Metagenomics Analysis of the Wheat Virome Identifies Novel Plant and Fungal-Associated Viral Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and Library Preparation
2.3. RNASeq Analysis and Virus Genome Identification
2.4. Viral Genome Sequence Validation
2.5. Rapid Amplification of cDNA Ends
2.6. Small RNA Deep Sequencing
2.7. Phylogenetic Analysis
3. Results
3.1. RNASeq Analysis
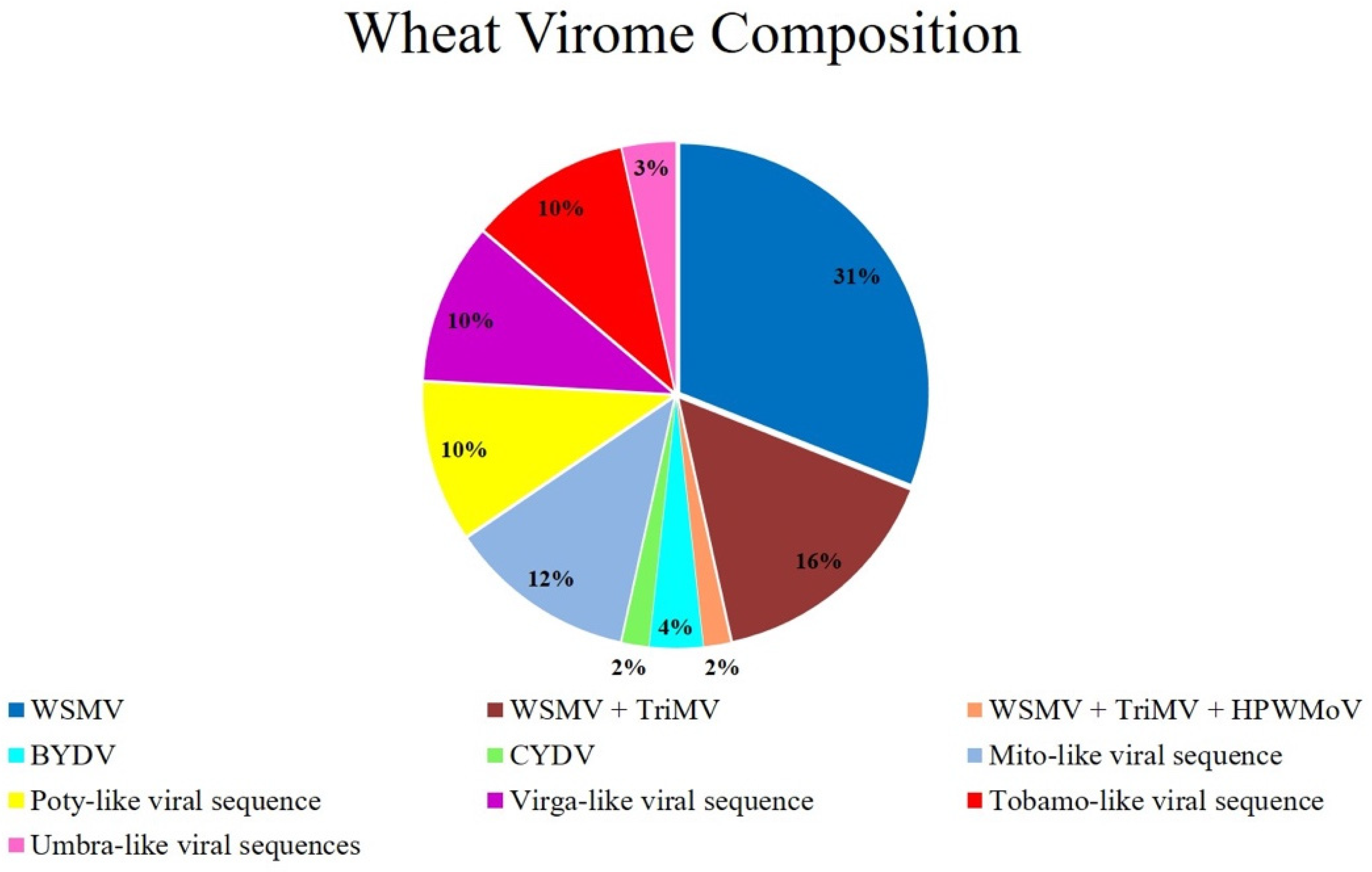
3.2. Wheat Virome Composition and Characterizing Novel Viral Sequences
3.2.1. Known Wheat Viruses
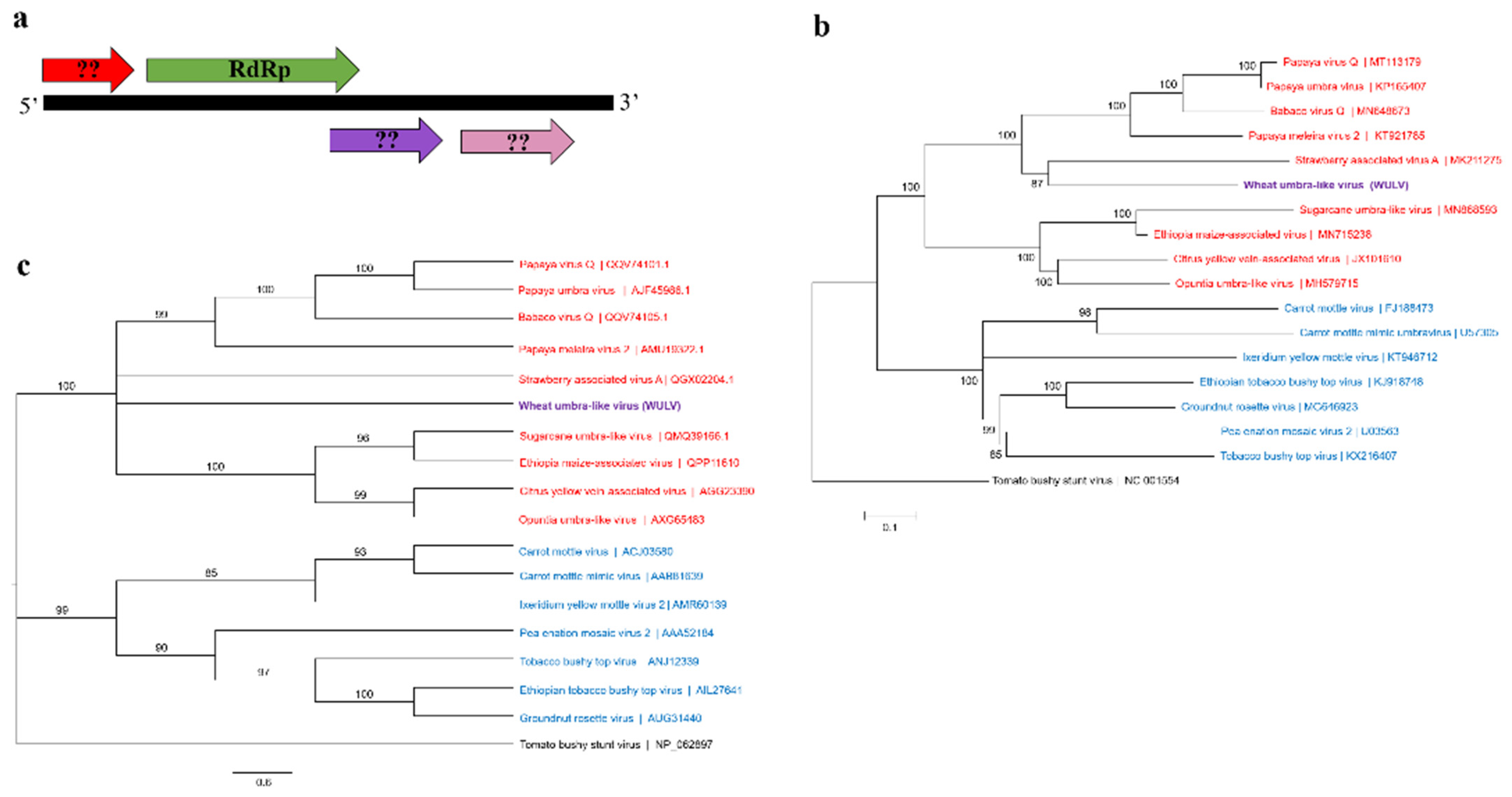
3.2.2. “Umbra-Like” Viral Sequences
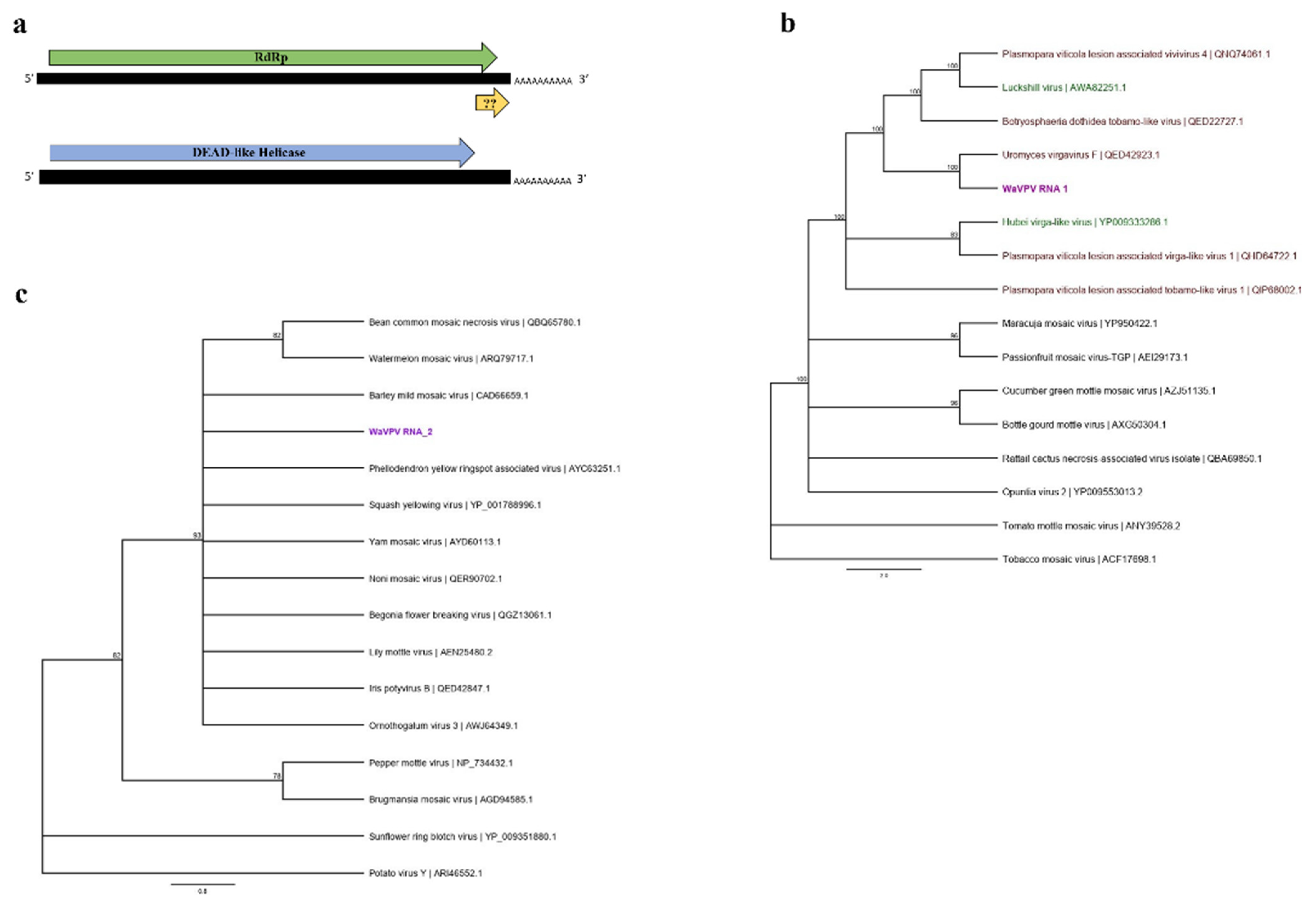
3.2.3. “Poty-Like” and “Virga-Like” Viral Sequences
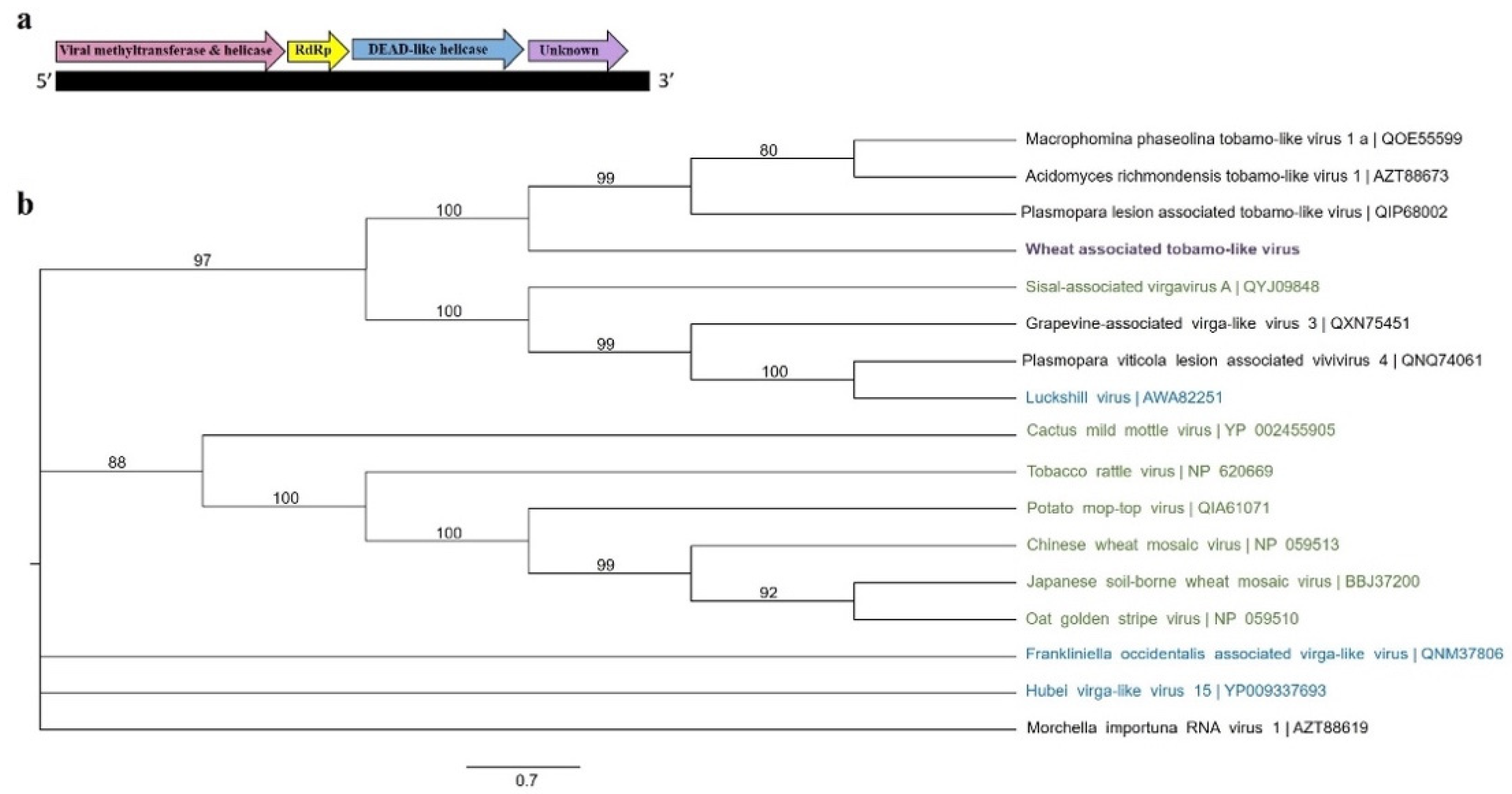
3.2.4. “Tobamo-Like” Viral Sequences
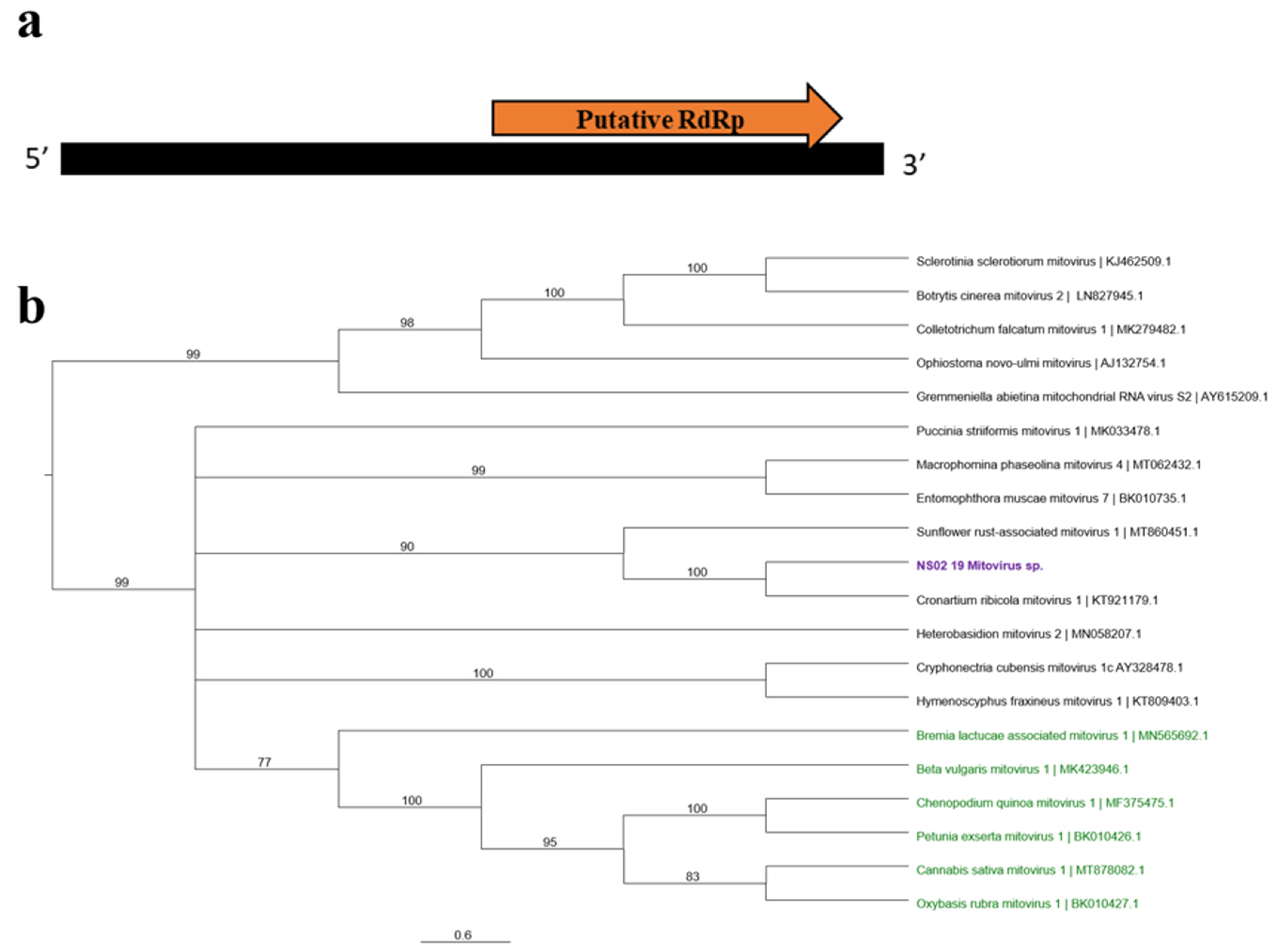
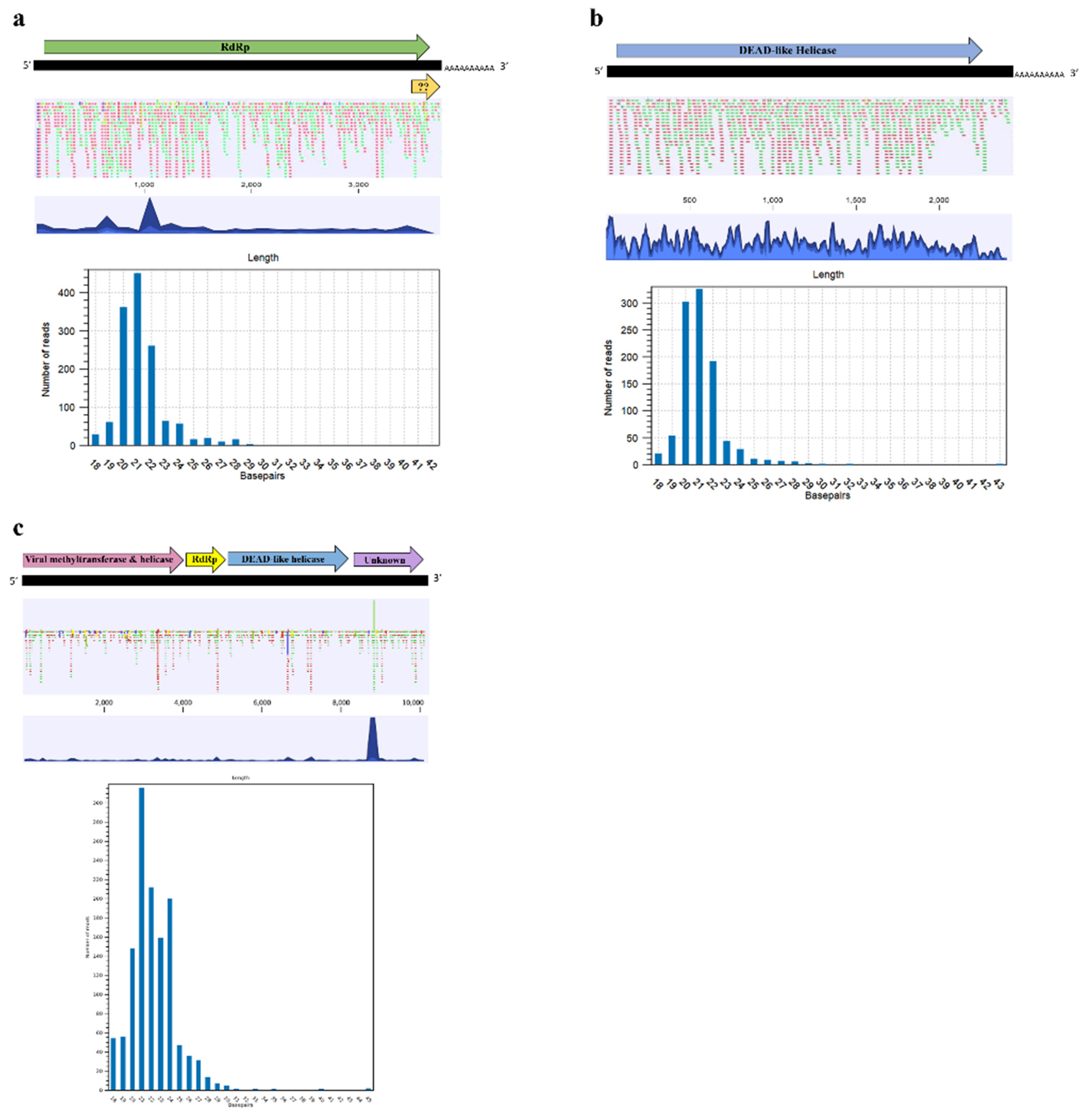
3.2.5. “Mito-Like” Viral Sequences
3.3. Small RNA Sequencing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, G.C.; Rosegrant, M.W.; Palazzo, A.; Gray, I.; Ingersoll, C.; Robertson, R.D.; Tokgoz, S.; Zhu, T.; Sulser, T.B.; Ringler, C.; et al. Food Security, Farming, and Climate Change to 2050: Scenarios; International Food Policy Research Institute: Washington, DC, USA, 2010; pp. 1–131. [Google Scholar]
- Hodge, B.A.; Paul, P.A.; Stewart, L.R. Occurrence and High-Throughput Sequencing of Viruses in Ohio Wheat. Plant Dis. 2020, 104, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Rotenberg, D.; Bockus, W.W.; Whitfield, A.E.; Hervey, K.; Baker, K.D.; Ou, Z.; Laney, A.G.; De Wolf, E.D.; Appel, J.A. Occurrence of Viruses and Associated Grain Yields of Paired Symptomatic and Nonsymptomatic Tillers in Kansas Winter Wheat Fields. Phytopathology 2016, 106, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Jarošová, J.; Fousek, J.; Chen, H.; Kundu, J.K. Virome Identification in Wheat in the Czech Republic Using Small RNA Deep Sequencing. J. Integr. Agric. 2020, 19, 1825–1833. [Google Scholar] [CrossRef]
- Hollandbeck, G.; DeWolf, E.; Todd, T. Kansas Cooperative Plant Disease Survey Report Preliminary 2017 Kansas Wheat Disease Loss Estimates. Plant Dis. Surv. Rep. 2017. Available online: https://agriculture.ks.gov/docs/default-source/pp-disease-reports-2012/2017-ks-wheat-disease-loss-estimates.pdf?sfvrsn=d49587c1_0 (accessed on 20 June 2020).
- Stenger, D.C.; Hall, J.S.; Choi, I.-R.; French, R. Phylogenetic Relationships Within the Family Potyviridae: Wheat Streak Mosaic Virus and Brome Streak Mosaic Virus Are Not Members of the Genus Rymovirus. Phytopathology 1998, 88, 782–787. [Google Scholar] [CrossRef]
- Seifers, D.L.; Martin, T.J.; Harvey, T.L.; Fellers, J.P.; Stack, J.P.; Ryba-White, M.; Haber, S.; Krokhin, O.; Spicer, V.; Lovat, N.; et al. Triticum Mosaic Virus: A New Virus Isolated from Wheat in Kansas. Plant Dis. 2008, 92, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Fellers, J.P.; Seifers, D.; Ryba-White, M.; Joe Martin, T. The Complete Genome Sequence of Triticum Mosaic Virus, a New Wheat-Infecting Virus of the High Plains. Arch. Virol. 2009, 154, 1511–1515. [Google Scholar] [CrossRef]
- Tatineni, S.; McMechan, A.J.; Wosula, E.N.; Wegulo, S.N.; Graybosch, R.A.; French, R.; Hein, G.L. An Eriophyid Mite-Transmitted Plant Virus Contains Eight Genomic RNA Segments with Unusual Heterogeneity in the Nucleocapsid Protein. J. Virol. 2014, 88, 11834–11845. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Hein, G.L. Genetics and Mechanisms Underlying Transmission of Wheat Streak Mosaic Virus by the Wheat Curl Mite. Curr. Opin. Virol. 2018, 33, 47–54. [Google Scholar] [CrossRef]
- Miller, W.A.; Rasochová, L. Barley Yellow Dwarf Viruses. Annu. Rev. Phytopathol. 1997, 35, 167–190. [Google Scholar] [CrossRef]
- Walls, J.; Rajotte, E.; Rosa, C. The Past, Present, and Future of Barley Yellow Dwarf Management. Agriculture 2019, 9, 23. [Google Scholar] [CrossRef]
- Bockus, W.W.; Appel, J.A.; Bowden, R.L.; Fritz, A.K.; Gill, B.S.; Martin, T.J.; Sears, R.G.; Seifers, D.L.; Brown-Guedira, G.L.; Eversmeyer, M.G. Success Stories: Breeding for Wheat Disease Resistance in Kansas. Plant Dis. 2001, 85, 453–461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Appel, J.; DeWolf, E.; Bockus, W.; Todd, T. Preliminary 2014 Kansas Wheat Disease Loss Estimates. Kansas Cooperative. Plant Dis. Surv. Rep. 2014. Available online: https://http://agriculture.ks.gov/docs/default-source/PP-Disease-Reports-2014/2014-ks-wheat-disease-loss-estimates.pdf (accessed on 20 June 2020).
- Gaunce, G.M.; Bockus, W.W. Estimating Yield Losses Due to Barley Yellow Dwarf on Winter Wheat in Kansas Using Disease Phenotypic Data. Plant. Health Prog. 2015, 16, 1–6. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, Z.; Hu, H.; Chen, Z.; Liu, P.; Gao, S.; Zhang, F.; He, L.; Jin, P.; Xu, M.; et al. Transcriptome-Wide N6-Methyladenosine (M6A) Profiling of Susceptible and Resistant Wheat Varieties Reveals the Involvement of Variety-Specific M6A Modification Involved in Virus-Host Interaction Pathways. Front. Microbiol. 2021, 12, 656302. [Google Scholar] [CrossRef] [PubMed]
- Nygren, J.; Shad, N.; Kvarnheden, A.; Westerbergh, A. Variation in Susceptibility to Wheat Dwarf Virus among Wild and Domesticated Wheat. PLoS ONE 2015, 10, e0121580. [Google Scholar] [CrossRef] [PubMed]
- Monier, A.; Claverie, J.-M.; Ogata, H. Taxonomic Distribution of Large DNA Viruses in the Sea. Genome Biol. 2008, 9, R106. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the Invertebrate RNA Virosphere. Nat. Cell Biol. 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Chen, X.; Tian, J.-H.; Chen, L.-J.; Li, K.; Wang, W.; Eden, J.-S.; Shen, J.-J.; Liu, L.; et al. The Evolutionary History of Vertebrate RNA Viruses. Nat. Cell Biol. 2018, 556, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Creager, A.N.H.; Scholthof, K.-B.G.; Citovsky, V.; Scholthof, H.B. Tobacco Mosaic Virus: Pioneering Research for a Century. Plant Cell 1999, 11, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-J.; Ju, H.-J.; Noh, J. A Review of Detection Methods for the Plant Viruses. Res. Plant Dis. 2014, 20, 173–181. [Google Scholar] [CrossRef]
- Zakrzewski, M.; Rašić, G.; Darbro, J.; Krause, L.; Poo, Y.S.; Filipović, I.; Parry, R.; Asgari, S.; Devine, G.; Suhrbier, A. Mapping the Virome in Wild-Caught Aedes Aegypti from Cairns and Bangkok. Sci. Rep. 2018, 8, 4690. [Google Scholar] [CrossRef] [PubMed]
- Murugan, M.; Cardona, P.S.; Duraimurugan, P.; Whitfield, A.E.; Schneweis, D.; Starkey, S.; Smith, C.M. Wheat Curl Mite Resistance: Interactions of Mite Feeding with Wheat Streak Mosaic Virus Infection. J. Econ. Èntom. 2011, 104, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Li, X.-L.; Wu, Y.-L.; Sun, Q.-Z.; Zhang, W.; Cao, M.; Wang, J.-J. RNA Virome Screening in Diverse but Ecologically Related Citrus Pests Reveals Potential Virus-Host Interactions. J. Invertebr. Pathol. 2020, 170, 107329. [Google Scholar] [CrossRef]
- Tatineni, S.; Alexander, J.; Gupta, A.K.; French, R. Asymmetry in Synergistic Interaction Between Wheat Streak Mosaic Virus and Triticum Mosaic Virus in Wheat. Mol. Plant-Microbe Interact. 2019, 32, 336–350. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant Virus Metagenomics: Advances in Virus Discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Mumo, N.N.; Mamati, G.E.; Ateka, E.M.; Rimberia, F.K.; Asudi, G.O.; Boykin, L.M.; Machuka, E.M.; Njuguna, J.N.; Pelle, R.; Stomeo, F. Metagenomic Analysis of Plant Viruses Associated With Papaya Ringspot Disease in Carica Papaya L. in Kenya. Front. Microbiol. 2020, 11, 205. [Google Scholar] [CrossRef]
- Adams, I.P.; Glover, R.H.; Monger, W.A.; Mumford, R.; Jackeviciene, E.; Navalinskiene, M.; Samuitiene, M.; Boonham, N. Next-Generation Sequencing and Metagenomic Analysis: A Universal Diagnostic Tool in Plant Virology. Mol. Plant Pathol. 2009, 10, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High Throughput Sequencing For Plant Virus Detection and Discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Chiapello, M.; Rodríguez-Romero, J.; Ayllón, M.A.; Turina, M. Analysis of the Virome Associated to Grapevine Downy Mildew Lesions Reveals New Mycovirus Lineages. Virus Evol. 2020, 6, veaa058. [Google Scholar] [CrossRef] [PubMed]
- Marzano, S.-Y.L.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of Diverse Mycoviruses through Metatranscriptomics Characterization of the Viromes of Five Major Fungal Plant Pathogens. J. Virol. 2016, 90, 6846–6863. [Google Scholar] [CrossRef] [PubMed]
- Otuka, A. Migration of Rice Planthoppers and Their Vectored Re-Emerging and Novel Rice Viruses in East Asia. Front. Microbiol. 2013, 4, 309. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, E.; Coletta-Filho, H.; Nouri, S.; Falk, B.; Nerva, L.; Oliveira, T.; Dorta, S.; Machado, M. Deep Sequencing Analysis of RNAs from Citrus Plants Grown in a Citrus Sudden Death-Affected Area Reveals Diverse Known and Putative Novel Viruses. Viruses 2017, 9, 92. [Google Scholar] [CrossRef]
- Olmedo-Velarde, A.; Park, A.C.; Sugano, J.; Uchida, J.Y.; Kawate, M.; Borth, W.B.; Hu, J.S.; Melzer, M.J. Characterization of Ti Ringspot-Associated Virus, a Novel Emaravirus Associated with an Emerging Ringspot Disease of Cordyline Fruticosa. Plant Dis. 2019, 103, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Ramos-González, P.; Chabi-Jesus, C.; Guerra-Peraza, O.; Breton, M.; Arena, G.; Nunes, M.; Kitajima, E.; Machado, M.; Freitas-Astúa, J. Phylogenetic and Molecular Variability Studies Reveal a New Genetic Clade of Citrus Leprosis Virus C. Viruses 2016, 8, 153. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 June 2020).
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L.; et al. Ensembl Genomes 2020—Enabling Non-Vertebrate Genomic Research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief. Bioinform. 2004, 5, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Redila, C.D.; Phipps, S.; Nouri, S. Full Genome Evolutionary Studies of Wheat Streak Mosaic-Associated Viruses Using High-Throughput Sequencing. Front. Microbiol. 2021, 12, 699078. [Google Scholar] [CrossRef] [PubMed]
- Burrows, M.; Franc, G.; Rush, C.; Blunt, T.; Ito, D.; Kinzer, K.; Olson, J.; O’Mara, J.; Price, J.; Tande, C.; et al. Occurrence of Viruses in Wheat in the Great Plains Region, 2008. Plant Health Prog. 2009, 10, 14. [Google Scholar] [CrossRef]
- Quito-Avila, D.F.; Alvarez, R.A.; Ibarra, M.A.; Martin, R.R. Detection and Partial Genome Sequence of a New Umbra-like Virus of Papaya Discovered in Ecuador. Eur. J. Plant Pathol. 2015, 143, 199–204. [Google Scholar] [CrossRef]
- Sá Antunes, T.F.; Amaral, R.J.V.; Ventura, J.A.; Godinho, M.T.; Amaral, J.G.; Souza, F.O.; Zerbini, P.A.; Zerbini, F.M.; Fernandes, P.M.B. The DsRNA Virus Papaya Meleira Virus and an SsRNA Virus Are Associated with Papaya Sticky Disease. PLoS ONE 2016, 11, e0155240. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Franco, J.F.; Flores, F.; Mollov, D.; Quito-Avila, D.F. An Umbra-Related Virus Found in Babaco (Vasconcellea × Heilbornii). Arch. Virol. 2021, 166, 2321–2324. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.N.; Bolus, S.; Grinstead, S.C.; McFarlane, S.A.; Mollov, D. A New Virus of the Family Tombusviridae Infecting Sugarcane. Arch. Virol. 2021, 166, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.-J.; Bodaghi, S.; Dang, T.; Gadhave, K.R.; Ho, T.; Osman, F.; Al Rwahnih, M.; Tzanetakis, I.E.; Simon, A.E.; Vidalakis, G. Complete Nucleotide Sequence, Genome Organization, and Comparative Genomic Analyses of Citrus Yellow-Vein Associated Virus (CYVaV). Front. Microbiol. 2021, 12, 683130. [Google Scholar] [CrossRef]
- Syller, J. Molecular and Biological Features of Umbraviruses, the Unusual Plant Viruses Lacking Genetic Information for a Capsid Protein. Physiol. Mol. Plant Pathol. 2003, 63, 35–46. [Google Scholar] [CrossRef]
- Taliansky, M.E.; Robinson, D.J. Molecular Biology of Umbraviruses: Phantom Warriors. J. Gen. Virol. 2003, 84, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Felker, P.; Bunch, R.; Russo, G.; Preston, K.; Tine, J.A.; Suter, B.; Mo, X.H.; Cushman, J.C.; Yim, W.C. Biology and chemistry of an Umbravirus like 2989 bp single stranded RNA as a possible causal agent for Opuntia stunting disease (engrosamiento de cladodios)—A Review. J. Prof. Assoc. Cactus Dev. 2019, 21, 1–31. [Google Scholar]
- Liu, J.; Carino, E.; Bera, S.; Gao, F.; May, J.P.; Simon, A.E. Structural Analysis and Whole Genome Mapping of a New Type of Plant Virus Subviral RNA: Umbravirus-Like Associated RNAs. Viruses 2021, 13, 646. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; White, S.; Layton, M.; Stenglein, M.; Haley, S.; Nachappa, P. Ecology and Epidemiology of Wheat Curl Mite and Mite-Transmissible Viruses in Colorado and Insights into the Wheat Virome. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cornejo-Franco, J.F.; Alvarez-Quinto, R.A.; Quito-Avila, D.F. Transmission of the Umbra-like Papaya Virus Q in Ecuador and Its Association with Meleira-Related Viruses from Brazil. Crop. Protect. 2018, 110, 99–102. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Nerva, L.; Forgia, M.; Chitarra, W.; Ayllón, M.A.; Turina, M. Putative New Plant Viruses Associated with Plasmopara Viticola -infected Grapevine Samples. Ann. Appl. Biol. 2020, 176, 180–191. [Google Scholar] [CrossRef]
- Nerva, L.; Varese, G.C.; Falk, B.W.; Turina, M. Mycoviruses of an Endophytic Fungus Can Replicate in Plant Cells: Evolutionary Implications. Sci. Rep. 2017, 7, 1908. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Wei, S.; Cao, C.; Salaipeth, L.; Kondo, H.; Sun, L. Phytopathogenic Fungus Hosts a Plant Virus: A Naturally Occurring Cross-Kingdom Viral Infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12267–12272. [Google Scholar] [CrossRef] [PubMed]
- Bian, R.; Andika, I.B.; Pang, T.; Lian, Z.; Wei, S.; Niu, E.; Wu, Y.; Kondo, H.; Liu, X.; Sun, L. Facilitative and Synergistic Interactions between Fungal and Plant Viruses. Proc. Natl. Acad. Sci. USA 2020, 117, 3779–3788. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.; Priyamvada, L.; Ende, Z.; Steel, J.; Lowen, A.C. Influenza Virus Reassortment Occurs with High Frequency in the Absence of Segment Mismatch. PLoS Pathog. 2013, 9, e1003421. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Holmes, E.C.; Joseph, U.; Fourment, M.; Su, Y.C.; Halpin, R.; Lee, R.T.; Deng, Y.-M.; Gunalan, V.; Lin, X.; et al. The Contrasting Phylodynamics of Human Influenza B Viruses. eLife 2015, 4, e05055. [Google Scholar] [CrossRef] [PubMed]
- Rastgou, M.; Habibi, M.K.; Izadpanah, K.; Masenga, V.; Milne, R.G.; Wolf, Y.I.; Koonin, E.V.; Turina, M. Molecular Characterization of the Plant Virus Genus Ourmiavirus and Evidence of Inter-Kingdom Reassortment of Viral Genome Segments as Its Possible Route of Origin. J. Gen. Virol. 2009, 90, 2525–2535. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F.; Kreuze, J. Virgaviridae: A New Family of Rod-Shaped Plant Viruses. Arch. Virol. 2009, 154, 1967–1972. [Google Scholar] [CrossRef]
- Wang, J.; Ni, Y.; Liu, X.; Zhao, H.; Xiao, Y.; Xiao, X.; Li, S.; Liu, H. Divergent RNA Viruses in Macrophomina Phaseolina Exhibit Potential as Virocontrol Agents. Virus Evol. 2021, 7, veaa095. [Google Scholar] [CrossRef] [PubMed]
- Hillman, B.I.; Cai, G. The Family Narnaviridae. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 86, pp. 149–176. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA Silencing in Plants. Nat. Cell Biol. 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA Interference: Biology, Mechanism, and Applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Kuo, Y.-W.; Falk, B.W. RNA Interference Approaches for Plant Disease Control. BioTechniques 2020, 69, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Lee Marzano, S.-Y.; Neupane, A.; Domier, L. Transcriptional and Small RNA Responses of the White Mold Fungus Sclerotinia Sclerotiorum to Infection by a Virulence-Attenuating Hypovirus. Viruses 2018, 10, 713. [Google Scholar] [CrossRef]
- Pooggin, M.M. Small RNA-Omics for Plant Virus Identification, Virome Reconstruction, and Antiviral Defense Characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef] [PubMed]
- Özkan, S.; Mohorianu, I.; Xu, P.; Dalmay, T.; Coutts, R.H.A. Profile and Functional Analysis of Small RNAs Derived from Aspergillus Fumigatus Infected with Double-Stranded RNA Mycoviruses. BMC Genom. 2017, 18, 416. [Google Scholar] [CrossRef] [PubMed]
| Novel Viral Sequence | Positive Libraries | Known Virus Hits | Similarity (%) | Query Coverage (%) | E Value |
|---|---|---|---|---|---|
| Umbra-like viruses | GL01_19, GL20 | Unclassified umbraviruses | ≤60% | <55% | <0.00001 |
| Virga-like | NS02_19, FO19, EL19, KW19, GL01_19, ME19 | Plant-associated virga viruses | ≤26% | <27% | <0.00001 |
| Fungal-associated virga viruses | ≤30% | <40% | <0.00001 | ||
| Poty-like | NS02_19, FO19, EL19, KW19, GL01_19, ME19 | Potyviruses | ≤25% | >40% | <0.00001 |
| Tobamo-like | NS02_19, FO19, EL19, KW19, GL01_19, ME19 | Fungal-associated tobamo-like viruses and plant tobamoviruses | ≤40% | ~50% | <0.00001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redila, C.D.; Prakash, V.; Nouri, S. Metagenomics Analysis of the Wheat Virome Identifies Novel Plant and Fungal-Associated Viral Sequences. Viruses 2021, 13, 2457. https://doi.org/10.3390/v13122457
Redila CD, Prakash V, Nouri S. Metagenomics Analysis of the Wheat Virome Identifies Novel Plant and Fungal-Associated Viral Sequences. Viruses. 2021; 13(12):2457. https://doi.org/10.3390/v13122457
Chicago/Turabian StyleRedila, Carla Dizon, Ved Prakash, and Shahideh Nouri. 2021. "Metagenomics Analysis of the Wheat Virome Identifies Novel Plant and Fungal-Associated Viral Sequences" Viruses 13, no. 12: 2457. https://doi.org/10.3390/v13122457
APA StyleRedila, C. D., Prakash, V., & Nouri, S. (2021). Metagenomics Analysis of the Wheat Virome Identifies Novel Plant and Fungal-Associated Viral Sequences. Viruses, 13(12), 2457. https://doi.org/10.3390/v13122457







