson, S.J.; Evilevitch, A. Pressurized DNA state inside herpes capsids-A novel antiviral target. PLoS Pathog. 2020, 16, 1008604. [Google Scholar] [CrossRef]
- Gelbart, W.M.; Knobler, C.M. Virology. Pressurized viruses. Science 2009, 323, 1682–1683. [Google Scholar] [CrossRef]
- Sekine, E.; Schmidt, N.; Gaboriau, D.; O’Hare, P. Spatiotemporal dynamics of HSV genome nuclear entry and compaction state transitions using bioorthogonal chemistry and super-resolution microscopy. PLoS Pathog. 2017, 13, 1006721. [Google Scholar] [CrossRef] [PubMed]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef]
- Ye, F.; Zhou, F.; Bedolla, R.G.; Jones, T.; Lei, X.; Kang, T. Reactive oxygen species hydrogen peroxide mediates Kaposi’s sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog. 2011, 7, 1002054. [Google Scholar] [CrossRef]
- Ben-Yishay, R.; Mor, A.; Shraga, A.; Ashkenazy-Titelman, A.; Kinor, N.; Schwed-Gross, A. Imaging within single NPCs reveals NXF1’s role in mRNA export on the cytoplasmic side of the pore. J. Cell Biol. 2019, 218, 2962–2981. [Google Scholar] [CrossRef] [PubMed]
- Suntharalingam, M.; Wente, S.R. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev Cell 2003, 4, 775–789. [Google Scholar] [CrossRef]
- Finlay, D.R.; Newmeyer, D.D.; Price, T.M.; Forbes, D.J. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 1987, 104, 189–200. [Google Scholar] [CrossRef]
- Marchetti, M.; Mastromarino, P.; Rieti, S.; Seganti, L.; Orsi, N. Inhibition of herpes simplex, rabies and rubella viruses by lectins with different specificities. Res. Virol. 1995, 146, 211–215. [Google Scholar] [CrossRef]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes simplex virus type 1 entry into host cells: Reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; Suliman, S.; Ben-Yishay, R.; Yunger, S.; Brody, Y.; Shav-Tal, Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010, 12, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Kylberg, K.; Bjork, P.; Fomproix, N.; Ivarsson, B.; Wieslander, L.; Daneholt, B. Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp. Cell Res. 2010, 316, 1028–1038. [Google Scholar] [CrossRef]
- Flatt, J.W.; Greber, U.F. Viral mechanisms for docking and delivering at nuclear pore complexes. Semin. Cell Dev. Biol. 2017, 68, 59–71. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Breaching the Barrier-The Nuclear Envelope in Virus Infection. J. Mol. Biol. 2016, 428, 1949–1961. [Google Scholar] [CrossRef]
- Fay, N.; Pante, N. Nuclear entry of DNA viruses. Front Microbiol 2015, 6, 467. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Au, S.; Pante, N. How viruses access the nucleus. Biochim. Biophys. Acta 2011, 1813, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Etingov, I.; Pante, N. Effect of viral infection on the nuclear envelope and nuclear pore complex. Int. Rev. Cell Mol. Biol. 2012, 299, 117–159. [Google Scholar] [CrossRef]
- Strunze, S.; Engelke, M.F.; Wang, I.H.; Puntener, D.; Boucke, K.; Schleich, S. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Flatt, J.W.; Seiler, D.; Cardel, B.; Emmenlauer, M.; Boucke, K. The E3 Ubiquitin Ligase Mind Bomb 1 Controls Adenovirus Genome Release at the Nuclear Pore Complex. Cell Rep. 2019, 29, 3785–3795. [Google Scholar] [CrossRef]
- Cohen, S.; Pante, N. Pushing the envelope: Microinjection of Minute virus of mice into Xenopus oocytes causes damage to the nuclear envelope. J. Gen. Virol. 2005, 86, 3243–3252. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Marr, A.K.; Garcin, P.; Pante, N. Nuclear envelope disruption involving host caspases plays a role in the parvovirus replication cycle. J. Virol. 2011, 85, 4863–4874. [Google Scholar] [CrossRef]
- Yamada, M.; Kasamatsu, H. Role of nuclear pore complex in simian virus 40 nuclear targeting. J. Virol. 1993, 67, 119–130. [Google Scholar] [CrossRef]
- Kuksin, D.; Norkin, L.C. Disassociation of the SV40 genome from capsid proteins prior to nuclear entry. Virol. J. 2012, 9, 158. [Google Scholar] [CrossRef]
- Lee, K.; Ambrose, Z.; Martin, T.D.; Oztop, I.; Mulky, A.; Julias, J.G. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233. [Google Scholar] [CrossRef]
- Cros, J.F.; Garcia-Sastre, A.; Palese, P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 2005, 6, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.W.; Weaver, L.L.; Pante, N. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. J. Mol. Biol. 2007, 374, 910–916. [Google Scholar] [CrossRef]
- Muhlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J. Virol. 2015, 89, 6009–6021. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.M.; Wang, L.; Damania, B. Kaposi’s sarcoma-associated herpesvirus encodes a viral deubiquitinase. J. Virol. 2009, 83, 10224–10233. [Google Scholar] [CrossRef]
- Inn, K.S.; Lee, S.H.; Rathbun, J.Y.; Wong, L.Y.; Toth, Z.; Machida, K. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 2011, 85, 10899–10904. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Sollars, P.J.; Pickard, G.E.; Smith, G.A. Visualizing Herpesvirus Procapsids in Living Cells. J. Virol. 2016, 90, 10182–10192. [Google Scholar] [CrossRef] [PubMed]
- Sheaffer, A.K.; Newcomb, W.W.; Brown, J.C.; Gao, M.; Weller, S.K.; Tenney, D.J. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 2000, 74, 6838–6848. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Newcomb, W.W.; Cheng, N.; Aksyuk, A.; Winkler, D.C.; Steven, A.C. Internal Proteins of the Procapsid and Mature Capsids of Herpes Simplex Virus 1 Mapped by Bubblegram Imaging. J. Virol. 2016, 90, 5176–5186. [Google Scholar] [CrossRef][Green Version]
- Wang, I.H.; Burckhardt, C.J.; Yakimovich, A.; Greber, U.F. Imaging, Tracking and Computational Analyses of Virus Entry and Egress with the Cytoskeleton. Viruses 2018, 10, 166. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dünn-Kittenplon, D.; Ashkenazy-Titelman, A.; Kalt, I.; Lellouche, J.-P.; Shav-Tal, Y.; Sarid, R. The Portal Vertex of KSHV Promotes Docking of Capsids at the Nuclear Pores. Viruses 2021, 13, 597. https://doi.org/10.3390/v13040597
Dünn-Kittenplon D, Ashkenazy-Titelman A, Kalt I, Lellouche J-P, Shav-Tal Y, Sarid R. The Portal Vertex of KSHV Promotes Docking of Capsids at the Nuclear Pores. Viruses. 2021; 13(4):597. https://doi.org/10.3390/v13040597
Chicago/Turabian StyleDünn-Kittenplon, Daniela, Asaf Ashkenazy-Titelman, Inna Kalt, Jean-Paul Lellouche, Yaron Shav-Tal, and Ronit Sarid. 2021. "The Portal Vertex of KSHV Promotes Docking of Capsids at the Nuclear Pores" Viruses 13, no. 4: 597. https://doi.org/10.3390/v13040597
APA StyleDünn-Kittenplon, D., Ashkenazy-Titelman, A., Kalt, I., Lellouche, J.-P., Shav-Tal, Y., & Sarid, R. (2021). The Portal Vertex of KSHV Promotes Docking of Capsids at the Nuclear Pores. Viruses, 13(4), 597. https://doi.org/10.3390/v13040597





