Abstract
Cellular restriction factors (RFs) act as important constitutive innate immune barriers against viruses. In 2006, the promyelocytic leukemia protein was described as the first RF against human cytomegalovirus (HCMV) infection which is antagonized by the viral immediate early protein IE1. Since then, at least 15 additional RFs against HCMV have been identified, including the chromatin regulatory protein SPOC1, the cytidine deaminase APOBEC3A and the dNTP triphosphohydrolase SAMHD1. These RFs affect distinct steps of the viral replication cycle such as viral entry, gene expression, the synthesis of progeny DNA or egress. This review summarizes our current knowledge on intrinsic immune mechanisms restricting HCMV replication as well as on the viral strategies to counteract the inhibitory effects of RFs. Detailed knowledge on the interplay between host RFs and antagonizing viral factors will be fundamental to develop new approaches to combat HCMV infection.
1. Introduction
Host organisms have evolved several layers of interconnected mechanisms to efficiently counteract viral infections. Conventional innate and adaptive immune mechanisms effectively target virus infections, however, both need to be evoked through diverse inducible pathways which delays the response [1]. In contrast, constitutive innate immune mechanisms have the advantage of an immediate host defense against invading pathogens. A large number of constitutive mechanisms of innate immunity have been described including chemical barriers, antimicrobial peptides, basal autophagy or proteasomal degradation [2]. One drawback of these mechanisms is a lack of specificity. Recent research emphasizes the importance of a subgroup of constitutive innate immune mechanisms mediated by host restriction factors (RFs). RFs, alternatively called cell intrinsic immune factors, are characterized by several hallmarks: RFs (i) are dominantly and autonomously acting factors which form a front-line defense against viral infections, (ii) are constitutively expressed in specific cells but may undergo profound upregulation by interferons, (iii) employ unique and virus-specific mechanisms to interfere with viral replication, (iv) exhibit high interspecies diversity due to co-evolution of hosts with different viruses and (v) are often antagonized by viral proteins or virus-specific adaptation mechanisms [1,3].
Intrinsic immunity has been extensively studied in the past decades, which led to the discovery of several different host cell effector proteins involved in the intrinsic defense against various viruses. For HIV-1 (human immunodeficiency virus type 1) the most prominent RFs are the E3 ubiquitin ligase TRIM5α and the cytidine deaminase APOBEC3G, as well as SAMHD1 and tetherin, which efficiently restrict distinct steps during HIV-1’s life cycle [4,5,6,7,8,9]. Some of these RFs as exemplified by TRIM5α are well known to act as an efficient barrier for cross-species transmission [10]. Other examples of RFs are the MxA protein and the IFIT (interferon-induced proteins with tetratricopeptide repeats) and IFITM (interferon-induced transmembrane) family members, which were identified as important players in the defense against influenza virus infection [9,11,12,13,14].
HCMV (human cytomegalovirus), a ubiquitous β-herpesvirus, can cause severe disease in individuals with immature or compromised immune systems and in congenitally infected children. The outcome of HCMV infection is determined by a number of factors, some related to the virus, others to the host, which include immunogenetic factors [15]. Its double-stranded DNA genome contains >200 protein-coding genes and a large number of HCMV gene products have been found to function in immune evasion including the antagonization of host RFs [16,17,18]. In 2006, the PML (promyelocytic leukemia) protein, the defining constituent of the nuclear domain 10 (ND10), was described as the first RF against HCMV [19,20]. Since then, numerous additional host RFs have been identified acting at distinct steps of the viral replication cycle. This short review summarizes our current knowledge on intrinsic immunity defending against HCMV infections. In addition, we also provide a description of the viral mechanisms used to modulate and counteract host RFs.
2. Galectin-9 Restricts the Entry of HCMV into Host Cells
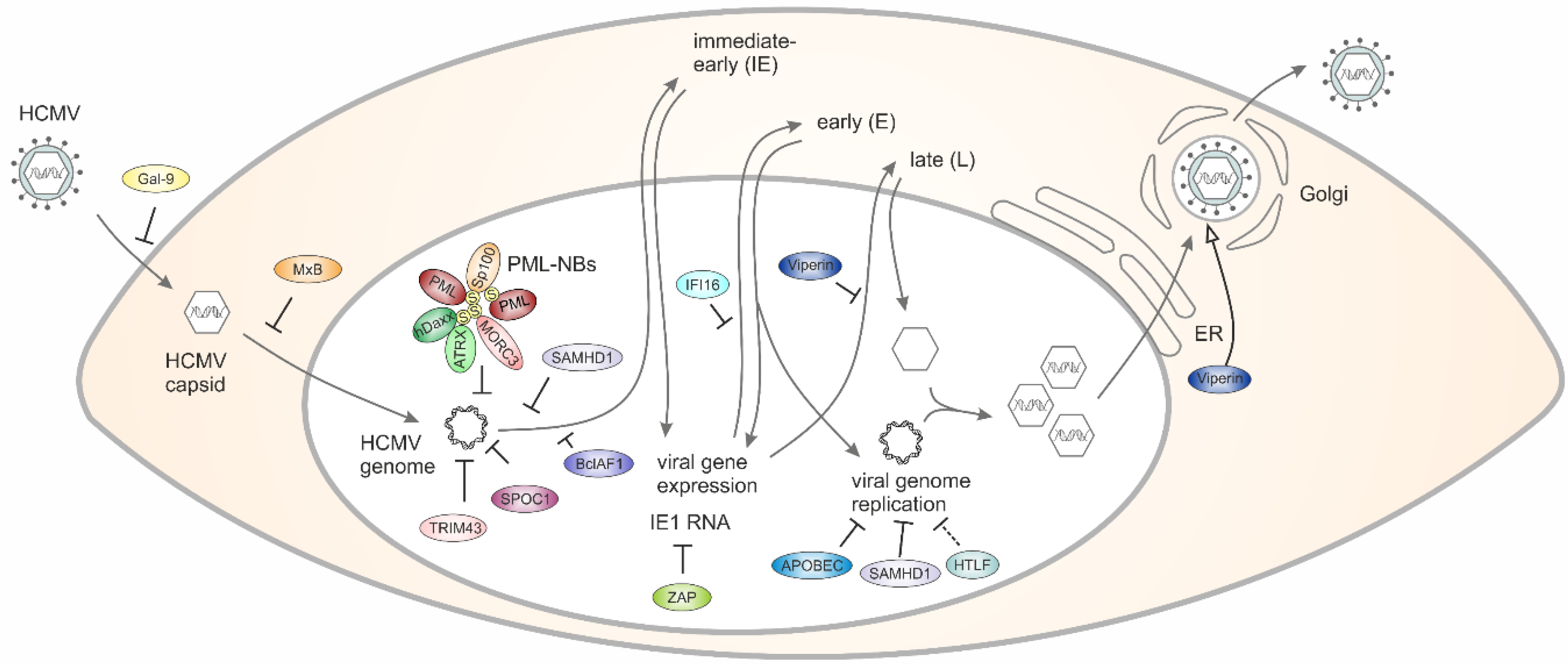
Galectin-9 (Gal-9), a member of a family of secreted glycan-binding proteins with a conserved carbohydrate recognition domain (CRD), is known for its pleiotropic immune-regulatory properties [21]. Galectins are able to bind glycan structures on the surface of cells and this can modulate a variety of diverse functions including cell–cell interactions, cell–matrix adhesion, or transmembrane signaling. First evidence for a role of Gal-9 during HCMV infection was obtained by monitoring blood samples of hematopoietic stem cell recipients for Gal-9 mRNA levels. This revealed significantly increased expression of Gal-9 mRNA in patients with HCMV reactivation [22]. Results obtained by in vitro infection of primary human fibroblasts demonstrated that a soluble factor mediates the upregulation of Gal-9 and finally interferon (IFN)-β was identified as a mediator of Gal-9 induction during HCMV infection [22]. In an attempt to evaluate the impact of Gal-9 on HCMV replication, primary human fibroblasts as well as retinal pigment epithelial cells were infected in the presence of increasing concentrations of Gal-9. This revealed a dose-dependent downregulation of HCMV infection which was specific for Gal-9 and could be observed in a cell-type and HCMV-strain-independent manner [23]. Mechanistically, downregulation required Gal-9 lectin binding and was mediated primarily via an interaction with virions. Furthermore, the authors provided evidence for HCMV entry inhibition by Gal-9 occurring at the level of virus–cell fusion [23]. Importantly, HCMV-dependent upregulation of soluble Gal-9 could be demonstrated in hematopoietic stem cell transplant patients suggesting that Gal-9 may function as an important antiviral defense effector molecule in vivo [23]. In summary, Gal-9 constitutes the first cellular RF against HCMV which operates via entry inhibition (Figure 1, Table 1).
Figure 1.
Schematic outline of the strategies used by host RFs (restriction factors) to counteract HCMV (human cytomegalovirus) replication.

Table 1.
Overview of host RFs targeting HCMV.
3. MxB Interferes with the Nuclear Delivery of HCMV Genomes
Mx proteins are IFN-induced dynamin-like GTPases that consist of a globular GTPase domain connected to an extended stalk. While MxA is well known for its restriction of a large panel of positive- and negative-strand RNA and certain DNA viruses but, notably, not herpesviruses or HIV-1 (reviewed in [24]), initial studies suggested that MxB lacks antiviral activity. A first report connecting this GTPase with antiviral restriction of herpesviruses identified MxB in a screen for IFN-induced genes conferring resistance to murine gammaherpesvirus 68 (MHV68) [25]. In 2018, Crameri et al. provided direct evidence that ectopic MxB expression restricts lytic replication of HSV (herpes simplex virus)-1 and HSV-2 as well as latent infection of KSHV [26]. Further experiments revealed that MxB interferes with the delivery of herpesviral genomes into the nucleus, a step that requires the GTPase function of MxB [26]. Shortly thereafter, two further studies confirmed and even extended the anti-herpesviral activity of MxB to HCMV and MCMV [27,28]. Moreover, it could be demonstrated that, in addition to the GTPase domain, the stalk region is required for MxB’s anti-herpesviral activity presumably by mediating dimerization or even oligomerization via self-assembly [27]. Altogether, all three studies published in 2018 collectively identified MxB as a novel pan-herpesvirus RF that acts as a postentry inhibitor interfering with the delivery of herpesviral genomes into the host cell nucleus (see Figure 1) [29].
4. PML Nuclear Bodies Associate with Parental HCMV Genomes to Induce Epigenetic Silencing
PML protein, a member of the tripartite motif (TRIM) protein family, represents the key component of subnuclear structures known as PML nuclear bodies (PML-NBs) or nuclear domain 10 (ND10). Since PML-NBs consist of numerous permanently or transiently associated proteins, they have been implicated in the regulation of diverse cellular processes including cell cycle, apoptosis, senescence, stress and DNA damage responses [30]. The role of PML-NBs in intrinsic immunity was discovered many years ago and has been extensively characterized in the context of herpesviral infections. As shown for other herpesviruses, HCMV genomes become associated with PML-NBs after entering the cell nucleus (Figure 1) [31,32]. This association results in epigenetic silencing of HCMV genomes and, thus, blocks the initiation of lytic replication (Table 1). Employment of knockdown or overexpression techniques in numerous studies has identified several PML-NB components, including PML, Sp100, hDaxx and ATRX as RFs that contribute to the silencing of HCMV DNA by recruiting chromatin-modifying enzymes (reviewed in [20,33]).
During lytic infection, HCMV expresses two regulatory proteins that act in a sequential manner to efficiently antagonize PML-NB-based repression. Initially, the tegument-delivered protein pp71 is imported into the nucleus where it induces dissociation of ATRX from PML-NBs [34]. This is followed by pp71-induced degradation of hDaxx, which occurs in a ubiquitin-independent but proteasome-dependent manner and facilitates initiation of viral IE gene expression [35,36]. Subsequently, the immediate early protein IE1 is expressed and induces a complete dispersal of PML-NBs [37]. Mechanistic studies have revealed that IE1 directly interacts with PML through its α-helical core domain and blocks the SUMOylation of PML, which is essential for PML-NB integrity [38,39]. More recently, a novel antagonist of PML-NBs has been identified, which is expressed during HCMV latency [40]. The latency-associated gene product LUNA, which shows deSUMOylase activity, was reported to induce PML deSUMOylation and dispersal of ND10 bodies, thereby promoting HCMV reactivation in response to external stimuli.
Of note, the PML protein emerges as a mediator of both intrinsic and innate immune defenses. In particular, PML was identified as a positive regulator of the IFN pathway, which can enhance the expression of IFN-β and, additionally, is required for an efficient transcription of type I and type II IFN-stimulated genes (ISGs) [41,42,43,44]. Consequently, viral proteins that modulate PML-NBs may not only antagonize intrinsic but also innate immune responses, which has already been demonstrated for IE1 of HCMV [43,44]. Controversial results, however, were provided by a recent study characterizing a recombinant HCMV that expresses a PML-binding deficient mutant of IE1 [45]. Since lower instead of higher cytokine and ISG expression was observed upon infection with the recombinant virus, the authors speculated that disruption of PML bodies may be linked to immune activation. Thus, further experimentation will be required to detect possible virus strain- and cell-type-specific differences in innate immune regulation and to define the exact role of PML during HCMV infection.
5. MORC3 Affects the Recruitment of PML-NB Components to Viral DNA
In 2016, the protein Microorchidia 3 (MORC3) was described as a novel RF against HSV-1 and HCMV [46]. MORC3 is a SUMOylated nuclear matrix protein with ATPase activity that has been reported to localize to PML-NBs via a SUMO-SIM mediated interaction with PMLI in uninfected cells [47]. Upon infection with HSV-1, MORC3 was found to be recruited to sites associated with HSV-1 genomes [46]. Viral plaque assays revealed antiviral activity, albeit only against ICP0-null mutant HSV-1 since restriction by MORC3 is efficiently antagonized by an ICP0-mediated degradation in a manner that depends on its RING finger domain [46,48]. Additionally, MORC3’s antiviral role extends to HCMV infection as its plaque-forming efficiency increased in MORC3-depleted cells [46]. For HCMV, data on a viral protein antagonizing MORC3 are currently not available. However, a proteomic screen to quantify protein degradation during the early phase of HCMV infection identified MORC3 to be degraded in a proteasomal manner, suggesting the activity of a viral countermeasure [49]. Depletion experiments revealed that MORC3 is required for efficient recruitment of PML, Sp100, hDaxx and γH2AX to viral DNA [46]. Since a recent publication suggested that MORC3 is capable of forming liquid-like nuclear condensates through phase separation, depending on its ATPase activity, one may speculate that MORC3 facilitates the formation of local nuclear structures fueling the epigenetic silencing of herpesvirus genomes [50].
6. TRIM43 Represses Active Viral Chromatin States via Ubiquitination and Degradation of Pericentrin
Besides PML, another TRIM protein emerges as a potent, herpesvirus-specific antiviral factor: by performing an RNAi screen, Full et al. identified 15 TRIM proteins that suppress KSHV reactivation. Among them, the centrosomal protein TRIM43 was distinguished by its role as RF for a broad range of herpesviruses, including HCMV [51]. TRIM43 expression is induced by herpesvirus infection, which displays a hallmark of antiviral factors. In contrast to many other RFs, however, this upregulation does not depend on IFN signaling but is part of a germline-specific transcriptional program mediated by the transcription factor DUX4. Mechanistically, the authors provided evidence that TRIM43 ubiquitinates the centrosomal protein pericentrin by utilizing its RING E3 ligase activity, thereby targeting it for proteasomal degradation [51]. This subsequently induces alterations of the nuclear lamina and leads to a repression of active viral chromatin states. Since an increase of HCMV IE gene expression was observed upon TRIM43 knockdown, it can be concluded that one of the first steps in the HCMV replication cycle is affected via the TRIM43–pericentrin–lamin axis.
7. SPOC1 Associates with the HCMV Major Immediate Early Promoter to Induce Chromatin Compaction
SPOC1 (survival-time associated PHD protein in ovarian cancer 1), also known as PHF13 (PHD finger 13), was first described in 2005 as a novel cellular protein with a single plant homeodomain (PHD) showing high expression in ovarian carcinoma patients which correlated with poor prognosis [52]. Subsequent studies revealed that this protein acts as a regulator of the DNA damage response and of chromatin structure via binding to H3K4me2/3-containing chromatin. This recruits corepressors such as KAP1 and histone methyltransferase SETDB1 to promote chromatin compaction [53,54]. Apart from its cellular regulatory functions, studies on human adenovirus type 5 (HAdV5) provided first evidence that SPOC1 contributes to the intrinsic defense against viral infections [55]. Schreiner and colleagues observed that HAdV5 gene expression was diminished upon overexpression of SPOC1, suggesting that repression takes place at the transcriptional level. Furthermore, human adenoviruses antagonize this repression by inducing the proteasomal degradation of SPOC1 early after infection [55]. For HCMV, a highly specific association of SPOC1 with the major immediate early promoter (MIEP) could be detected by chromatin immunoprecipitation sequencing, suggesting that SPOC1 represses HCMV replication by MIEP binding and recruitment of heterochromatin-building factors [56]. Consistently, SPOC1 overexpression severely impaired HCMV replication while SPOC1 depleted cells displayed an augmented initiation of viral immediate early gene expression. In contrast to adenovirus infection, SPOC1 was not degraded during infection but a transient upregulation of SPOC1 during the early phase of HCMV infection was observed [56]. However, since only high SPOC1 levels at the start of infection mediate efficient repression, HCMV may have evolved alternative antagonistic mechanisms operating during later phases of viral replication that require further investigation. Since SPOC1 expression levels exhibit a considerable variation between cell types, one may assume that this protein endows specific tissues of an organism with an additional RF against HCMV acting via chromatin compaction.
8. IFI16 Targets Incoming HCMV DNA to Regulate Viral Promoter Activities
IFI16 is an IFN-inducible and predominantly nuclear protein that, as a member of the PYHIN protein family, contains an N-terminal pyrin domain (PYD) and two partially conserved 200-residue domains (HIN domains) in the C-terminus. While the α-helical PYD promotes homotypic protein interactions with other PYD-containing proteins, the HIN domains allow an interaction with both dsDNA and ssDNA in a sequence-independent manner [57,58,59]. During HCMV infection, the viral dsDNA genome is recognized by IFI16 resulting in a colocalization inside the cell nucleus [60]. Since live-cell studies revealed a re-localization of IFI16 to nuclear peripheral foci within the first hours of HCMV infection, it can be assumed that IFI16 rapidly targets incoming HCMV DNA. The authors furthermore reported that IFI16 oligomerizes at sites of herpesviral DNA deposition in order to regulate viral gene expression and limit viral replicative capacity [61]. Whereas the recruitment of IFI16 appears to activate the major immediate early promoter (MIEP) immediately upon HCMV infection, IFI16 displays a restrictive activity at later times as it suppresses expression of the HCMV DNA polymerase (UL54) and its processivity factor (UL44), which are required for viral DNA synthesis [62,63,64]. The molecular mechanism of the latter, antiviral activity of IFI16 is based on its ability to bind and block Sp1-like transcription factors on viral promoters [64]. Notably, the interaction of HCMV tegument protein pp65 (pUL83) with IFI16 was shown to be required for both the positive and negative regulation of HCMV promoter activity [62,63]. At late stages of infection, pp65 additionally modulates IFI16 function as it induces a re-localization into the cytoplasm thereby antagonizing the antiviral activity of IFI16 inside the nucleus [62]. As a second viral regulator, the HCMV phosphoprotein UL97 has been demonstrated to bind and phosphorylate IFI16, which promotes the nucleo-cytoplasmic re-localization of this protein and finally results in translocation to the virus assembly complex and incorporation into newly formed virions [60].
Besides its role as HCMV RF, IFI16 participates in the innate immune defense by acting as a pattern recognition receptor: IFI16 binds nuclear HCMV dsDNA and triggers expression of antiviral cytokines via the STING/TBK1/IRF signaling pathway [65]. Again, this response is blocked by pp65, which sequesters the IFI16 PYD and blocks nuclear IFI16 oligomerization and subsequent immune signaling [65]. Thus, the interaction with pp65 appears critical for the outcome of HCMV infection as it modulates the intrinsic and innate immune activities of IFI16.
10. HCMV Evades ZAP Detection by Suppressing CpG Dinucleotides in the Major Immediate Early 1 RNA
To avoid recognition as foreign nucleic acids, genomes of mammalian RNA and small DNA viruses mimic the composition of their host genomes by significantly suppressing CpG dinucleotide frequencies, whose artificial increase, however, results in considerable attenuation of virus replication [70,71,72,73]. Recently, the IFN-inducible Zinc finger antiviral protein (ZAP) was identified as host factor responsible for sensing CpG in viral RNA, through direct binding and possibly downstream targeting for degradation [74]. A recent arrayed ISG expression screening performed by Lin and colleagues identified ZAP to restrict HCMV in a manner that is independent of IRF3. Overexpression and knockdown experiments displayed decreased or increased virus replication, respectively, further confirming ZAP as a RF against HCMV [75]. For herpesviruses, the pattern of CpG dinucleotide frequencies is distinct: while the majority of alpha-herpesviruses demonstrate little or no CpG suppression, gamma-herpesviruses exhibit substantial suppression across the genome. Interestingly, beta-herpesviruses display the most striking pattern, with suppression of CpG dinucleotides confined to gene regions expressed with immediate early kinetics [76]. Consistently, scanning analysis demonstrates that the IE1 gene is the only region of the HCMV genome that is suppressed for CpG content [76]. In accordance, Lin et al. demonstrated that HCMV transcripts with high CpG content are specifically targeted by ZAP, while the CpG-suppressed IE1 transcript remains unaffected [75]. However, artificially increasing the IE1 CpG content by introducing mutations into the IE1 coding region renders IE1 accessible to ZAP inhibition. Interestingly, subsequent analyses revealed that endogenous ZAP is induced during HCMV infection but its expression is mutually exclusive to acute virus progression. The authors further speculated that higher levels of CpG in viral genes expressed subsequent to IE1 result from the loss of ZAP-mediated pressure in infected cells [75].
11. HTLF is Degraded during HCMV Infection to Avoid Post-Replicative DNA Repair
Helicase-like transcription factor (HTLF), a member of the switch/sucrose non-fermenting (SWI/SNF) family, was first identified as a DNA binding protein interacting with motifs of the SV40 enhancer and HIV-1 promoter [49]. Subsequent studies associated this cellular factor with the prognosis of a number of cancer types leading to the hypothesis that HTLF may act as a tumor suppressor [77]. Mechanistically, HLTF was shown to function as a ubiquitin ligase for DNA replication processivity factor PCNA to ensure error-free post-replication repair of damaged DNA replication forks [78]. A recently performed high-definition analysis to quantify protein stability during HCMV infection detected that HTLF is targeted for degradation at very early times of the replicative cycle starting from 4 h [49]. The authors postulated that HLTF might undergo early degradation in order to annihilate antiviral restriction instituted by this protein. In order to demonstrate antiviral restriction by HLTF, shRNA-mediated knockdown was performed revealing enhanced HCMV infection in HLTF-depleted cells under conditions of low multiplicity of infection. Furthermore, it could be shown that the viral protein UL145 recruits the cullin E3 ligase complex to target HTLF for proteasomal degradation [49]. Of note, this resembles the Vpr protein of HIV-1 which reprograms the CRL4DCAF1 E3 ubiquitin ligase to degrade HLTF [79]. Interestingly, a recent publication demonstrated restriction of HIV-1 replication in T-cells by HLTF which was dependent on a functional HIRAN domain suggesting that sensing and processing of fork-like branched DNA structures by HLTF interfere with ordered progression of plus strand synthesis [80]. Although not formally proven, one may speculate that a similar mechanism of restriction by the HLTF DNA helicase may also apply to HCMV which replicates through a double-stranded DNA intermediate.
12. APOBEC3 Proteins Induce Cytidine-Deaminase Mediated Hypermutation of the HCMV Genome in a Cell-Type Specific Manner
The apolipoprotein B editing enzyme catalytic subunit 3 (APOBEC3) family of proteins is known for more than a decade for its strong antiviral activity against HIV-1 [3]. APOBEC3G constitutes the prototype antiretroviral cytidine deaminase which acts during reverse transcription to preferentially deaminate the third cytosine of the single-stranded DNA sequence 5′-CCCA-3′ [81]. This disrupts the coding potential of the viral genome, generally rendering it replication defective [3]. Other APOBEC3 proteins can also restrict lentiviruses via their hypermutation activity but differ in their domain organization, sequence preferences and their propensity to utilize deaminase-independent mechanisms [82]. In addition to retroviruses, the replication of a number of DNA viruses including hepatitis B virus and human papillomaviruses were reported to undergo hypermutation-induced inhibition by APOBEC3 proteins [82]. Only recently, APOBEC3 proteins were also implicated in the restriction of HCMV infection: using a unique ex vivo organ culture model of native human decidual tissue, Weisblum and colleagues observed that the APOBEC3A isoform is profoundly upregulated in HCMV-infected decidual cells. Since this was not the case either in chorionic villi or in HCMV permissive cell lines, upregulation of APOBEC3A may constitute a specific antiviral restriction mechanism of the maternal–fetal interface [83]. Importantly, cytidine deamination editing of the HCMV genome was found to be required for inhibition of HCMV replication both in cell culture models and in vivo during congenital infection, strongly suggesting that APOBEC3A acts as an important anti-HCMV host factor in the maternal–fetal interface [83]. In contrast to HIV-1, which encodes the Vif protein as a well-characterized antagonist of APOBEC3-mediated deamination, no antagonistic proteins are known for HCMV so far [84]. However, a recent study by Pautasso and colleagues, which described an IFN-β mediated induction of APOBEC3G in HCMV infected fibroblasts, suggested that HCMV may have evolved mutational robustness by limiting the presence of APOBEC3G hot spots in essential open reading frames [85]. This may constitute a novel viral strategy to evade the cellular restriction of APOBEC3 proteins.
13. SAMHD1 Restricts HCMV Replication by Limiting NF-kB Activation and Intracellular Deoxynucleoside Triphosphate Pools
The sterile alpha motif (Sam) and histidine-aspartate (HD) domain-containing protein 1 (SAMHD1) is best known and most extensively characterized for its ability to restrict HIV-1 particularly in non-dividing myeloid cells such as macrophages and dendritic cells (DCs) as well as resting CD4+ T cells [86,87,88]. In the meantime, further viruses restricted by SAMHD1 have been identified, among them HSV-1, HCMV and EBV [89,90,91,92]. For HSV-1, the authors observed increased viral DNA replication in the absence of SAMHD1 in primary human monocyte-derived macrophages and in differentiated macrophage cell lines, thereby providing a mechanism of restriction that relies on SAMHD1’s dNTP triphosphohydrolase activity leading to depletion of intracellular dNTPs [89,90]. While the addition of exogenous deoxynucleosides partially overcomes the restriction, the absence of viral genes that are involved in dNTP metabolism such as thymidine kinase leads to a more potent suppression by SAMHD1 [89,90]. Interestingly, studies performed by the Weitzman group revealed an additional mechanism of HCMV restriction in permissive fibroblasts and conditionally permissive myeloid cells [93]. They observed that HCMV is restricted by SAMHD1 through inhibition of viral gene expression, which is achieved by inhibiting nuclear factor κB (NF-κB) activation [93].
In the last year, mechanisms of SAMHD1 antagonization during herpesvirus infection were identified by several independent groups: conserved herpesvirus protein kinases from all beta- and gamma-herpesviruses (HHV-6/7 U69, HCMV UL97, EBV BGLF4 and KSHV ORF36) were shown to induce a strong phosphorylation of SAMHD1 thereby converting it to its inactive form [91,92]. An additional study utilizing a SAMHD1 knockout mouse model further revealed that MCMV is targeted by SAMHD1 in vitro and that SAMHD1 restricts the replication of MCMV in vivo, thereby underlining the important role of SAMHD1 for CMV restriction [94]. In accordance to findings on HCMV, the authors could demonstrate that MCMV likewise developed countermeasure mechanisms involving the viral kinase M97 [94]. Only recently, the Ahn group provided an additional mechanism of viral countermeasure against SAMHD1-mediated intrinsic defense. They observed that the steady-state SAMHD1 protein level is reduced at late times of infection through a mechanism dependent on Cullin-RING-E3 ligase complexes [95].
14. HCMV Redirects the RF Viperin to Enhance Viral Infectivity
Viperin (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible), also known as RSAD2 (cig-5), is an IFN-inducible protein that exerts antiviral activity against different viruses. It belongs to the radical S-adenosylmethionine (SAM) superfamily of enzymes [96]. Recent studies demonstrate that viperin catalyzes the transformation of cytidine triphosphate to its analogue 3′deoxy-3′,4′-didehydro-CTP which inhibits the NAD+ dependent activity of metabolic enzymes [97]. Initially, it was described as a RF against HCMV, as stable expression of the protein in fibroblasts inhibits productive infection by downregulating the expression of several structural proteins (gB, pp28 and pp65) known to be indispensable for viral assembly and maturation [98]. Interestingly, viperin is not only induced by IFN but also by HCMV infection and by the HCMV envelope protein, glycoprotein B (gB) [98]. Moreover, it has been shown that HCMV infection causes the redistribution of the induced viperin from its normal endoplasmic reticulum association, first to the Golgi apparatus and then to cytoplasmic vacuoles containing gB and pp28, thereby potentially evading the antiviral effects of viperin [98]. Surprisingly, a later study proposed that HCMV even co-opts viperin to enhance its own infectivity [99]. This is achieved by disrupting the metabolism of infected cells: viperin interacts with the HCMV-encoded viral mitochondrion-localized inhibitor of apoptosis (vMIA) causing the re-localization of viperin from the endoplasmic reticulum to the mitochondria [99]. There, viperin interacts with and blocks the function of the mitochondrial trifunctional protein that mediates β-oxidation of fatty acids to generate adenosine triphosphate (ATP) resulting in reduced cellular ATP levels. This leads to actin cytoskeleton disruption and enhancement of infection [99]. Later on, the Creswell group further proved that viperin, presumably as a major effector, regulates cellular lipid metabolism during HCMV infection [100]. They demonstrated that decreased ATP levels activate the enzyme AMP-activated protein kinase, thereby inducing a cascade of events starting with expression of the glucose transporter GLUT4. As a consequence, increased glucose import and activation of glucose-regulated transcription factor ChREBP were observed. This culminated in increased transcription of genes responsible for lipid synthesis, which boosts viral envelopment [100].
15. Conclusions
During the last few years, several new RFs against HCMV have been identified. The present panel of host intrinsic immune proteins comprises well known players like SAMHD1 or APOBEC3 family members but also factors like HTLF which are so far unique for HCMV. For many RFs, the exact mechanism of restriction has been only partially elucidated. For instance, while Gal-9 restricts HCMV by inhibition of virus–cell fusion, the lectin interacting with Gal-9 on the surface of virions remains to be identified. Similarly, MxB affects the nuclear import of viral DNA but the viral structures or processes targeted by MxB are unknown so far. Another open question is the relative contribution of individual RFs to the overall defense which may vary depending on the infected tissue or organ. For instance, several of the identified RFs (e.g., PML-NBs, SPOC1, TRIM43, BclAF1) converge on chromatin compaction leading to a silencing of gene expression, however, the role of individual silencing mechanisms for the overall antiviral defense remains to be determined. Finally, significant progress has been made concerning the characterization of evasion strategies identifying a number of viral proteins as antagonists of host RFs. However, further research will be necessary to characterize molecular interactions in detail. This will be a prerequisite to develop novel therapeutic agents either boosting host RFs or interfering with viral antagonistic factors.
Author Contributions
Conceptualization, E.-M.S. and T.S.; writing—original draft preparation, E.-M.S. and M.S.; writing—review and editing, E.-M.S. and M.S.; visualization, M.S.; supervision, T.S.; project administration, T.S.; funding acquisition, T.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (STA357/7-1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
References
- Bieniasz, P.D. Intrinsic immunity: A front-line defense against viral attack. Nat. Immunol. 2004, 5, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Pradeu, T.; Masters, S.L.; Mogensen, T.H. Constitutive immune mechanisms: Mediators of host defence and immune regulation. Nat. Rev. Immunol. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Venkatesh, S.; Bieniasz, P.D. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 2012, 37, 399–411. [Google Scholar] [CrossRef]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011, 474, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Simon, V.; Bloch, N.; Landau, N.R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 2015, 16, 546–553. [Google Scholar] [CrossRef]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Yan, N.; Chen, Z.J. Intrinsic antiviral immunity. Nat. Immunol. 2012, 13, 214–222. [Google Scholar] [CrossRef]
- Nakayama, E.E.; Shioda, T. TRIM5α and Species Tropism of HIV/SIV. Front. Microbiol. 2012, 3, 13. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, J.; Haller, O.; Staeheli, P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 1992, 66, 2564–2569. [Google Scholar] [CrossRef] [PubMed]
- Shapira, S.D.; Gat-Viks, I.; Shum, B.O.; Dricot, A.; de Grace, M.M.; Wu, L.; Gupta, P.B.; Hao, T.; Silver, S.J.; Root, D.E.; et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 2009, 139, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Aiello, A.; Accardi, G.; Candore, G.; Caruso, C.; Colomba, C.; Di Bona, D.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Pandey, J.P. Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing. Int. J. Mol. Sci. 2019, 20, 685. [Google Scholar] [CrossRef]
- Jackson, S.E.; Mason, G.M.; Wills, M.R. Human cytomegalovirus immunity and immune evasion. Virus Res. 2011, 157, 151–160. [Google Scholar] [CrossRef]
- Berry, R.; Watson, G.M.; Jonjic, S.; Degli-Esposti, M.A.; Rossjohn, J. Modulation of innate and adaptive immunity by cytomegaloviruses. Nat. Rev. Immunol. 2020, 20, 113–127. [Google Scholar] [CrossRef]
- Mocarski, E.S., Jr. Betaherpes viral genes and their functions. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University PressCopyright © Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Tavalai, N.; Papior, P.; Rechter, S.; Leis, M.; Stamminger, T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 2006, 80, 8006–8018. [Google Scholar] [CrossRef]
- Tavalai, N.; Stamminger, T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar] [CrossRef]
- John, S.; Mishra, R. Galectin-9: From cell biology to complex disease dynamics. J. Biosci. 2016, 41, 507–534. [Google Scholar] [CrossRef]
- McSharry, B.P.; Forbes, S.K.; Cao, J.Z.; Avdic, S.; Machala, E.A.; Gottlieb, D.J.; Abendroth, A.; Slobedman, B. Human cytomegalovirus upregulates expression of the lectin galectin 9 via induction of beta interferon. J. Virol. 2014, 88, 10990–10994. [Google Scholar] [CrossRef] [PubMed]
- Machala, E.A.; Avdic, S.; Stern, L.; Zajonc, D.M.; Benedict, C.A.; Blyth, E.; Gottlieb, D.J.; Abendroth, A.; McSharry, B.P.; Slobedman, B. Restriction of Human Cytomegalovirus Infection by Galectin-9. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Sanchez, D.J.; Aliyari, R.; Lu, S.; Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. USA 2012, 109, 4239–4244. [Google Scholar] [CrossRef]
- Crameri, M.; Bauer, M.; Caduff, N.; Walker, R.; Steiner, F.; Franzoso, F.D.; Gujer, C.; Boucke, K.; Kucera, T.; Zbinden, A.; et al. MxB is an interferon-induced restriction factor of human herpesviruses. Nat. Commun. 2018, 9, 1980. [Google Scholar] [CrossRef] [PubMed]
- Schilling, M.; Bulli, L.; Weigang, S.; Graf, L.; Naumann, S.; Patzina, C.; Wagner, V.; Bauersfeld, L.; Goujon, C.; Hengel, H.; et al. Human MxB Protein Is a Pan-herpesvirus Restriction Factor. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Jaguva Vasudevan, A.A.; Bähr, A.; Grothmann, R.; Singer, A.; Häussinger, D.; Zimmermann, A.; Münk, C. MXB inhibits murine cytomegalovirus. Virology 2018, 522, 158–167. [Google Scholar] [CrossRef]
- Staeheli, P.; Haller, O. Human MX2/MxB: A Potent Interferon-Induced Postentry Inhibitor of Herpesviruses and HIV-1. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef]
- Ishov, A.M.; Maul, G.G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 1996, 134, 815–826. [Google Scholar] [CrossRef]
- Ishov, A.M.; Stenberg, R.M.; Maul, G.G. Human cytomegalovirus immediate early interaction with host nuclear structures: Definition of an immediate transcript environment. J. Cell Biol. 1997, 138, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Full, F.; Ensser, A. Early Nuclear Events after Herpesviral Infection. J. Clin. Med. 2019, 8, 1408. [Google Scholar] [CrossRef] [PubMed]
- Lukashchuk, V.; McFarlane, S.; Everett, R.D.; Preston, C.M. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 2008, 82, 12543–12554. [Google Scholar] [CrossRef] [PubMed]
- Saffert, R.T.; Kalejta, R.F. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 2006, 80, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Kalejta, R.F. Human cytomegalovirus protein pp71 induces Daxx SUMOylation. J. Virol. 2009, 83, 6591–6598. [Google Scholar] [CrossRef] [PubMed]
- Korioth, F.; Maul, G.G.; Plachter, B.; Stamminger, T.; Frey, J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 1996, 229, 155–158. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Klingl, S.; Sevvana, M.; Otto, V.; Schilling, E.M.; Stump, J.D.; Müller, R.; Reuter, N.; Sticht, H.; Muller, Y.A.; et al. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLoS Pathog. 2014, 10, e1004512. [Google Scholar] [CrossRef]
- Schilling, E.M.; Scherer, M.; Reuter, N.; Schweininger, J.; Muller, Y.A.; Stamminger, T. The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Poole, E.L.; Kew, V.G.; Lau, J.C.H.; Murray, M.J.; Stamminger, T.; Sinclair, J.H.; Reeves, M.B. A Virally Encoded DeSUMOylase Activity Is Required for Cytomegalovirus Reactivation from Latency. Cell Rep. 2018, 24, 594–606. [Google Scholar] [CrossRef]
- Ulbricht, T.; Alzrigat, M.; Horch, A.; Reuter, N.; von Mikecz, A.; Steimle, V.; Schmitt, E.; Krämer, O.H.; Stamminger, T.; Hemmerich, P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J. Cell Biol. 2012, 199, 49–63. [Google Scholar] [CrossRef]
- El Asmi, F.; Maroui, M.A.; Dutrieux, J.; Blondel, D.; Nisole, S.; Chelbi-Alix, M.K. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014, 10, e1003975. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Ahn, J.H. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 2015, 11, e1004785. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Otto, V.; Stump, J.D.; Klingl, S.; Müller, R.; Reuter, N.; Muller, Y.A.; Sticht, H.; Stamminger, T. Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses. J. Virol. 2016, 90, 1190–1205. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Harwardt, T.; Walter, B.; Marxreiter, A.; Zenger, M.; Reuschel, E.; Nevels, M.M. Revisiting promyelocytic leukemia protein targeting by human cytomegalovirus immediate-early protein 1. PLoS Pathog. 2020, 16, e1008537. [Google Scholar] [CrossRef]
- Sloan, E.; Orr, A.; Everett, R.D. MORC3, a Component of PML Nuclear Bodies, Has a Role in Restricting Herpes Simplex Virus 1 and Human Cytomegalovirus. J. Virol. 2016, 90, 8621–8633. [Google Scholar] [CrossRef]
- Mimura, Y.; Takahashi, K.; Kawata, K.; Akazawa, T.; Inoue, N. Two-step colocalization of MORC3 with PML nuclear bodies. J. Cell Sci. 2010, 123 Pt 12, 2014–2024. [Google Scholar] [CrossRef]
- Sloan, E.; Tatham, M.H.; Groslambert, M.; Glass, M.; Orr, A.; Hay, R.T.; Everett, R.D. Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 2015, 11, e1005059. [Google Scholar] [CrossRef]
- Nightingale, K.; Lin, K.M.; Ravenhill, B.J.; Davies, C.; Nobre, L.; Fielding, C.A.; Ruckova, E.; Fletcher-Etherington, A.; Soday, L.; Nichols, H.; et al. High-Definition Analysis of Host Protein Stability during Human Cytomegalovirus Infection Reveals Antiviral Factors and Viral Evasion Mechanisms. Cell Host Microbe 2018, 24, 447–460.e11. [Google Scholar] [CrossRef]
- Zhang, Y.; Bertulat, B.; Tencer, A.H.; Ren, X.; Wright, G.M.; Black, J.; Cardoso, M.C.; Kutateladze, T.G. MORC3 Forms Nuclear Condensates through Phase Separation. iScience 2019, 17, 182–189. [Google Scholar] [CrossRef]
- Full, F.; van Gent, M.; Sparrer, K.M.J.; Chiang, C.; Zurenski, M.A.; Scherer, M.; Brockmeyer, N.H.; Heinzerling, L.; Stürzl, M.; Korn, K.; et al. Centrosomal protein TRIM43 restricts herpesvirus infection by regulating nuclear lamina integrity. Nat. Microbiol. 2019, 4, 164–176. [Google Scholar] [CrossRef]
- Mohrmann, G.; Hengstler, J.G.; Hofmann, T.G.; Endele, S.U.; Lee, B.; Stelzer, C.; Zabel, B.; Brieger, J.; Hasenclever, D.; Tanner, B.; et al. SPOC1, a novel PHD-finger protein: Association with residual disease and survival in ovarian cancer. Int. J. Cancer 2005, 116, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Mund, A.; Schubert, T.; Staege, H.; Kinkley, S.; Reumann, K.; Kriegs, M.; Fritsch, L.; Battisti, V.; Ait-Si-Ali, S.; Hoffbeck, A.S.; et al. SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response. Nucleic Acids Res. 2012, 40, 11363–11379. [Google Scholar] [CrossRef] [PubMed]
- Kinkley, S.; Staege, H.; Mohrmann, G.; Rohaly, G.; Schaub, T.; Kremmer, E.; Winterpacht, A.; Will, H. SPOC1: A novel PHD-containing protein modulating chromatin structure and mitotic chromosome condensation. J. Cell Sci. 2009, 122 Pt 16, 2946–2956. [Google Scholar] [CrossRef]
- Schreiner, S.; Kinkley, S.; Bürck, C.; Mund, A.; Wimmer, P.; Schubert, T.; Groitl, P.; Will, H.; Dobner, T. SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog. 2013, 9, e1003775. [Google Scholar] [CrossRef] [PubMed]
- Reichel, A.; Stilp, A.C.; Scherer, M.; Reuter, N.; Lukassen, S.; Kasmapour, B.; Schreiner, S.; Cicin-Sain, L.; Winterpacht, A.; Stamminger, T. Chromatin-Remodeling Factor SPOC1 Acts as a Cellular Restriction Factor against Human Cytomegalovirus by Repressing the Major Immediate Early Promoter. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Park, Y.H.; Jeong, M.S.; Jang, S.B. Structural insights of homotypic interaction domains in the ligand-receptor signal transduction of tumor necrosis factor (TNF). BMB Rep. 2016, 49, 159–166. [Google Scholar] [CrossRef]
- Yan, H.; Dalal, K.; Hon, B.K.; Youkharibache, P.; Lau, D.; Pio, F. RPA nucleic acid-binding properties of IFI16-HIN200. Biochim. Biophys. Acta 2008, 1784, 1087–1097. [Google Scholar] [CrossRef]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Sirois, C.M.; Jin, T.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef]
- Dell’Oste, V.; Gatti, D.; Gugliesi, F.; De Andrea, M.; Bawadekar, M.; Lo Cigno, I.; Biolatti, M.; Vallino, M.; Marschall, M.; Gariglio, M.; et al. Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J. Virol. 2014, 88, 6970–6982. [Google Scholar] [CrossRef]
- Diner, B.A.; Lum, K.K.; Toettcher, J.E.; Cristea, I.M. Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection. mBio 2016, 7. [Google Scholar] [CrossRef]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; von Einem, J.; Marschall, M.; Plachter, B.; Gariglio, M.; De Andrea, M.; Landolfo, S. Regulatory Interaction between the Cellular Restriction Factor IFI16 and Viral pp65 (pUL83) Modulates Viral Gene Expression and IFI16 Protein Stability. J. Virol. 2016, 90, 8238–8250. [Google Scholar] [CrossRef] [PubMed]
- Cristea, I.M.; Moorman, N.J.; Terhune, S.S.; Cuevas, C.D.; O’Keefe, E.S.; Rout, M.P.; Chait, B.T.; Shenk, T. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 2010, 84, 7803–7814. [Google Scholar] [CrossRef] [PubMed]
- Gariano, G.R.; Dell’Oste, V.; Bronzini, M.; Gatti, D.; Luganini, A.; De Andrea, M.; Gribaudo, G.; Gariglio, M.; Landolfo, S. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012, 8, e1002498. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, J.; Cristea, I.M. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 2013, 14, 591–599. [Google Scholar] [CrossRef]
- Kasof, G.M.; Goyal, L.; White, E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 1999, 19, 4390–4404. [Google Scholar] [CrossRef]
- Sarras, H.; Alizadeh Azami, S.; McPherson, J.P. In search of a function for BCLAF1. Sci. World J. 2010, 10, 1450–1461. [Google Scholar] [CrossRef]
- Lee, S.H.; Kalejta, R.F.; Kerry, J.; Semmes, O.J.; O’Connor, C.M.; Khan, Z.; Garcia, B.A.; Shenk, T.; Murphy, E. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2012, 109, 9575–9580. [Google Scholar] [CrossRef]
- Qin, C.; Zhang, R.; Lang, Y.; Shao, A.; Xu, A.; Feng, W.; Han, J.; Wang, M.; He, W.; Yu, C.; et al. Bclaf1 critically regulates the type I interferon response and is degraded by alphaherpesvirus US3. PLoS Pathog. 2019, 15, e1007559. [Google Scholar] [CrossRef]
- Karlin, S.; Doerfler, W.; Cardon, L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994, 68, 2889–2897. [Google Scholar] [CrossRef]
- Rima, B.K.; McFerran, N.V. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol. 1997, 78 Pt 11, 2859–2870. [Google Scholar] [CrossRef]
- Simmonds, P.; Xia, W.; Baillie, J.K.; McKinnon, K. Modelling mutational and selection pressures on dinucleotides in eukaryotic phyla--selection against CpG and UpA in cytoplasmically expressed RNA and in RNA viruses. BMC Genom. 2013, 14, 610. [Google Scholar] [CrossRef]
- Gaunt, E.; Wise, H.M.; Zhang, H.; Lee, L.N.; Atkinson, N.J.; Nicol, M.Q.; Highton, A.J.; Klenerman, P.; Beard, P.M.; Dutia, B.M.; et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. eLife 2016, 5, e12735. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.P. Evolution: Zapping viral RNAs. Nature 2017, 550, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Chiweshe, S.; McCormick, D.; Raper, A.; Wickenhagen, A.; DeFillipis, V.; Gaunt, E.; Simmonds, P.; Wilson, S.J.; Grey, F. Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog. 2020, 16, e1008844. [Google Scholar] [CrossRef] [PubMed]
- Honess, R.W.; Gompels, U.A.; Barrell, B.G.; Craxton, M.; Cameron, K.R.; Staden, R.; Chang, Y.N.; Hayward, G.S. Deviations from expected frequencies of CpG dinucleotides in herpesvirus DNAs may be diagnostic of differences in the states of their latent genomes. J. Gen. Virol. 1989, 70 Pt 4, 837–855. [Google Scholar] [CrossRef]
- Dhont, L.; Mascaux, C.; Belayew, A. The helicase-like transcription factor (HLTF) in cancer: Loss of function or oncomorphic conversion of a tumor suppressor? Cell. Mol. Life Sci. CMLS 2016, 73, 129–147. [Google Scholar] [CrossRef]
- Unk, I.; Hajdú, I.; Fátyol, K.; Hurwitz, J.; Yoon, J.H.; Prakash, L.; Prakash, S.; Haracska, L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. USA 2008, 105, 3768–3773. [Google Scholar] [CrossRef]
- Zhou, X.; DeLucia, M.; Hao, C.; Hrecka, K.; Monnie, C.; Skowronski, J.; Ahn, J. HIV-1 Vpr protein directly loads helicase-like transcription factor (HLTF) onto the CRL4-DCAF1 E3 ubiquitin ligase. J. Biol. Chem. 2017, 292, 21117–21127. [Google Scholar] [CrossRef]
- Yan, J.; Shun, M.C.; Zhang, Y.; Hao, C.; Skowronski, J. HIV-1 Vpr counteracts HLTF-mediated restriction of HIV-1 infection in T cells. Proc. Natl. Acad. Sci. USA 2019, 116, 9568–9577. [Google Scholar] [CrossRef]
- Yu, Q.; König, R.; Pillai, S.; Chiles, K.; Kearney, M.; Palmer, S.; Richman, D.; Coffin, J.M.; Landau, N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004, 11, 435–442. [Google Scholar] [CrossRef]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Oiknine-Djian, E.; Zakay-Rones, Z.; Vorontsov, O.; Haimov-Kochman, R.; Nevo, Y.; Stockheim, D.; Yagel, S.; Panet, A.; Wolf, D.G. APOBEC3A Is Upregulated by Human Cytomegalovirus (HCMV) in the Maternal-Fetal Interface, Acting as an Innate Anti-HCMV Effector. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Goila-Gaur, R.; Strebel, K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 2008, 5, 51. [Google Scholar] [CrossRef]
- Pautasso, S.; Galitska, G.; Dell’Oste, V.; Biolatti, M.; Cagliani, R.; Forni, D.; De Andrea, M.; Gariglio, M.; Sironi, M.; Landolfo, S. Strategy of Human Cytomegalovirus To Escape Interferon Beta-Induced APOBEC3G Editing Activity. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, H.M.; Pan, X.; Erikson, E.; Schmidt, S.; Daddacha, W.; Burggraf, M.; Schenkova, K.; Ambiel, I.; Wabnitz, G.; Gramberg, T.; et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012, 18, 1682–1687. [Google Scholar] [CrossRef]
- Descours, B.; Cribier, A.; Chable-Bessia, C.; Ayinde, D.; Rice, G.; Crow, Y.; Yatim, A.; Schwartz, O.; Laguette, N.; Benkirane, M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 2012, 9, 87. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Hollenbaugh, J.A.; Gee, P.; Baker, J.; Daly, M.B.; Amie, S.M.; Tate, J.; Kasai, N.; Kanemura, Y.; Kim, D.H.; Ward, B.M.; et al. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 2013, 9, e1003481. [Google Scholar] [CrossRef]
- Kim, E.T.; White, T.E.; Brandariz-Núñez, A.; Diaz-Griffero, F.; Weitzman, M.D. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 2013, 87, 12949–12956. [Google Scholar] [CrossRef]
- Businger, R.; Deutschmann, J.; Gruska, I.; Milbradt, J.; Wiebusch, L.; Gramberg, T.; Schindler, M. Human cytomegalovirus overcomes SAMHD1 restriction in macrophages via pUL97. Nat. Microbiol. 2019, 4, 2260–2272. [Google Scholar] [CrossRef]
- Zhang, K.; Lv, D.W.; Li, R. Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication. Cell Rep. 2019, 28, 449–459.e5. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.T.; Roche, K.L.; Kulej, K.; Spruce, L.A.; Seeholzer, S.H.; Coen, D.M.; Diaz-Griffero, F.; Murphy, E.A.; Weitzman, M.D. SAMHD1 Modulates Early Steps during Human Cytomegalovirus Infection by Limiting NF-κB Activation. Cell Rep. 2019, 28, 434–448.e6. [Google Scholar] [CrossRef] [PubMed]
- Deutschmann, J.; Schneider, A.; Gruska, I.; Vetter, B.; Thomas, D.; Kießling, M.; Wittmann, S.; Herrmann, A.; Schindler, M.; Milbradt, J.; et al. A viral kinase counteracts in vivo restriction of murine cytomegalovirus by SAMHD1. Nat. Microbiol. 2019, 4, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, S.; Lee, M.K.; Kim, Y.E.; Lee, G.M.; Ahn, J.H. Degradation of SAMHD1 Restriction Factor Through Cullin-Ring E3 Ligase Complexes During Human Cytomegalovirus Infection. Front. Cell. Infect. Microbiol. 2020, 10, 391. [Google Scholar] [CrossRef]
- Honarmand Ebrahimi, K. A unifying view of the broad-spectrum antiviral activity of RSAD2 (viperin) based on its radical-SAM chemistry. Met. Integr. Biometal Sci. 2018, 10, 539–552. [Google Scholar] [CrossRef]
- Honarmand Ebrahimi, K.; Vowles, J.; Browne, C.; McCullagh, J.; James, W.S. ddhCTP produced by the radical-SAM activity of RSAD2 (viperin) inhibits the NAD(+) -dependent activity of enzymes to modulate metabolism. FEBS Lett. 2020, 594, 1631–1644. [Google Scholar] [CrossRef]
- Chin, K.C.; Cresswell, P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2001, 98, 15125–15130. [Google Scholar] [CrossRef]
- Seo, J.Y.; Yaneva, R.; Hinson, E.R.; Cresswell, P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 2011, 332, 1093–1097. [Google Scholar] [CrossRef]
- Seo, J.Y.; Cresswell, P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013, 9, e1003497. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).