Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement
Abstract
1. Introduction
2. Material and Methods
2.1. Cells and Viruses
2.2. Antibody Purification
2.3. Normal Human Serum
2.4. Plaque Assay
2.5. Serum-Sensitivity Assay
2.6. RT-PCR
2.7. Virus Capture Assay
2.8. Flow Cytometric Analysis
2.9. Western Blot
2.10. Statistical Analyses
3. Results
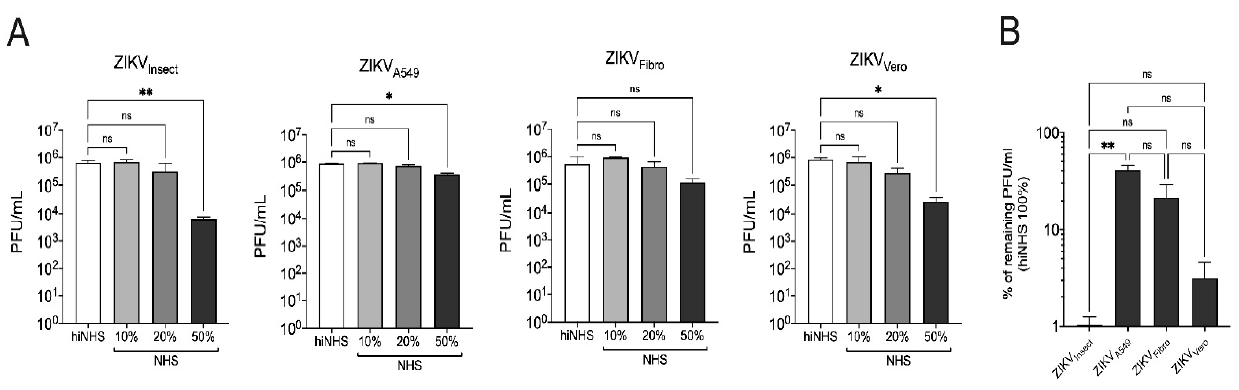
3.1. Cell Line-Dependent ZIKV Lysis
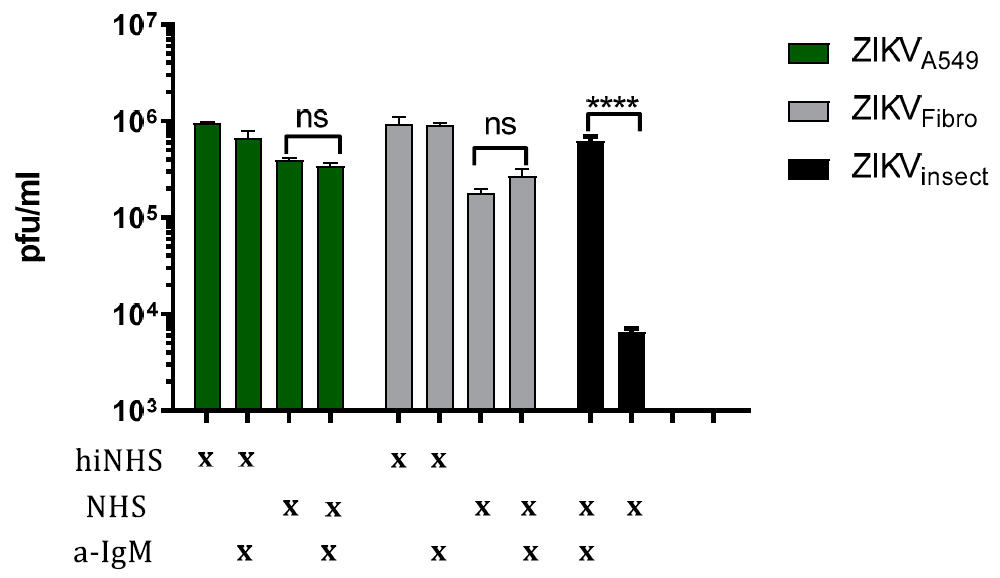
3.2. Complement Activation Is Not Driven by IgM
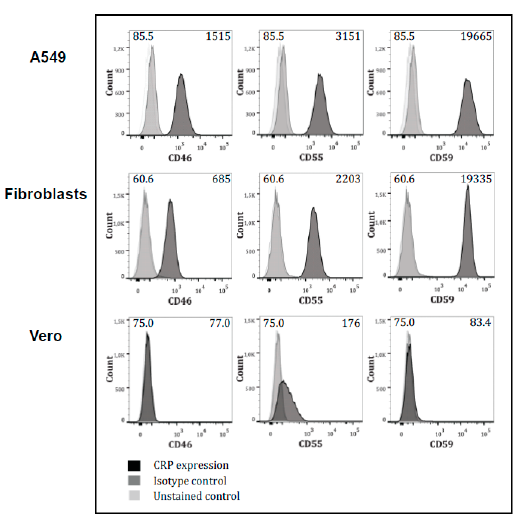
3.3. Expression of Complement-Regulators on the Cells
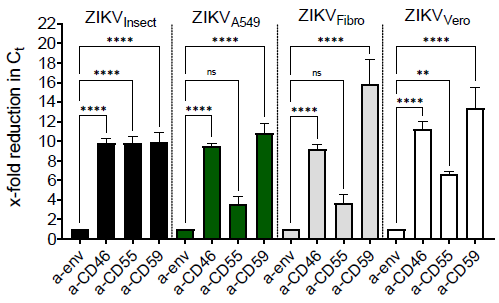
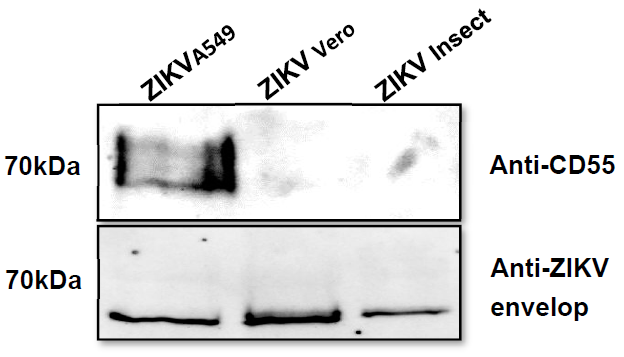
3.4. Incorporation of Complement-Regulator CD55 into the Viral Envelope
3.5. Membrane-Bound Regulators Provide Additionally Serum Resistance
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Faye, O.; Freire, C.C.; Iamarino, A.; Faye, O.; de Oliveira, J.V.; Diallo, M.; Zanotto, P.M. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl. Trop. Dis. 2014, 8, e2636. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; DuBray, C.; et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef]
- Subissi, L.; Dub, T.; Besnard, M.; Mariteragi-Helle, T.; Nhan, T.; Lutringer-Magnin, D.; Barboza, P.; Gurry, C.; Brindel, P.; Nilles, E.J.; et al. Zika Virus Infection during Pregnancy and Effects on Early Childhood Development, French Polynesia, 2013–2016. Emerg. Infect. Dis. 2018, 24, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Krow-Lucal, E.R.; de Andrade, M.R.; Cananéa, J.N.A.; Moore, C.A.; Leite, P.L.; Biggerstaff, B.J.; Cabral, C.M.; Itoh, M.; Percio, J.; Wada, M.Y.; et al. Association and birth prevalence of microcephaly attributable to Zika virus infection among infants in Paraíba, Brazil, in 2015–2016: A case-control study. Lancet Child. Adolesc. Health 2018, 2, 205–213. [Google Scholar] [CrossRef]
- Musso, D.; Ko, A.I.; Baud, D. Zika Virus Infection-After the Pandemic. Reply. N. Engl. J. Med. 2020, 382, e3. [Google Scholar]
- Hasan, S.S.; Sevvana, M.; Kuhn, R.J.; Rossmann, M.G. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018, 25, 13–20. [Google Scholar] [CrossRef]
- Guo, M.; Hui, L.; Nie, Y.; Tefsen, B.; Wu, Y. ZIKV viral proteins and their roles in virus-host interactions. Sci. China Life Sci. 2020, 14, 1–11. [Google Scholar] [CrossRef]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.A.; et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir. Res. 2010, 87, 125–148. [Google Scholar] [CrossRef]
- Avirutnan, P.; Mehlhop, E.; Diamond, M.S. Complement and its role in protection and pathogenesis of flavivirus infections. Vaccine 2008, 26 (Suppl. S8), I100–I107. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.N.; Silva, E.M.; Barbosa, A.S.; Mohana-Borges, R. The Complement System in Flavivirus Infections. Front. Microbiol. 2017, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Schiela, B.; Bernklau, S.; Malekshahi, Z.; Deutschmann, D.; Koske, I.; Banki, Z.; Thielens, N.M.; Würzner, R.; Speth, C.; Weiss, G.; et al. Active Human Complement Reduces the Zika Virus Load via Formation of the Membrane-Attack Complex. Front. Immunol. 2018, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Malekshahi, Z.; Schiela, B.; Bernklau, S.; Banki, Z.; Würzner, R.; Stoiber, H. Interference of the Zika Virus E-Protein With the Membrane Attack Complex of the Complement System. Front. Immunol. 2020, 11, 69549. [Google Scholar] [CrossRef]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part I-Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6, 62. [Google Scholar] [CrossRef]
- Merle, N.S.; Noe, R.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6, 57. [Google Scholar] [CrossRef]
- Rus, H.; Cudrici, C.; Niculescu, F. The role of the complement system in innate immunity. Immunol. Res. 2005, 33, 103–112. [Google Scholar] [CrossRef]
- Thurman, J.M.; Holers, V.M. The central role of the alternative complement pathway in human disease. J. Immunol. 2006, 176, 1305–1310. [Google Scholar] [CrossRef]
- Bayly-Jones, C.; Bubeck, D.; Dunstone, M.A. The mystery behind membrane insertion: A review of the complement membrane attack complex. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160221. [Google Scholar] [CrossRef]
- Zipfel, P.F.; Skerka, C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009, 9, 729–740. [Google Scholar] [CrossRef]
- Lambris, J.D.; Ricklin, D.; Geisbrecht, B.V. Complement evasion by human pathogens. Nat. Rev. Microbiol. 2008, 6, 132–142. [Google Scholar] [CrossRef]
- Mellors, J.; Tipton, T.; Longet, S.; Carroll, M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front. Immunol. 2020, 11, 450. [Google Scholar] [CrossRef]
- Stoiber, H.; Pruenster, M.; Ammann, C.G.; Dierich, M.P. Complement-opsonized HIV: The free rider on its way to infection. Mol. Immunol. 2005, 42, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, U.; Sutter, K.; Kassiotis, G.; Zelinskyy, G.; Bánki, Z.; Stoiber, H.; Santiago, M.L.; Hasenkrug, K.J. Friend retrovirus studies reveal complex interactions between intrinsic, innate and adaptive immunity. FEMS Microbiol. Rev. 2019, 43, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Rosengard, A.M.; Liu, Y.; Nie, Z.; Jimenez, R. Variola virus immune evasion design: Expression of a highly efficient inhibitor of human complement. Proc. Natl. Acad. Sci. USA 2002, 99, 8808–8813. [Google Scholar] [CrossRef]
- Sfyroera, G.; Katragadda, M.; Morikis, D.; Isaacs, S.N.; Lambris, J.D. Electrostatic Modeling Predicts the Activities of Orthopoxvirus Complement Control Proteins. J. Immunol. 2005, 174, 2143–2151. [Google Scholar] [CrossRef]
- Mullick, J.; Bernet, J.; Singh, A.K.; Lambris, J.D.; Sahu, A. Kaposi’s Sarcoma-Associated Herpesvirus (Human Herpesvirus 8) Open Reading Frame 4 Protein (Kaposica) Is a Functional Homolog of Complement Control Proteins. J. Virol. 2003, 77, 3878–3881. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, A.; Steinmann, E.; Bánki, Z.; Khalid, A.S.; Lengauer, S.; Wilhelm, C.; Zoller, H.; Schloegl, A.; Steinmann, J. Specific acquisition of functional CD59 but not CD46 or CD55 by hepatitis C virus. PLoS ONE 2012, 7, e45770. [Google Scholar] [CrossRef]
- Conde, J.N.; da Silva, E.M.; Allonso, D.; Coelho, D.R.; Andrade, I.D.S.; de Medeiros, L.N.; Barbosa, A.S.; Mohana-Borges, R. Inhibition of the Membrane Attack Complex by Dengue Virus NS1 through Interaction with Vitronectin and Terminal Complement Proteins. J. Virol. 2016, 90, 9570–9581. [Google Scholar] [CrossRef]
- Pipperger, L.; Koske, I.; Wild, N.; Müllauer, B.; Krenn, D.; Stoiber, H.; Wollmann, G.; Kimpel, J.; Von Laer, D.; Bánki, Z. Xenoantigen-Dependent Complement-Mediated Neutralization of Lymphocytic Choriomeningitis Virus Glycoprotein-Pseudotyped Vesicular Stomatitis Virus in Human Serum. J. Virol. 2019, 93, e00567-19. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.L.; Lublin, D.M.; Morgan, B.P. Efficient generation of monoclonal antibodies for specific protein domains using recombinant immunoglobulin fusion proteins: Pitfalls and solutions. J. Immunol. Methods 2002, 268, 245–258. [Google Scholar] [CrossRef]
- Bonnon, C.; Wendeler, M.W.; Paccaud, J.P.; Hauri, H.P. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 2010, 123 Pt 10, 1705–1715. [Google Scholar] [CrossRef]
- Amet, T.; Ghabril, M.; Chalasani, N.; Byrd, D.; Hu, N.; Grantham, A.; Liu, Z.; Qin, X.; He, J.J.; Yu, Q. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology 2012, 55, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, B.; Kim, H.; Meyer, K.; Bose, S.K.; Di Bisceglie, A.M.; Ray, R.B.; Diamond, M.S.; Atkinson, J.P. Hepatitis C virus infection upregulates CD55 expression on the hepatocyte surface and promotes association with virus particles. J. Virol. 2013, 87, 7902–7910. [Google Scholar] [CrossRef]
- Johnson, J.B.; Grant, K.; Parks, G.D. The Paramyxoviruses Simian Virus 5 and Mumps Virus Recruit Host Cell CD46 To Evade Complement-Mediated Neutralization. J. Virol. 2009, 83, 7602–7611. [Google Scholar] [CrossRef]
- Saifuddin, M.; Hedayati, T.; Atkinson, J.P.; Holguin, M.H.; Parker, C.J.; Spear, G.T. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. J. Gen. Virol. 1997, 78 Pt 8, 1907–1911. [Google Scholar] [CrossRef]
- Montefiori, D.C.; Cornell, R.J.; Zhou, J.Y.; Zhou, J.T.; Hirsch, V.M.; Johnson, P.R. Complement control proteins, CD46, CD55, and CD59, as common surface constituents of human and simian immunodeficiency viruses and possible targets for vaccine protection. Virology 1994, 205, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Marschang, P.; Sodroski, J.; Wurzner, R.; Dierich, M.P. Decay-accelerating factor (CD55) protects human immunodeficiency virus type 1 from inactivation by human complement. Eur. J. Immunol. 1995, 25, 285–290. [Google Scholar] [CrossRef]
- Nakagawa, M.; Mizuno, M.; Kawada, M.; Uesu, T.; Nasu, J.; Takeuchi, K.; Okada, H.; Endo, Y.; Fujita, T.; Tsuji, T. Polymorphic expression of decay-accelerating factor in human colorectal cancer. J. Gastroenterol. Hepatol. 2001, 16, 184–189. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malekshahi, Z.; Bernklau, S.; Schiela, B.; Koske, I.; Banki, Z.; Stiasny, K.; Harris, C.L.; Würzner, R.; Stoiber, H. Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement. Viruses 2021, 13, 510. https://doi.org/10.3390/v13030510
Malekshahi Z, Bernklau S, Schiela B, Koske I, Banki Z, Stiasny K, Harris CL, Würzner R, Stoiber H. Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement. Viruses. 2021; 13(3):510. https://doi.org/10.3390/v13030510
Chicago/Turabian StyleMalekshahi, Zahra, Sarah Bernklau, Britta Schiela, Iris Koske, Zoltan Banki, Karin Stiasny, Claire L. Harris, Reinhard Würzner, and Heribert Stoiber. 2021. "Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement" Viruses 13, no. 3: 510. https://doi.org/10.3390/v13030510
APA StyleMalekshahi, Z., Bernklau, S., Schiela, B., Koske, I., Banki, Z., Stiasny, K., Harris, C. L., Würzner, R., & Stoiber, H. (2021). Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement. Viruses, 13(3), 510. https://doi.org/10.3390/v13030510





