Improving the Inhibitory Effect of Phages against Pseudomonas aeruginosa Isolated from a Burn Patient Using a Combination of Phages and Antibiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. P. aeruginosa Strains
2.2. Minimum Inhibitory Concentration
2.3. Isolation of Phages
2.4. Storage and Stability of Phages
2.4.1. Temperature and pH
2.4.2. Burn Ointments
2.5. Single-Step Growth Curve
2.6. Adsorption Degree
2.7. Phage–Antibiotic Combination
2.8. DNA Extraction and Sequencing
2.9. Genome Assembly and Annotation
2.10. Phylogenetic Analyses
2.11. Transmission Electron Microscopy (TEM)
3. Results
3.1. Phage Isolation and Selection
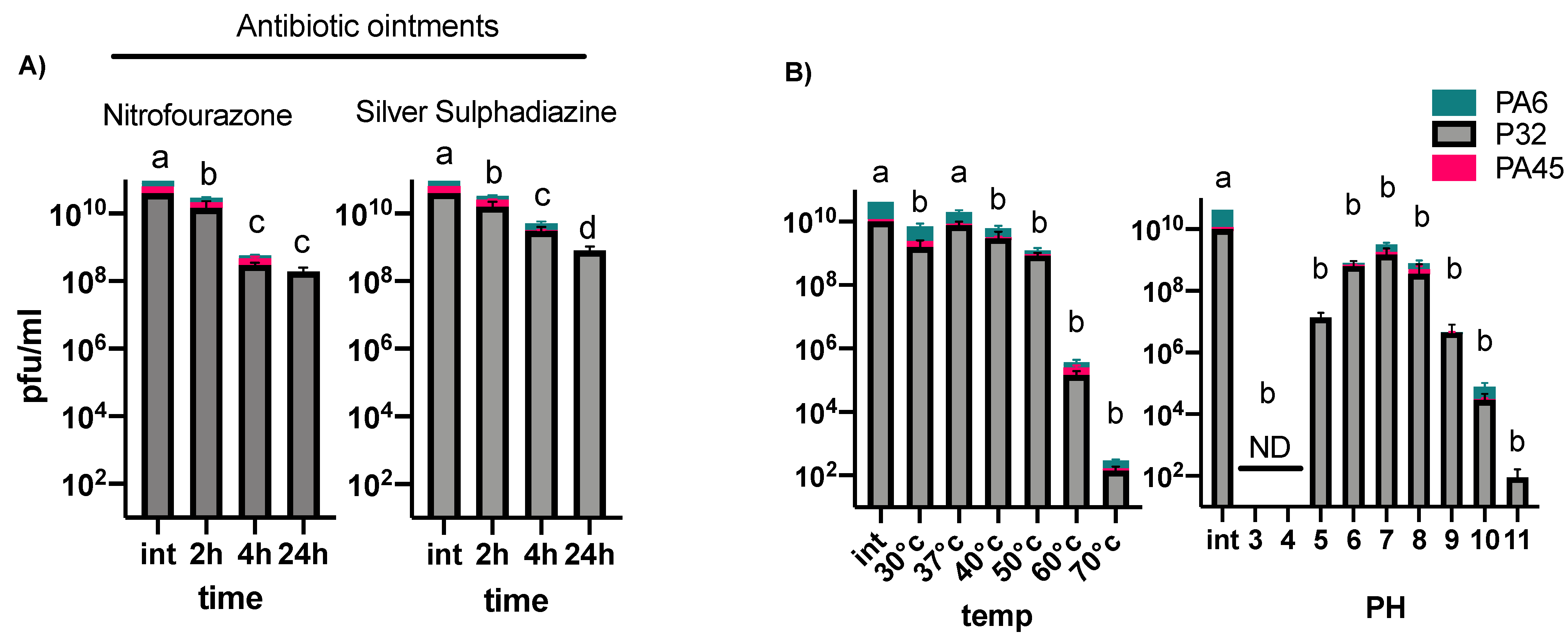
3.2. Phage Stability under Different Temperatures, pH Conditions, and in Burn Ointments
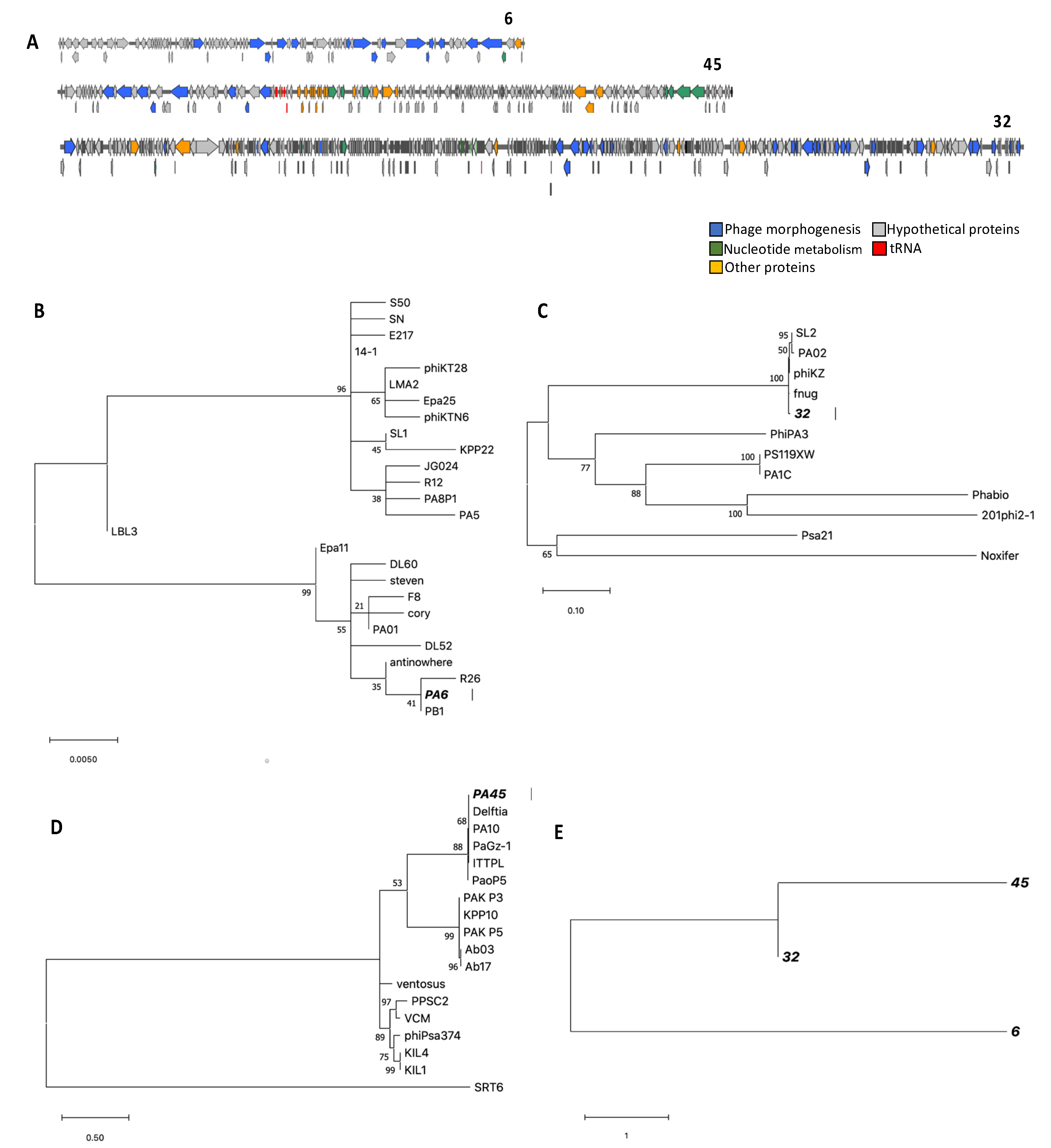
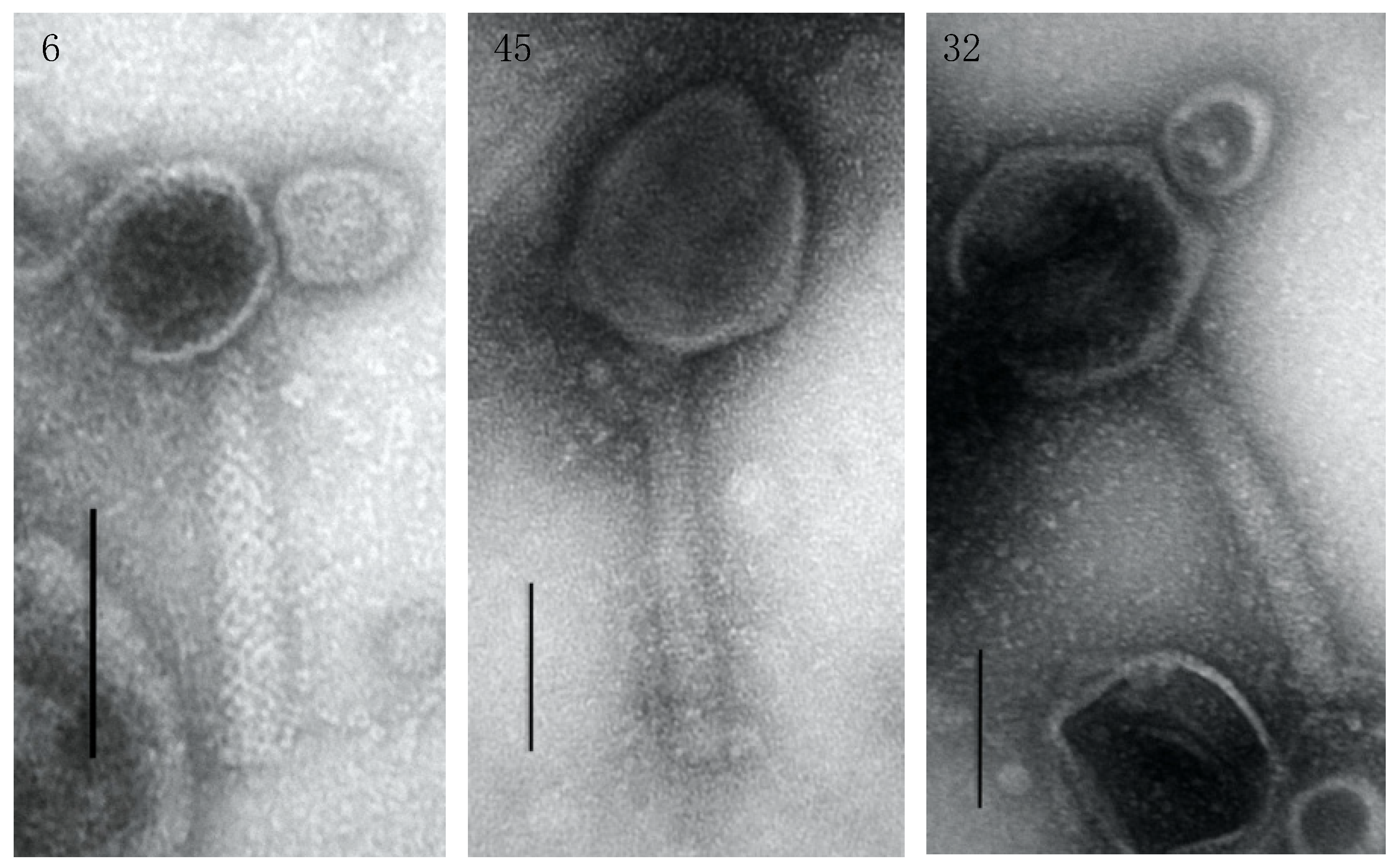
3.3. Genome Characterization, Annotation, DNA Homology, Phylogeny, and Morphology
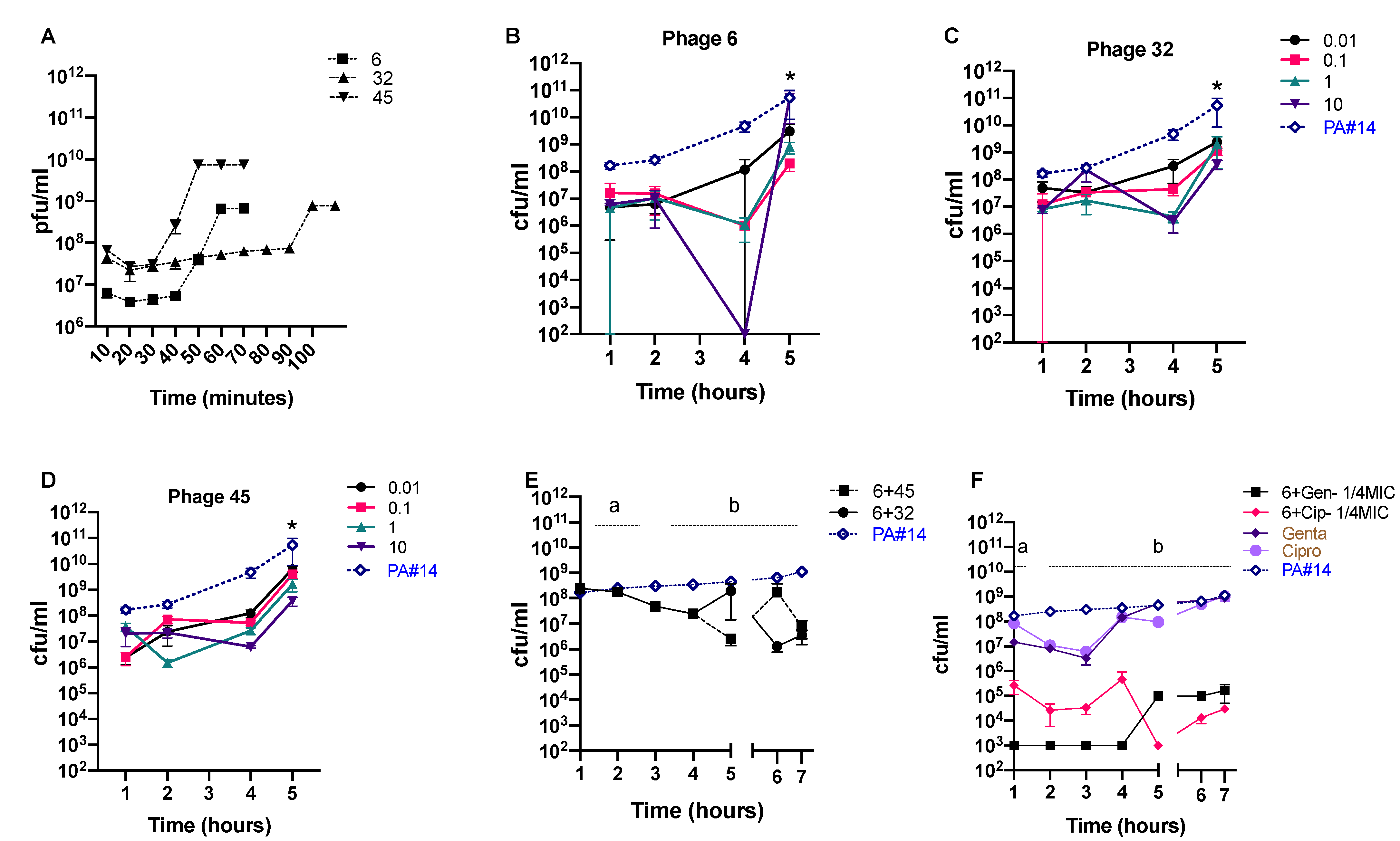
3.4. In Vitro Phage Efficacy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The PLOS Medicine Editors. Antimicrobial Resistance: Is the World UNprepared? PLoS Med. 2016, 13, e1002130. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Genet. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Church, D.; Elsayed, S.; Reid, O.; Winston, B.; Lindsay, R. Burn Wound Infections. Clin. Microbiol. Rev. 2006, 19, 403–434. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 1–18. [Google Scholar] [CrossRef]
- Tümmler, B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Research 2019, 8, 1371. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Maurice, C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Xue, J.; Costa, R.; Ru, J.; Schulz, S.; Taranu, Z.E.; Deng, L. Challenges of Studying the Human Virome–Relevant Emerging Technologies. Trends Microbiol. 2021, 29, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Altamirano, F.L.G.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Khan, A.A.; Ghosh, P.; Taranu, Z.E.; Taguer, M.; Ru, J.; Chowdhury, R.; Kabir, M.; Deng, L.; Mondal, D.; et al. Bacteriophages Isolated from Stunted Children Can Regulate Gut Bacterial Communities in an Age-Specific Manner. Cell Host Microbe 2020, 27, 199–212.e5. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Abedon, S.T. Pharmacologically Aware Phage Therapy: Pharmacodynamic and Pharmacokinetic Obsta-cles to Phage Antibacterial Action in Animal and Human Bodies. Microbiol. Mol. Biol. Rev. 2019, 83, e00012-19. [Google Scholar] [CrossRef]
- Nilsson, A.S. Pharmacological limitations of phage therapy. Upsala J. Med. Sci. 2019, 124, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.W.; Harper, D.; et al. Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and Safety Requirements for Sustainable Phage Therapy Products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef]
- Forti, F.; Roach, D.R.; Cafora, M.; Pasini, M.E.; Horner, D.S.; Fiscarelli, E.V.; Rossitto, M.; Cariani, L.; Briani, F.; Debarbieux, L.; et al. Design of a Broad-Range Bacteriophage Cocktail That ReducesPseudomonas aeruginosaBiofilms and Treats Acute Infections in Two Animal Models. Antimicrob. Agents Chemother. 2018, 62, e02573-17. [Google Scholar] [CrossRef] [PubMed]
- Khan Mirzaei, M.; Haileselassie, Y.; Navis, M.; Cooper, C.; Sverremark-Ekström, E.; Nilsson, A.S. Morphologically Distinct Escherichia coli Bacteriophages Differ in Their Efficacy and Ability to Stimulate Cytokine Release In Vitro. Front. Microbiol. 2016, 7, 437. [Google Scholar]
- Abedon, S.T. Use of phage therapy to treat long-standing, persistent, or chronic bacterial infections. Adv. Drug Deliv. Rev. 2019, 145, 18–39. [Google Scholar] [CrossRef]
- Li, G.; Leung, C.Y.; Wardi, Y.; Debarbieux, L.; Weitz, J.S. Optimizing the Timing and Composition of Therapeutic Phage Cocktails: A Control-Theoretic Approach. Bull. Math. Biol. 2020, 82, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepiński, P.; Douce, G.R.; Clokie, M.R. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Kimberly, C.; Gilmour, J.S.; Deborah, J.-S.; Robert, T.S.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacte-rium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage-Antibiotic Combination Treatments: Antagonistic Impacts of Antibiotics on the Pharmacodynamics of Phage Therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Liu, C.G.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Minakhin, L.; Van den Bossche, A.; Yakunina, M.; Klimuk, E.; Blasdel, B.; Lavigne, R. Development of giant bacteriophage ϕKZ is independent of the host transcription apparatus. J. Virol. 2014, 88, 10501–10510. [Google Scholar] [CrossRef]
- Uchiyama, J.; Shigehisa, R.; Nasukawa, T.; Mizukami, K.; Takemura-Uchiyama, I.; Ujihara, T.; Murakami, H.; Imanishi, I.; Nishifuji, K.; Sakaguchi, M.; et al. Piperacillin and ceftazidime produce the strongest synergistic phage–antibiotic effect in Pseudomonas aeruginosa. Arch. Virol. 2018, 163, 1941–1948. [Google Scholar] [CrossRef]
- Coulter, L.B.; McLean, R.J.C.; Rohde, R.E.; Aron, G.M. Effect of Bacteriophage Infection in Combination with Tobramycin on the Emergence of Resistance in Escherichia coli and Pseudomonas aeruginosa Biofilms. Viruses 2014, 6, 3778–3786. [Google Scholar] [CrossRef] [PubMed]
- Hayward, R.S.; Harding, J.; Molloy, R.; Land, L.; Longcroft-Neal, K.; Moore, D.; Ross, J.D.C. Adverse effects of a single dose of gentamicin in adults: A systematic review. Br. J. Clin. Pharmacol. 2017, 84, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Hernandez, A.; Plucain, J.; Gori, F.; Pena-Miller, R.; Reding, C.; Jansen, G.; Schulenburg, H.; Gudelj, I.; Beardmore, R. Using a Sequential Regimen to Eliminate Bacteria at Sublethal Antibiotic Dosages. PLoS Biol. 2015, 13, e1002104. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Futur. Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Merabishvili, M.; Monserez, R.; Van Belleghem, J.; Rose, T.; Jennes, S.; De Vos, D.; Verbeken, G.; Vaneechoutte, M.; Pirnay, J.-P. Stability of bacteriophages in burn wound care products. PLoS ONE 2017, 12, e0182121. [Google Scholar] [CrossRef]
- Karami, P.; Mohajeri, P.; Mashouf, R.Y.; Karami, M.; Yaghoobi, M.H.; Dastan, D.; Alikhani, M.Y. Molecular characterization of clinical and environmental Pseudomonas aeruginosa isolated in a burn cen-ter. Saudi J. Biol. Sci. 2019, 26, 1731–1736. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Dieases (ESCMID). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2000, 6, 509–515. [Google Scholar] [CrossRef]
- Lim, L.M.; Ly, N.; Anderson, D.; Yang, J.C.; Macander, L.; Jarkowski, A.; Forrest, A.; Bulitta, J.B.; Tsuji, B.T. Resurgence of Colistin: A Review of Resistance, Toxicity, Pharmacodynamics, and Dosing. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2010, 30, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Gutema, G.; Engidawork, E. Affordability of commonly prescribed antibiotics in a large tertiary teaching hospital in Ethiopia: A challenge for the national drug policy objective. Bmc Res. Notes 2018, 11, 925. [Google Scholar] [CrossRef]
- Schäfermann, S.; Neci, R.; Ndze, E.N.; Nyaah, F.; Pondo, V.B.; Heide, L. Availability, prices and affordability of selected antibiotics and medicines against non-communicable diseases in western Cameroon and northeast DR Congo. PLoS ONE 2020, 15, e0227515. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Van Twest, R.; Kropinski, A.M. Bacteriophage Enrichment from Water and Soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Eriksson, H.; Kasuga, K.; Haggård-Ljungquist, E.; Nilsson, A.S. Genomic, Proteomic, Morphological, and Phylogenetic Analyses of vB_EcoP_SU10, a Podoviridae Phage with C3 Morphology. PLoS ONE 2014, 9, e116294. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics Act with vB_AbaP_AGC01 Phage against Acinetobacter baumannii in Human Heat-Inactivated Plasma Blood and Galleria mellonella Models. Int. J. Mol. Sci. 2020, 21, 4390. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Practical Methods for Determining Phage Growth Parameters. Springer Protoc. Handb. 2009, 501, 175–202. [Google Scholar] [CrossRef]
- Magoč, T.; Magoč, M.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers EC Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glim-mer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar] [CrossRef]
- WHO|No Time to Wait: Securing the future from drug-resistant infections. Available online: http://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/ (accessed on 18 November 2020).
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef]
- Altamirano, F.G.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef]
- Sato, Y.; Unno, Y.; Ubagai, T.; Ono, Y. Sub-minimum inhibitory concentrations of colistin and polymyxin B promote Aci-netobacter baumannii biofilm formation. PLoS ONE 2018, 13, e0194556. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K. Phage therapy: What factors shape phage pharmacokinetics and bioavailability? Systematic and critical re-view. Med. Res. Rev. 2019, 39, 2000–2025. [Google Scholar]
- Jeon, J.; Park, J.-H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. Bmc Microbiol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Majewska, J.; Kaźmierczak, Z.; Lahutta, K.; Lecion, D.; Szymczak, A.; Miernikiewicz, P.; Drapała, J.; Harhala, M.; Marek-Bukowiec, K.; Jędruchniewicz, N.; et al. Induction of Phage-Specific Antibodies by Two Therapeutic Staphylococcal Bacteriophages Administered per os. Front. Immunol. 2019, 10, 2607. [Google Scholar] [CrossRef]
- McClean, D.; Friman, V.; Finn, A.; Salzberg, L.I.; Donohue, I. Coping with multiple enemies: Pairwise interactions do not predict evolutionary change in complex multitrophic communities. Oikos 2019, 128, 1588–1599. [Google Scholar] [CrossRef]
- Wright, R.C.; Friman, V.-P.; Smith, M.C.; Brockhurst, M.A. Resistance Evolution against Phage Combinations De-pends on the Timing and Order of Exposure. mBio 2019, 10, e01652-19. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Bull, J.J.; Levin, B.R.; Molineux, I.J. Promises and Pitfalls of In Vivo Evolution to Improve Phage Therapy. Viruses 2019, 11, 1083. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-H.; Kuo, C.-F.; Wang, C.-H.; Wu, C.-M.; Tsao, N. Experimental phage therapy in treating Klebsiella pneumoni-ae-mediated liver abscesses and bacteremia in mice. Antimicrob. Agents Chemother. 2011, 55, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Betts, A.; Gray, C.; Zelek, M.; MacLean, R.C.; King, K.C. High parasite diversity accelerates host adaptation and diversi-fication. Science 2018, 360, 907–911. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Turner, P.E. The evolution of life history trade-offs in viruses. Curr. Opin. Virol. 2014, 8, 79–84. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Gabard, J.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeru-ginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Peivandi, A.; Tian, L.; Mahabir, R.; Hosseinidoust, Z. Hierarchically Structured, Self-Healing, Fluorescent, Bioactive Hy-drogels with Self-Organizing Bundles of Phage Nanofilaments. Chem. Mater. 2019, 31, 5442–5449. [Google Scholar] [CrossRef]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 2016, 215, 703–712. [Google Scholar] [CrossRef]
- Jeon, J.; Yong, D. Two Novel Bacteriophages Improve Survival in Galleria mellonella Infection and Mouse Acute Pneu-monia Models Infected with Extensively Drug-Resistant Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2019, 85, e02900-18. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Wojciuk, B.; Roszak, M.; Łubowska, N.; Błażejczak, P.; Jursa-Kulesza, J. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter bau-mannii Biofilm in a Human Urine Model. Microb Drug Resist. 2021, 27, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Dąbrowska, K.; Vaneechoutte, M.; Barr, J.J.; Bollyky, P.L. Interactions between Bacteriophage, Bac-teria, and the Mammalian Immune System. Viruses 2018, 11, 10. [Google Scholar] [CrossRef]
- Lourenço, M.; Chaffringeon, L.; Lamy-Besnier, Q.; Pédron, T.; Campagne, P.; Eberl, C.; Bérard, M.; Stecher, B.; Debarbieux, L.; De Sordi, L. The Spatial Heterogeneity of the Gut Limits Predation and Fosters Coexistence of Bacteria and Bacteriophages. Cell Host Microbe 2020, 28, 390–401.e5. [Google Scholar] [CrossRef]
- Abdullahi, A.; Amini-Nik, S.; Jeschke, M.G. Animal models in burn research. Cell. Mol. Life Sci. 2014, 71, 3241–3255. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aghaee, B.L.; Khan Mirzaei, M.; Alikhani, M.Y.; Mojtahedi, A.; Maurice, C.F. Improving the Inhibitory Effect of Phages against Pseudomonas aeruginosa Isolated from a Burn Patient Using a Combination of Phages and Antibiotics. Viruses 2021, 13, 334. https://doi.org/10.3390/v13020334
Aghaee BL, Khan Mirzaei M, Alikhani MY, Mojtahedi A, Maurice CF. Improving the Inhibitory Effect of Phages against Pseudomonas aeruginosa Isolated from a Burn Patient Using a Combination of Phages and Antibiotics. Viruses. 2021; 13(2):334. https://doi.org/10.3390/v13020334
Chicago/Turabian StyleAghaee, Bahareh Lashtoo, Mohammadali Khan Mirzaei, Mohammad Yousef Alikhani, Ali Mojtahedi, and Corinne F. Maurice. 2021. "Improving the Inhibitory Effect of Phages against Pseudomonas aeruginosa Isolated from a Burn Patient Using a Combination of Phages and Antibiotics" Viruses 13, no. 2: 334. https://doi.org/10.3390/v13020334
APA StyleAghaee, B. L., Khan Mirzaei, M., Alikhani, M. Y., Mojtahedi, A., & Maurice, C. F. (2021). Improving the Inhibitory Effect of Phages against Pseudomonas aeruginosa Isolated from a Burn Patient Using a Combination of Phages and Antibiotics. Viruses, 13(2), 334. https://doi.org/10.3390/v13020334





