Spatiotemporal Associations and Molecular Evolution of Highly Pathogenic Avian Influenza A H7N9 Virus in China from 2017 to 2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Virus Isolation
2.2. RNA Extraction and Genome Sequencing
2.3. Data Collection and Down-Sample
2.4. Phylogenetic Analysis
2.5. Molecular Evolutionary Analysis
2.6. Geographic Trait Association Test (BaTS)
2.7. Adaptation and Parallel Evolution
3. Results
3.1. HP H7N9 AIV Isolation and Sequencing
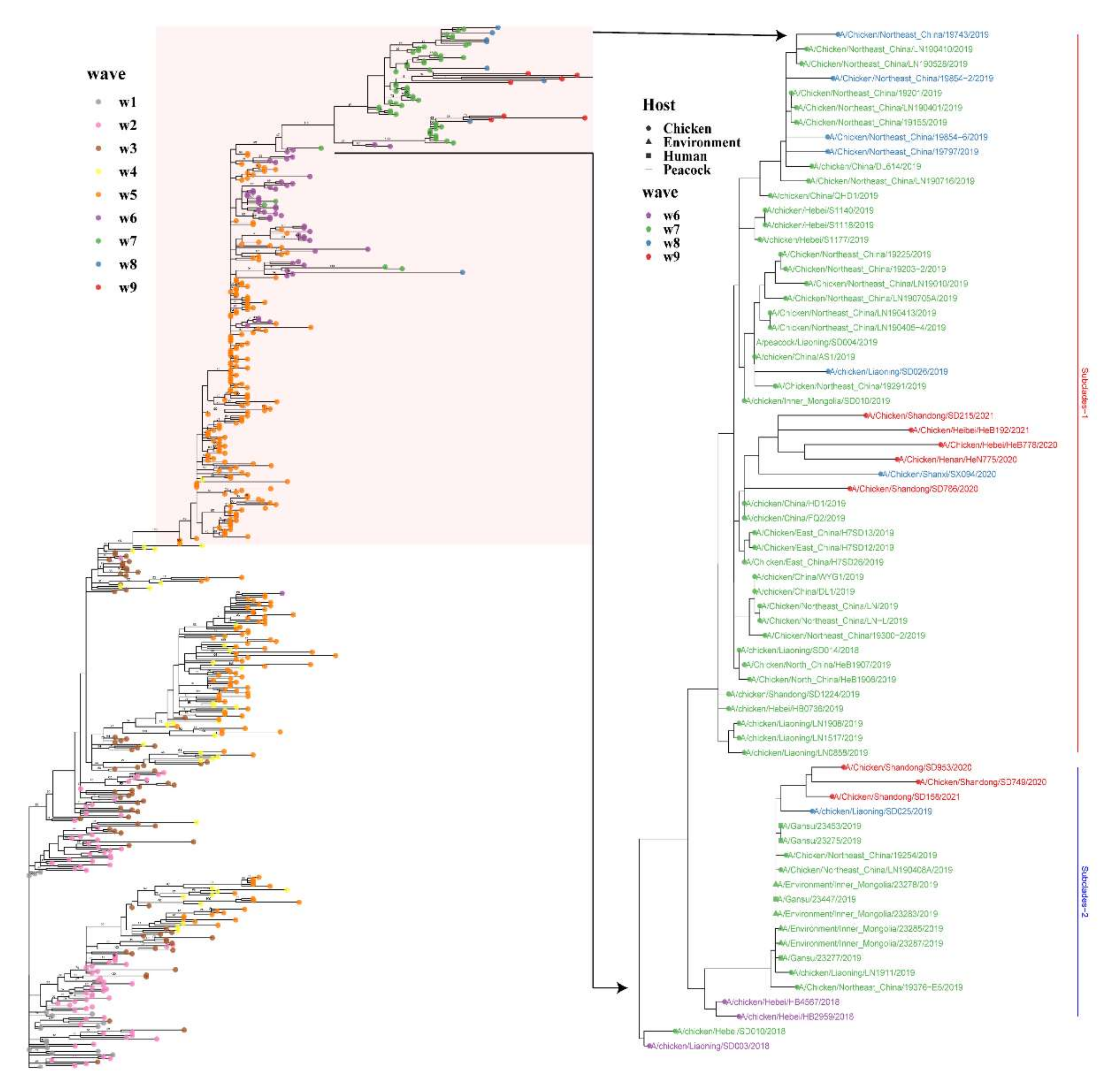
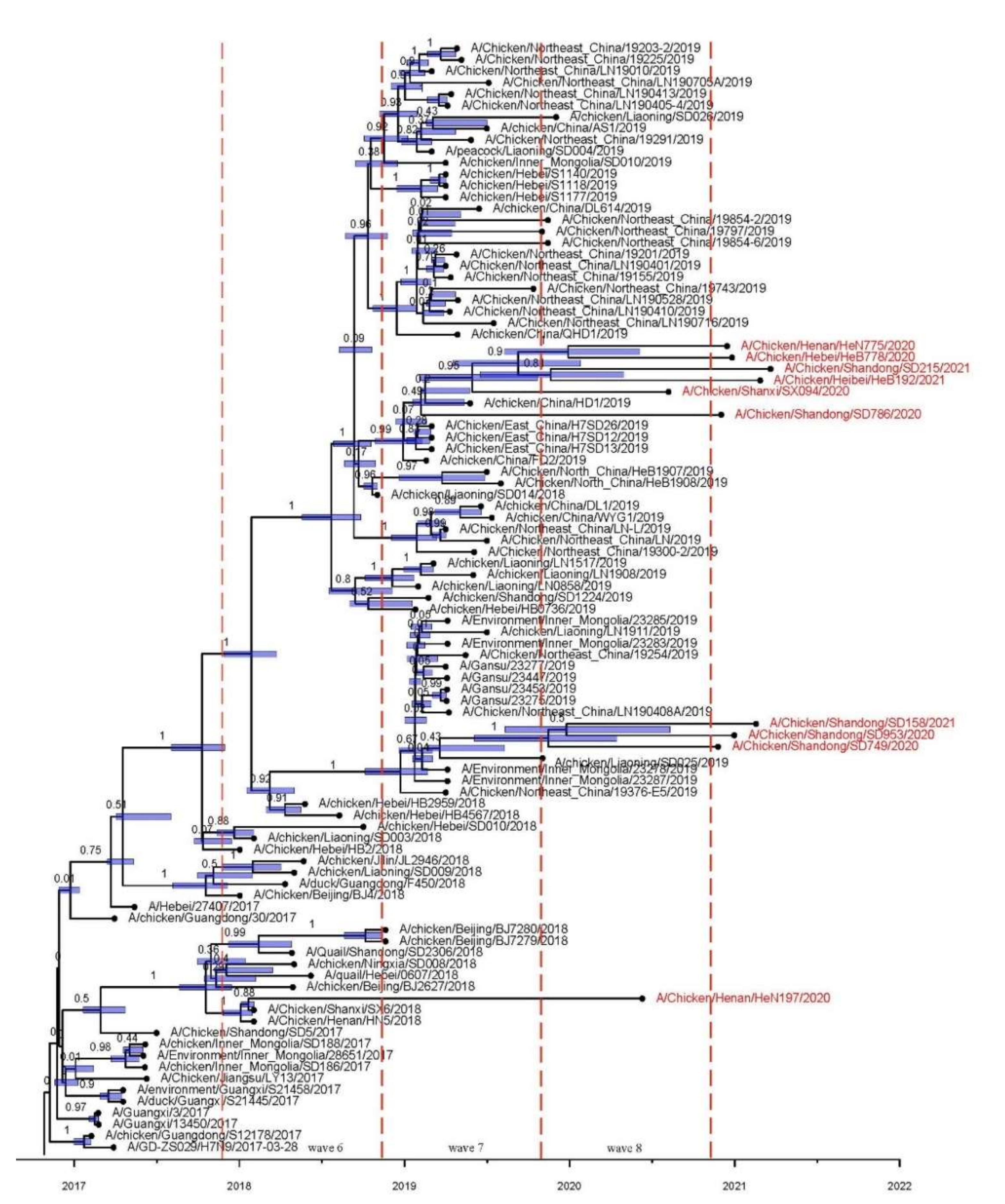
3.2. Phylogenetic and Population Dynamic Analyses
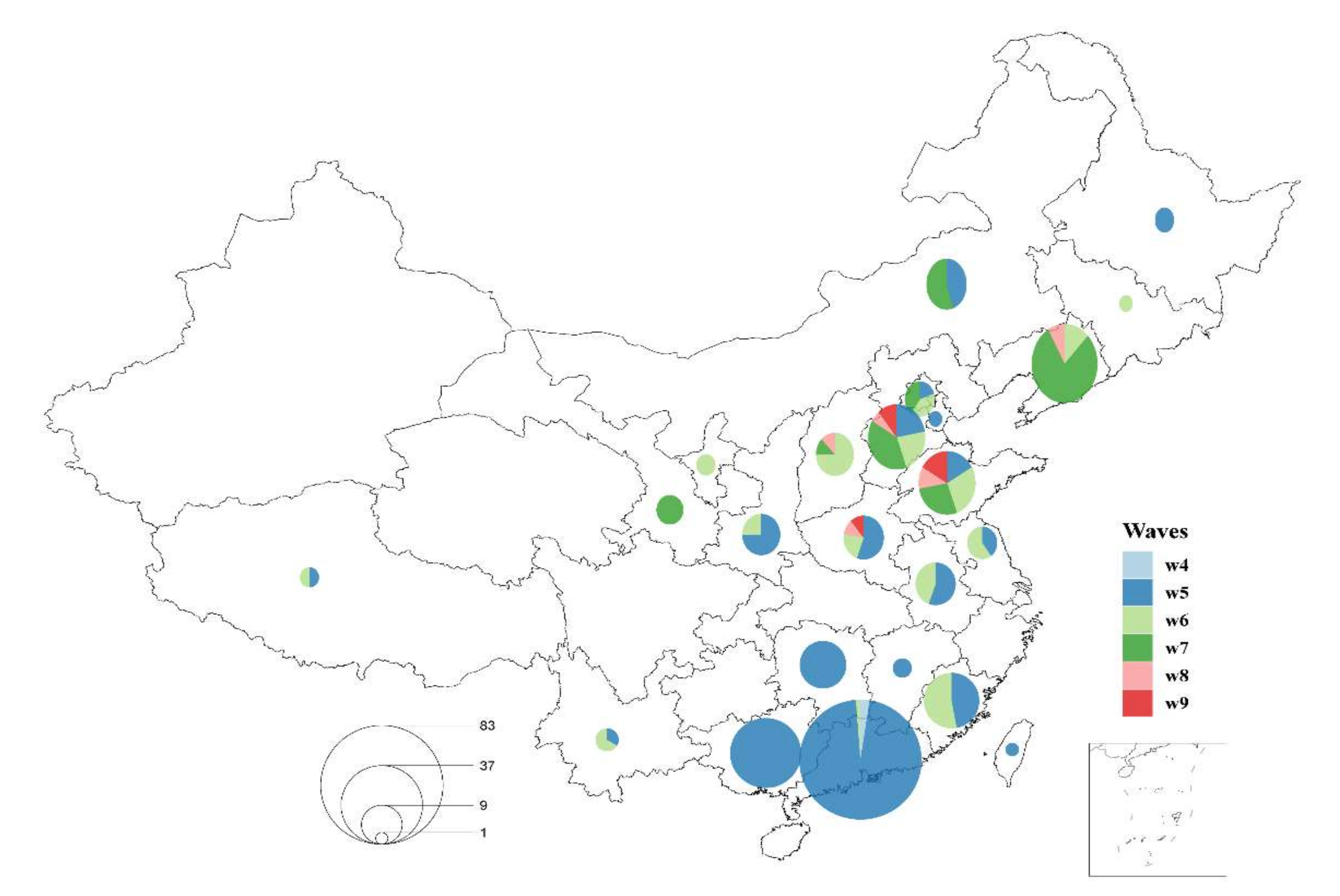
3.3. Spatial and Temporal Dynamics of HP H7N9 AIV
3.4. Natural Selection and Parallel Evolution
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ke, C.; Mok, C.K.P.; Zhu, W.; Zhou, H.; He, J.; Guan, W.; Wu, J.; Song, W.; Wang, D.; Liu, J.; et al. Human Infection with Highly Pathogenic Avian Influenza A(H7N9) Virus, China. Emerg. Infect. Dis. 2017, 23, 1332–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Tan, Y.; Kang, M.; Liu, F.; Ren, R.; Wang, Y.; Chen, T.; Yang, Y.; Li, C.; Wu, J.; et al. Preliminary Epidemiology of Human Infections with Highly Pathogenic Avian Influenza A(H7N9) Virus, China, 2017. Emerg. Infect. Dis. 2017, 23, 1355–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Chen, H. H7N9 Influenza Virus in China. Cold Spring Harb. Perspect. Med. 2021, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, W.; Jia, W.; Liu, D.; Li, J.; Bi, Y.; Xie, S.; Li, B.; Hu, T.; Du, Y.; Xing, L.; et al. Emergence and Adaptation of a Novel Highly Pathogenic H7N9 Influenza Virus in Birds and Humans from a 2013 Human-Infecting Low-Pathogenic Ancestor. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhang, J.; Zhu, W.; Zhang, Y.; Tan, H.; Liu, M.; Cai, M.; Shen, J.; Ly, H.; Chen, J. First genome report and analysis of chicken H7N9 influenza viruses with poly-basic amino acids insertion in the hemagglutinin cleavage site. Sci. Rep. 2017, 7, 9972. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Zhao, X.; Lemey, P.; Suchard, M.A.; Bi, Y.; Shi, W.; Liu, D.; Qi, W.; Zhang, G.; Stenseth, N.C.; et al. Assessing the role of live poultry trade in community-structured transmission of avian influenza in China. Proc. Natl. Acad. Sci. USA 2020, 117, 5949–5954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, R.; Sikkema, R.S.; Munnink, B.B.O.; Li, C.R.; Wu, J.; Zou, L.; Jing, Y.; Lu, J.; Yuan, R.Y.; Liao, M.; et al. Exploring utility of genomic epidemiology to trace origins of highly pathogenic influenza A/H7N9 in Guangdong. Virus Evol. 2020, 6, veaa097. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, J.; Faria, N.R.; Zeng, X.; Song, Y.; Zou, L.; Yi, L.; Liang, L.; Ni, H.; Kang, M.; et al. Effect of Live Poultry Market Interventions on Influenza A(H7N9) Virus, Guangdong, China. Emerg. Infect. Dis. 2016, 22, 2104–2112. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Yang, L.; Shu, Y. Did the Highly Pathogenic Avian Influenza A(H7N9) Viruses Emerged in China Raise Increased Threat to Public Health? Vector Borne Zoonotic Dis. 2019, 19, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Hou, G.; Li, J.; Peng, C.; Wang, S.; Liu, S.; Zhuang, Q.; Yuan, L.; Yu, X.; Li, Y.; et al. Antigenic Variant of Highly Pathogenic Avian Influenza A(H7N9) Virus, China, 2019. Emerg. Infect. Dis. 2020, 26, 379–380. [Google Scholar] [CrossRef] [Green Version]
- Ministry of Agriculture and Rural Affairs of China. Available online: http://www.moa.gov.cn/govpublic/SYJ/201707/t20170711_5744436.htm (accessed on 21 May 2021).
- Zhang, J.; Ye, H.; Li, H.; Ma, K.; Qiu, W.; Chen, Y.; Qiu, Z.; Li, B.; Jia, W.; Liang, Z.; et al. Evolution and Antigenic Drift of Influenza A (H7N9) Viruses, China, 2017-2019. Emerg. Infect. Dis. 2020, 26, 1906–1911. [Google Scholar] [CrossRef]
- Wei, X.; Cui, J. Why were so few people infected with H7N9 influenza A viruses in China from late 2017 to 2018? Sci. China Life Sci. 2018, 61, 1442–1444. [Google Scholar] [CrossRef]
- Wu, J.; Ke, C.; Lau, E.H.Y.; Song, Y.; Cheng, K.L.; Zou, L.; Kang, M.; Song, T.; Peiris, M.; Yen, H.-L. Influenza H5/H7 Virus Vaccination in Poultry and Reduction of Zoonotic Infections, Guangdong Province, China, 2017–18. Emerg. Infect. Dis. 2019, 25, 116–118. [Google Scholar] [CrossRef]
- Zeng, X.; Tian, G.; Shi, J.; Deng, G.; Li, C.; Chen, H. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Sci. China Life Sci. 2018, 61, 1465–1473. [Google Scholar] [CrossRef]
- Yin, X.; Deng, G.; Zeng, X.; Cui, P.; Hou, Y.; Liu, Y.; Fang, J.; Pan, S.; Wang, D.; Chen, X.; et al. Genetic and biological properties of H7N9 avian influenza viruses detected after application of the H7N9 poultry vaccine in China. PLoS Pathog. 2021, 17, e1009561. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hu, J.; Jin, X.; Li, X.; Wang, J.; Zhang, M.; Chen, J.; Xie, S.; Qi, W.; Liao, M.; et al. Accelerated Evolution of H7N9 Subtype Influenza Virus under Vaccination Pressure. Virol. Sin. 2021, 35, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Gu, J.; Gu, M.; Wu, H.; Li, J.; Zhan, T.; Chen, Y.; Xu, N.; Ge, Z.; Wang, G.; et al. Genetic and antigenic diversity of H7N9 highly pathogenic avian influenza virus in China. Infect. Genet. Evol. 2021, 93, 104993. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Zhu, W.; Zhang, Y.; Zou, S.; Bo, H.; Gao, R.; Dong, J.; Huang, W.; Guo, J.; et al. Two Outbreak Sources of Influenza A (H7N9) Viruses Have Been Established in China. J. Virol. 2016, 90, 5561–5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Shi, W.; Zhang, L.; Xu, Y.; Xu, J.; Li, S.; Zhang, J.; Hu, K.; Ma, C.; Zhao, X.; et al. Westward Spread of Highly Pathogenic Avian Influenza A(H7N9) Virus among Humans, China. Emerg. Infect. Dis. 2018, 24, 1095–1098. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Zhang, W.; Wang, F.; Qi, J.; Wu, Y.; Song, H.; Gao, F.; Bi, Y.; Zhang, Y.; Fan, Z.; et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 2013, 342, 243–247. [Google Scholar] [CrossRef]
- Xu, R.; de Vries, R.P.; Zhu, X.; Nycholat, C.M.; McBride, R.; Yu, W.; Paulson, J.C.; Wilson, I.A. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 2013, 342, 1230–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Peng, R.; Zhang, W.; Qi, J.; Song, H.; Liu, S.; Wang, H.; Wang, M.; Xiao, H.; Fu, L.; et al. Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin. Cell Rep. 2019, 29, 2217–2228. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, B.; Escalera-Zamudio, M.; Pybus, O.G. Parallel molecular evolution and adaptation in viruses. Curr. Opin. Virol. 2019, 34, 90–96. [Google Scholar] [CrossRef]
- Escalera-Zamudio, M.; Golden, M.; Gutiérrez, B.; Thézé, J.; Keown, J.R.; Carrique, L.; Bowden, T.A.; Pybus, O.G. Publisher Correction: Parallel evolution in the emergence of highly pathogenic avian influenza A viruses. Nat. Commun. 2020, 11, 6070. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shen, X.; Pu, Z.; Irwin, D.M.; Liao, M.; Shen, Y. Convergent Evolution of Human-Isolated H7N9 Avian Influenza A Viruses. J. Infect. Dis. 2018, 217, 1699–1707. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.-J.; Qiu, Y.; Pu, Y.; Huang, X.; Ge, X.-Y. BioAider: An efficient tool for viral genome analysis and its application in tracing SARS-CoV-2 transmission. Sustain. Cities Soc. 2020, 63, 102466. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, V.; Baele, G. Bayesian estimation of past population dynamics in BEAST 1.10 using the Skygrid coalescent model. Mol. Biol. Evol. 2019, 36, 2620–2628. [Google Scholar] [CrossRef] [Green Version]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegrant Graphics for Data Analysis/Hadley Wickham, 2nd ed.; With Contributions by Carson Sievert; Springer: Basel, Switzerland, 2016; ISBN 9783319242750. [Google Scholar]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect. Genet. Evol. 2008, 8, 239–246. [Google Scholar] [CrossRef]
- Yuan, F.; Nguyen, H.; Graur, D. ProtParCon: A Framework for Processing Molecular Data and Identifying Parallel and Convergent Amino Acid Replacements. Genes 2019, 10, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spielman, S.J.; Weaver, S.; Shank, S.D.; Magalis, B.R.; Li, M.; Kosakovsky Pond, S.L. Evolution of Viral Genomes: Interplay between Selection, Recombination, and Other Forces; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödinger, L.L. The PyMOL Molecular Graphics System, version 1.8; Academic Publisher: Cambridge, MA, USA, 2015. [Google Scholar]
- Zhou, X.; Li, Y.; Wang, Y.; Edwards, J.; Guo, F.; Clements, A.C.A.; Huang, B.; Magalhaes, R.J.S. The role of live poultry movement and live bird market biosecurity in the epidemiology of influenza A (H7N9): A cross-sectional observational study in four eastern China provinces: A cross-sectional observational study in four eastern China provinces. J. Infect. 2015, 71, 470–479. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhu, W.; Li, X.; Chen, M.; Wu, J.; Yu, P.; Qi, S.; Huang, Y.; Shi, W.; Dong, J.; et al. Genesis and Spread of Newly Emerged Highly Pathogenic H7N9 Avian Viruses in Mainland China. J. Virol. 2017, 91, e01277-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naguib, M.M.; Verhagen, J.H.; Mostafa, A.; Wille, M.; Li, R.; Graaf, A.; Järhult, J.D.; Ellström, P.; Zohari, S.; Lundkvist, Å.; et al. Global patterns of avian influenza A (H7): Virus evolution and zoonotic threats. FEMS Microbiol. Rev. 2019, 43, 608–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millman, A.J.; Havers, F.; Iuliano, A.D.; Davis, C.T.; Sar, B.; Sovann, L.; Chin, S.; Corwin, A.L.; Vongphrachanh, P.; Douangngeun, B.; et al. Detecting Spread of Avian Influenza A(H7N9) Virus Beyond China. Emerg. Infect. Dis. 2015, 21, 741–749. [Google Scholar] [CrossRef] [Green Version]
- Pantin-Jackwood, M.J.; Miller, P.J.; Spackman, E.; Swayne, D.E.; Susta, L.; Costa-Hurtado, M.; Suarez, D.L. Role of poultry in the spread of novel H7N9 influenza virus in China. J. Virol. 2014, 88, 5381–5390. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Wang, J.; Su, W.; Gao, S.; Luo, J.; Zhang, M.; Xie, L.; Liu, S.; Liu, X.; Chen, Y.; et al. Relationship between domestic and wild birds in live poultry market and a novel human H7N9 virus in China. J. Infect. Dis. 2014, 209, 34–37. [Google Scholar] [CrossRef] [Green Version]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-H.; Erazo, E.M.; Ishcol, M.R.C.; Lin, C.-Y.; Assavalapsakul, W.; Thitithanyanont, A.; Wang, S.-F. Virus-induced pathogenesis, vaccine development, and diagnosis of novel H7N9 avian influenza A virus in humans: A systemic literature review: A systemic literature review. J. Int. Med. Res. 2020, 48, 300060519845488. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Shi, Y.; Lu, X.; Shu, Y.; Qi, J.; Gao, G.F. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science 2013, 340, 1463–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, C.A.; Fonville, J.M.; Brown, A.E.X.; Burke, D.F.; Smith, D.L.; James, S.L.; Herfst, S.; van Boheemen, S.; Linster, M.; Schrauwen, E.J.; et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 2012, 336, 1541–1547. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Li, N.; Li, W.; Zhou, J.; Ning, R.; Hou, M.; Liu, L. New hemagglutinin dual-receptor-binding pattern of a human-infecting influenza A (H7N9) virus isolated after fifth epidemic wave. Virus Evol. 2020, 6, veaa021. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stevens, D.J.; Haire, L.F.; Walker, P.A.; Coombs, P.J.; Russell, R.J.; Gamblin, S.J.; Skehel, J.J. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl. Acad. Sci. USA 2009, 106, 17175–17180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, Y.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 2003, 309, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Qi, J.; Xiao, H.; Bi, Y.; Zhang, W.; Xu, Y.; Wang, F.; Shi, Y.; Gao, G.F. Avian-to-Human Receptor-Binding Adaptation by Influenza A Virus Hemagglutinin H4. Cell Rep. 2017, 20, 1201–1214. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Peng, W.; Grant, O.C.; Thompson, A.J.; Zhu, X.; Bouwman, K.M.; de La Pena, A.T.T.; van Breemen, M.J.; Ambepitiya Wickramasinghe, I.N.; de Haan, C.A.M.; et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog. 2017, 13, e1006390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.; Sealy, J.E.; Sadeyen, J.-R.; Iqbal, M. Amino Acid Residue 217 in the Hemagglutinin Glycoprotein Is a Key Mediator of Avian Influenza H7N9 Virus Antigenicity. J. Virol. 2019, 93, e01627-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, T.; Nie, J.; Huang, W.; Li, C.; Li, X.; Liu, Q.; Zhao, H.; Wang, Y. Antigenic Drift of Influenza A(H7N9) Virus Hemagglutinin. J. Infect. Dis. 2019, 219, 19–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Full Name | Cleavage Site | Wave | Province | Location | Vaccine | Avian Species | Source | Date |
|---|---|---|---|---|---|---|---|---|
| A/Chicken/Henan/HeN197/2020 | KRTARG | w8 | Henan | Farm | unknown | Layer | Diagnosis | 9 June 2020 |
| A/Chicken/Shanxi/SX094/2020 | KRTARG | w8 | Shanxi | Farm | unknown | Layer | Diagnosis | 6 August 2020 |
| A/Chicken/Shandong/SD749/2020 | KRTARG | w9 | Shandong | Farm | unknown | Layer | Diagnosis | 24 November 2020 |
| A/Chicken/Shandong/SD786/2020 | KRTARG | w9 | Shandong | Farm | H5+ H7 | Layer | Diagnosis | 1 December 2020 |
| A/Chicken/Henan/HeN775/2020 | KRTARG | w9 | Henan | Farm | unknown | Broiler | Diagnosis | 14 December 2020 |
| A/Chicken/Hebei/HeB778/2020 | KRTARG | w9 | Hebei | Farm | unknown | Layer | Diagnosis | 25 December 2020 |
| A/Chicken/Shandong/SD953/2020 | KRTARG | w9 | Shandong | Farm | unknown | Layer | Diagnosis | 30 December 2020 |
| A/Chicken/Shandong/SD158/2021 | KRTARG | w9 | Shandong | Farm | H5 + H7 | Layer | Diagnosis | 16 February 2021 |
| A/Chicken/Hebei/HeB192/2021 | KRTARG | w9 | Hebei | Farm | H5 + H7 | Layer | Diagnosis | 25 February 2021 |
| A/Chicken/Shandong/SD215/2021 | KRTARG | w9 | Shandong | Farm | unknown | Layer | Diagnosis | 20 March 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, D.; Gu, M.; Wang, X.; Wang, X.; Li, G.; Yan, Y.; Gu, J.; Zhan, T.; Wu, H.; Hao, X.; et al. Spatiotemporal Associations and Molecular Evolution of Highly Pathogenic Avian Influenza A H7N9 Virus in China from 2017 to 2021. Viruses 2021, 13, 2524. https://doi.org/10.3390/v13122524
He D, Gu M, Wang X, Wang X, Li G, Yan Y, Gu J, Zhan T, Wu H, Hao X, et al. Spatiotemporal Associations and Molecular Evolution of Highly Pathogenic Avian Influenza A H7N9 Virus in China from 2017 to 2021. Viruses. 2021; 13(12):2524. https://doi.org/10.3390/v13122524
Chicago/Turabian StyleHe, Dongchang, Min Gu, Xiyue Wang, Xiaoquan Wang, Gairu Li, Yayao Yan, Jinyuan Gu, Tiansong Zhan, Huiguang Wu, Xiaoli Hao, and et al. 2021. "Spatiotemporal Associations and Molecular Evolution of Highly Pathogenic Avian Influenza A H7N9 Virus in China from 2017 to 2021" Viruses 13, no. 12: 2524. https://doi.org/10.3390/v13122524
APA StyleHe, D., Gu, M., Wang, X., Wang, X., Li, G., Yan, Y., Gu, J., Zhan, T., Wu, H., Hao, X., Wang, G., Hu, J., Hu, S., Liu, X., Su, S., Ding, C., & Liu, X. (2021). Spatiotemporal Associations and Molecular Evolution of Highly Pathogenic Avian Influenza A H7N9 Virus in China from 2017 to 2021. Viruses, 13(12), 2524. https://doi.org/10.3390/v13122524






