Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
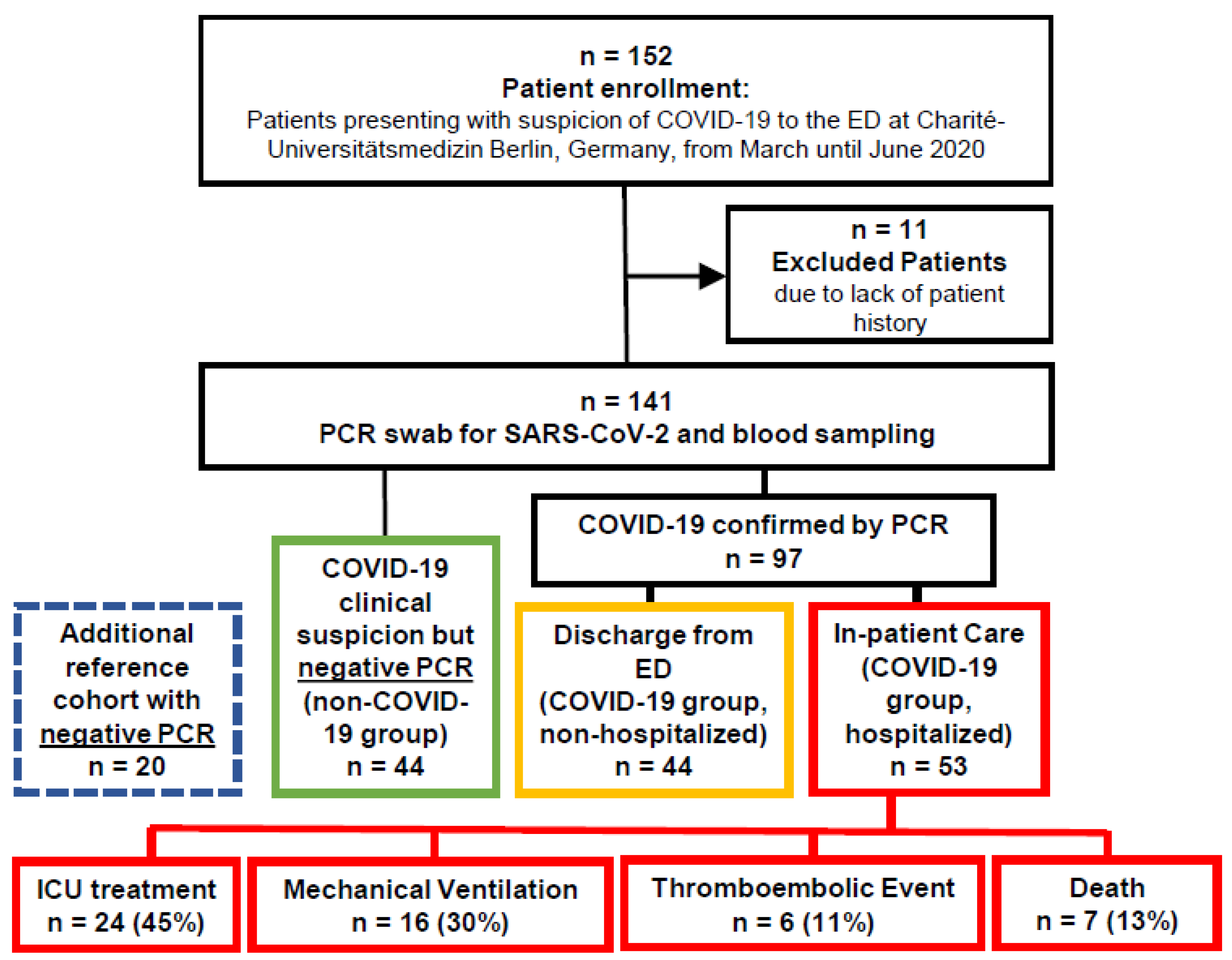
2.1. Study Population
2.2. Standard of Care Blood Analyses
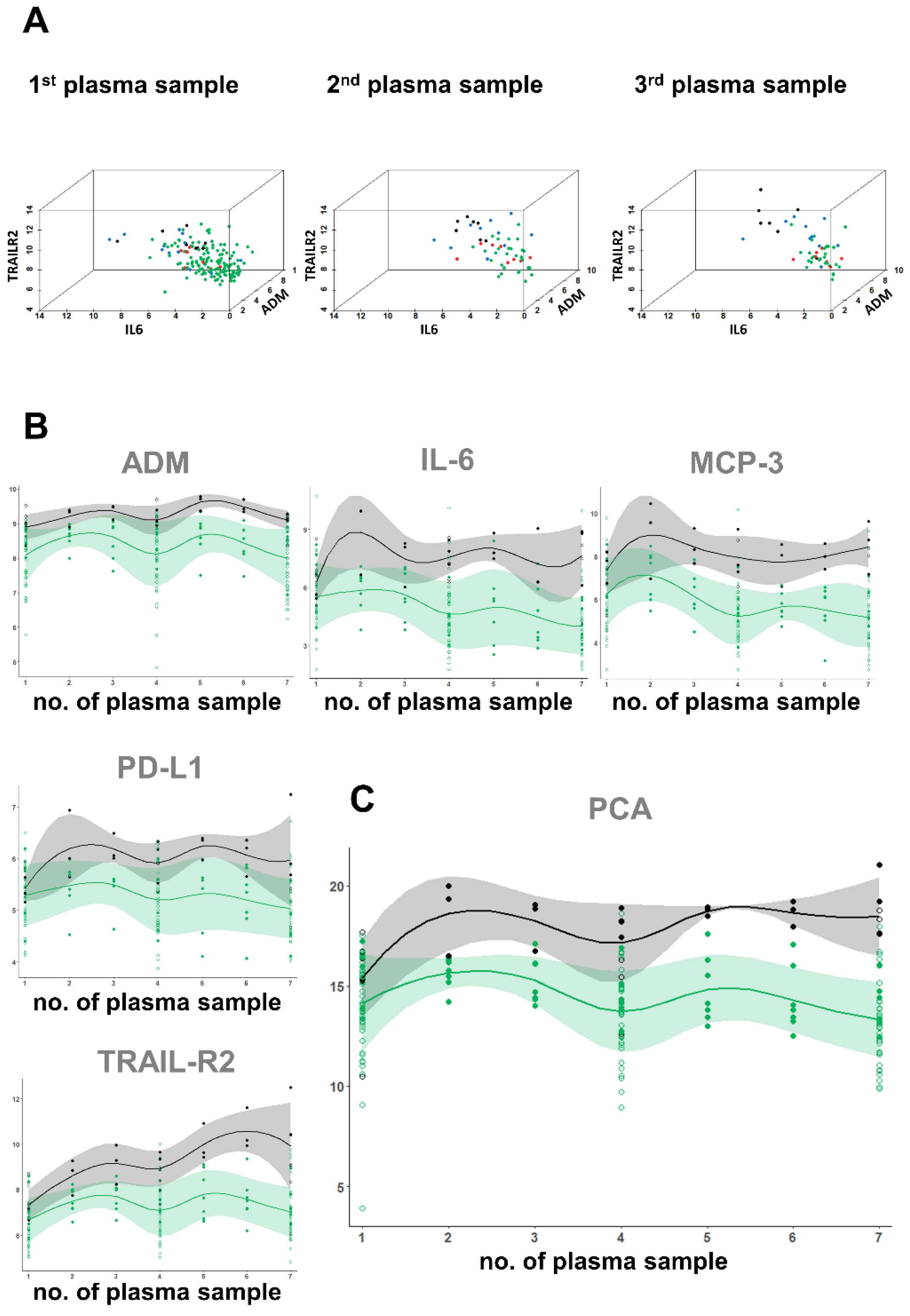
2.3. Fingerprinting the Circulating Proteome
2.4. Statistics
3. Results
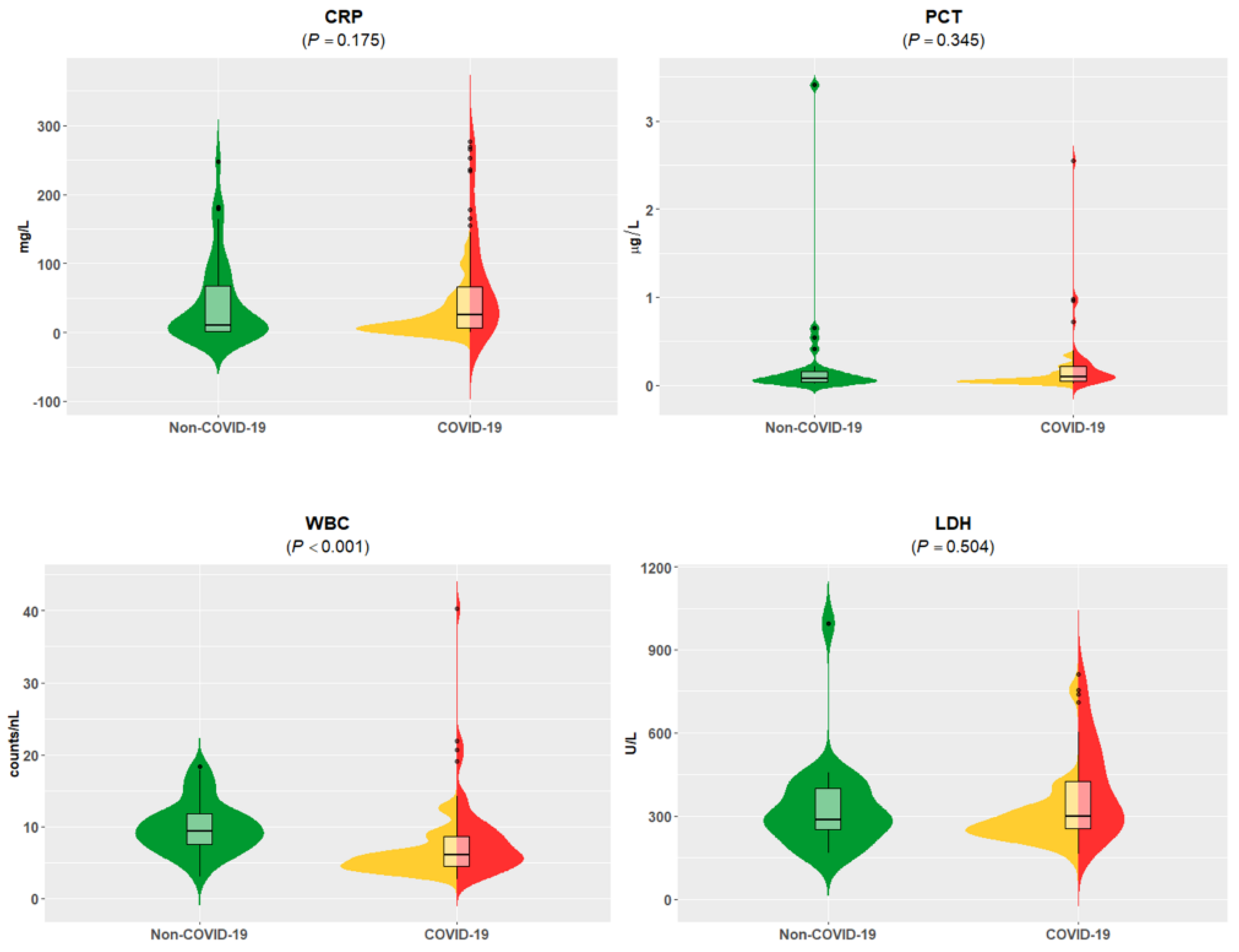
3.1. Patient Stratification, Clinical and Standard of Care Laboratory Assessment
3.2. Plasma Proteins Associated to COVID-19 Status
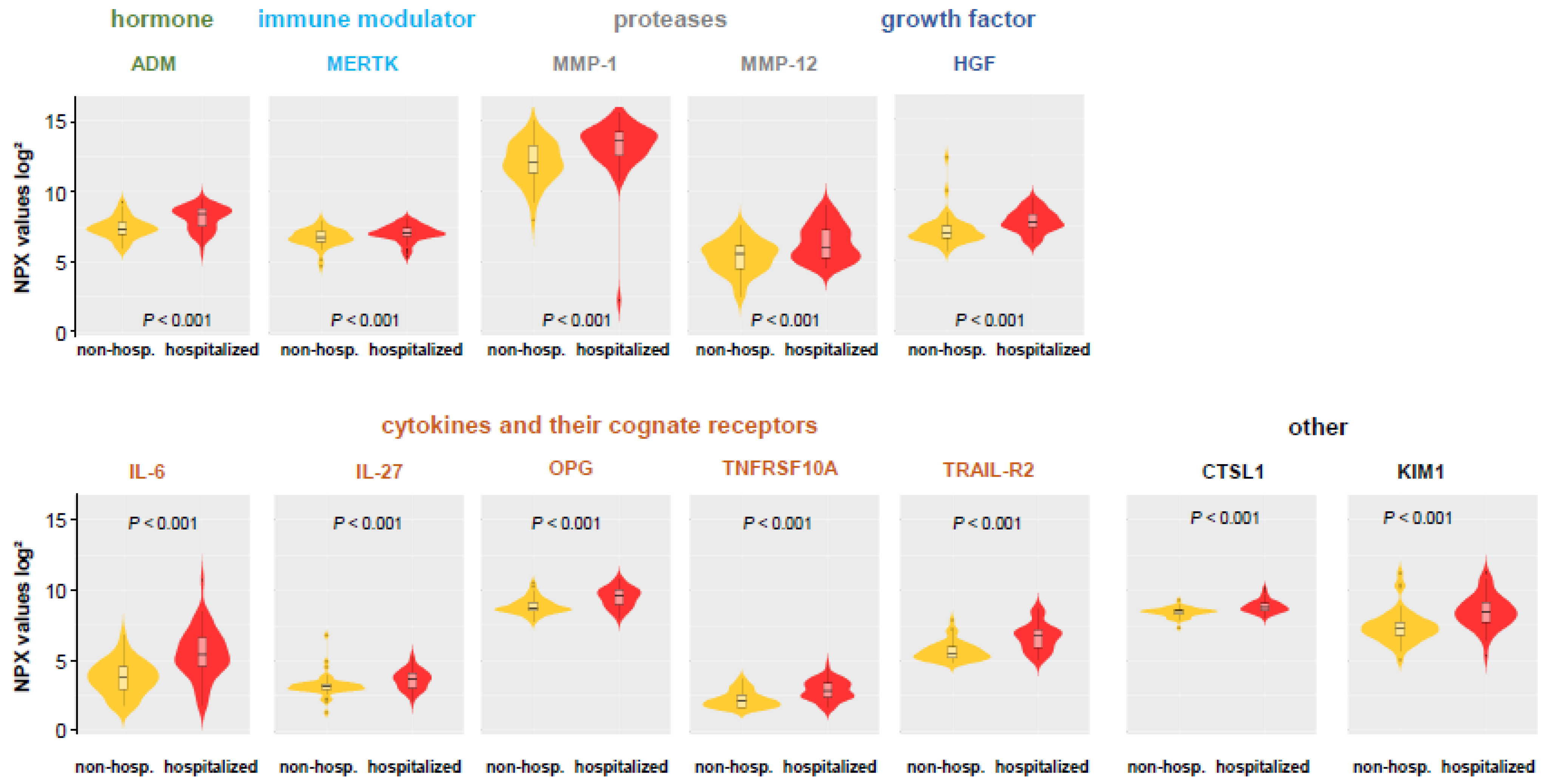
3.3. Comparison of Hospitalized and Non-Hospitalized COVID-19 Patients
3.4. Proteome Analyses in Hospitalized COVID-19 Patients with Regard to Clinical Outcome
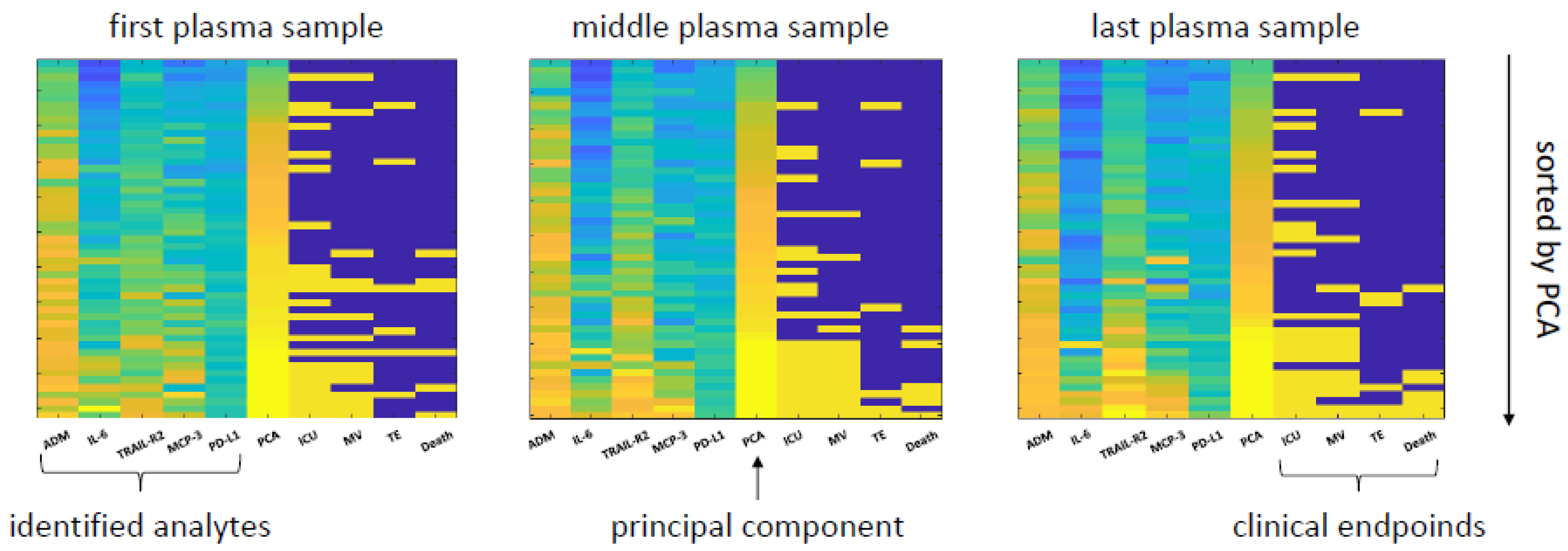
3.5. A Proteomic Fingerprint Predicts Outcome
4. Discussion
4.1. How Can Proteomic Fingerprinting Help Clinicians Better Understand COVID-19?
4.2. What Are the Advantages of Our Analysis Design?
4.3. What Distinguishes Patients with SARS-CoV-2 Infection from Patients with Similar Symptoms but Negative PCR Results?
4.4. Which Plasma Proteins Indicate a Severe Outcome in COVID-19 Patients?
4.5. What Are Clinical Implications and Do Possible Therapeutic Targets Exist?
4.6. Can the Multiplexed Proteomics Technique Be Implemented in Clinical Routine as a Diagnostic Method?
4.7. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.A.; Debray, T.P.A.; et al. Prediction Models for Diagnosis and Prognosis of Covid-19 Infection: Systematic Review and Critical Appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [Green Version]
- Giorgi Rossi, P.; Marino, M.; Formisano, D.; Venturelli, F.; Vicentini, M.; Grilli, R.; Reggio Emilia, C.-W.G. Characteristics and Outcomes of a Cohort of Covid-19 Patients in the Province of Reggio Emilia, Italy. PLoS ONE 2020, 15, e0238281. [Google Scholar] [CrossRef] [PubMed]
- Goodman, K.E.; Magder, L.S.; Baghdadi, J.D.; Pineles, L.; Levine, A.R.; Perencevich, E.N.; Harris, A.D. Impact of Sex and Metabolic Comorbidities on Covid-19 Mortality Risk across Age Groups: 66,646 Inpatients across 613 U.S. Hospitals. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Gargan, S.; Lu, Y.; Stevenson, N.J. An Overview of Current Knowledge of Deadly Covs and Their Interface with Innate Immunity. Viruses 2021, 13, 560. [Google Scholar] [CrossRef] [PubMed]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe COVID-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Graham, N.R.; Whitaker, A.N.; Strother, C.A.; Miles, A.K.; Grier, D.; McElvany, B.D.; Bruce, E.A.; Poynter, M.E.; Pierce, K.K.; Kirkpatrick, B.D.; et al. Kinetics and Isotype Assessment of Antibodies Targeting the Spike Protein Receptor-Binding Domain of Severe Acute Respiratory Syndrome-Coronavirus-2 in Covid-19 Patients as a Function of Age, Biological Sex and Disease Severity. Clin. Transl. Immunol. 2020, 9, e1189. [Google Scholar] [CrossRef]
- Bauer, W.; Diehl-Wiesenecker, E.; Ulke, J.; Galtung, N.; Havelka, A.; Hegel, J.K.; Tauber, R.; Somasundaram, R.; Kappert, K. Outcome Prediction by Serum Calprotectin in Patients with Covid-19 in the Emergency Department. J. Infect. 2021, 82, 84–123. [Google Scholar] [CrossRef]
- Fakhroo, A.D.; Al Thani, A.A.; Yassine, H.M. Markers Associated with Covid-19 Susceptibility, Resistance, and Severity. Viruses 2020, 13, 45. [Google Scholar] [CrossRef]
- Stegeman, I.; Ochodo, E.A.; Guleid, F.; Holtman, G.A.; Yang, B.; Davenport, C.; Deeks, J.J.; Dinnes, J.; Dittrich, S.; Emperador, D.; et al. Routine Laboratory Testing to Determine If a Patient Has Covid-19. Cochrane Database Syst. Rev. 2020, 11, CD013787. [Google Scholar]
- Wu, D.; Yang, X.O. Dysregulation of Pulmonary Responses in Severe Covid-19. Viruses 2021, 13, 957. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with Covid-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Nie, X.; Qian, L.; Sun, R.; Huang, B.; Dong, X.; Xiao, Q.; Zhang, Q.; Lu, T.; Yue, L.; Chen, S.; et al. Multi-Organ Proteomic Landscape of Covid-19 Autopsies. Cell 2021, 184, 775–791.e714. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Chong, J.; Xia, J. Comprehensive Meta-Analysis of Covid-19 Global Metabolomics Datasets. Metabolites 2021, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Bassler, K.; Schlickeiser, S.; Zhang, B.; Kramer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe Covid-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e1423. [Google Scholar] [CrossRef] [PubMed]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of Covid-19 Infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef]
- Patel, H.; Ashton, N.J.; Dobson, R.J.B.; Andersson, L.M.; Yilmaz, A.; Blennow, K.; Gisslen, M.; Zetterberg, H. Proteomic Blood Profiling in Mild, Severe and Critical Covid-19 Patients. Sci. Rep. 2021, 11, 6357. [Google Scholar] [CrossRef]
- Lee, E.E.; Song, K.H.; Hwang, W.; Ham, S.Y.; Jeong, H.; Kim, J.H.; Oh, H.S.; Kang, Y.M.; Lee, E.B.; Kim, N.J.; et al. Pattern of Inflammatory Immune Response Determines the Clinical Course and Outcome of Covid-19: Unbiased Clustering Analysis. Sci Rep. 2021, 11, 8080. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhong, M.; Hong, K.; Yang, Q.; Zhang, E.; Zhou, D.; Xia, J.; Chen, Y.Q.; Sun, M.; Zhao, B.; et al. Characteristics of T-Cell Responses in Covid-19 Patients with Prolonged Sars-Cov-2 Positivity—A Cohort Study. Clin. Transl. Immunol. 2021, 10, e1259. [Google Scholar] [CrossRef] [PubMed]
- Djureinovic, D.; Ponten, V.; Landelius, P.; Al Sayegh, S.; Kappert, K.; Kamali-Moghaddam, M.; Micke, P.; Stahle, E. Multiplex Plasma Protein Profiling Identifies Novel Markers to Discriminate Patients with Adenocarcinoma of the Lung. BMC Cancer 2019, 19, 741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sozio, E.; Tascini, C.; Fabris, M.; D’Aurizio, F.; De Carlo, C.; Graziano, E.; Bassi, F.; Sbrana, F.; Ripoli, A.; Pagotto, A.; et al. Mr-Proadm as Prognostic Factor of Outcome in Covid-19 Patients. Sci. Rep. 2021, 11, 5121. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, E.; Kwiecien, I.; Zabicka, M.; Maliborski, A.; Raniszewska, A.; Klos, K.; Urbanska, W.; Klajnowicz, I.; Rzepecki, P.; Chcialowski, A. Cytokines and Leukocytes Subpopulations Profile in Sars-Cov-2 Patients Depending on the Ct Score Severity. Viruses 2021, 13, 880. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. Covid-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; El Jammal, T.; Walzer, T.; Francois, B.; Seve, P. Should We Stimulate or Suppress Immune Responses in Covid-19? Cytokine and Anti-Cytokine Interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Harrison, E.M.; Ho, A.; Docherty, A.B.; Knight, S.R.; van Smeden, M.; Abubakar, I.; Lipman, M.; Quartagno, M.; Pius, R.; et al. Development and Validation of the Isaric 4c Deterioration Model for Adults Hospitalised with Covid-19: A Prospective Cohort Study. Lancet Respir. Med. 2021, 9, 349–359. [Google Scholar] [CrossRef]
- Bronte, V.; Ugel, S.; Tinazzi, E.; Vella, A.; De Sanctis, F.; Cane, S.; Batani, V.; Trovato, R.; Fiore, A.; Petrova, V.; et al. Baricitinib Restrains the Immune Dysregulation in Patients with Severe Covid-19. J. Clin. Investig. 2020, 130, 6409–6416. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical Predictors of Mortality Due to Covid-19 Based on an Analysis of Data of 150 Patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.C.; Schefold, J.C.; Baldira, J.; Spinetti, T.; Saeed, K.; Elke, G. Adrenomedullin in Covid-19 Induced Endotheliitis. Crit. Care 2020, 24, 411. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics Meets Proteomics: Perspectives for Large Population-Based Studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. Fda Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 2018, 23, 943–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tharmarajah, E.; Buazon, A.; Patel, V.; Hannah, J.R.; Adas, M.; Allen, V.B.; Bechman, K.; Clarke, B.D.; Nagra, D.; Norton, S.; et al. Il-6 Inhibition in the Treatment of Covid-19: A Meta-Analysis and Meta-Regression. J. Infect. 2021, 82, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Guillen, L.; Padilla, S.; Fernandez, M.; Agullo, V.; Garcia, J.A.; Telenti, G.; Garcia-Abellan, J.; Botella, A.; Gutierrez, F.; Masia, M. Preemptive Interleukin-6 Blockade in Patients with Covid-19. Sci. Rep. 2020, 10, 16826. [Google Scholar] [CrossRef]
- Santos, J.; Brierley, S.; Gandhi, M.J.; Cohen, M.A.; Moschella, P.C.; Declan, A.B.L. Repurposing Therapeutics for Potential Treatment of Sars-Cov-2: A Review. Viruses 2020, 12, 705. [Google Scholar] [CrossRef] [PubMed]
- Roumier, M.; Paule, R.; Vallee, A.; Rohmer, J.; Ballester, M.; Brun, A.L.; Cerf, C.; Chabi, M.L.; Chinet, T.; Colombier, M.A.; et al. Tocilizumab for Severe Worsening Covid-19 Pneumonia: A Propensity Score Analysis. J. Clin. Immunol. 2021, 41, 303–314. [Google Scholar] [CrossRef]
- Tsai, A.; Diawara, O.; Nahass, R.G.; Brunetti, L. Impact of Tocilizumab Administration on Mortality in Severe Covid-19. Sci. Rep. 2020, 10, 19131. [Google Scholar] [CrossRef] [PubMed]
- Iring, A.; Jin, Y.J.; Albarran-Juarez, J.; Siragusa, M.; Wang, S.; Dancs, P.T.; Nakayama, A.; Tonack, S.; Chen, M.; Kunne, C.; et al. Shear Stress-Induced Endothelial Adrenomedullin Signaling Regulates Vascular Tone and Blood Pressure. J. Clin. Investig. 2019, 129, 2775–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geven, C.; Kox, M.; Pickkers, P. Adrenomedullin and Adrenomedullin-Targeted Therapy as Treatment Strategies Relevant for Sepsis. Front. Immunol. 2018, 9, 292. [Google Scholar] [CrossRef] [PubMed]
- Hupf, J.; Mustroph, J.; Hanses, F.; Evert, K.; Maier, L.S.; Jungbauer, C.G. Rna-Expression of Adrenomedullin Is Increased in Patients with Severe Covid-19. Crit. Care 2020, 24, 527. [Google Scholar] [CrossRef]
- Blet, A.; Deniau, B.; Geven, C.; Sadoune, M.; Caillard, A.; Kounde, P.R.; Polidano, E.; Pickkers, P.; Samuel, J.L.; Mebazaa, A. Adrecizumab, a Non-Neutralizing Anti-Adrenomedullin Antibody, Improves Haemodynamics and Attenuates Myocardial Oxidative Stress in Septic Rats. Intensive Care Med. Exp. 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.A.; Fossali, T.; Pandolfi, L.; Carsana, L.; Ottolina, D.; Frangipane, V.; Rech, R.; Tosoni, A.; Lopez, G.; Agarossi, A.; et al. Hypoalbuminemia in Covid-19: Assessing the Hypothesis for Underlying Pulmonary Capillary Leakage. J. Intern. Med. 2021, 289, 861–872. [Google Scholar] [CrossRef]
- Karakas, M.; Jarczak, D.; Becker, M.; Roedl, K.; Addo, M.M.; Hein, F.; Bergmann, A.; Zimmermann, J.; Simon, T.P.; Marx, G.; et al. Targeting Endothelial Dysfunction in Eight Extreme-Critically Ill Patients with Covid-19 Using the Anti-Adrenomedullin Antibody Adrecizumab (Ham8101). Biomolecules 2020, 10, 1171. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Honjo, T. The Pd-1-Pd-L Pathway in Immunological Tolerance. Trends Immunol. 2006, 27, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Schonrich, G.; Raftery, M.J. The Pd-1/Pd-L1 Axis and Virus Infections: A Delicate Balance. Front. Cell. Infect. Microbiol. 2019, 9, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, D.; Ao, X.; Yang, Y.; Chen, Z.; Xu, X. Soluble Immune Checkpoints in Cancer: Production, Function and Biological Significance. J. Immunother. Cancer 2018, 6, 132. [Google Scholar] [CrossRef] [Green Version]
- Haljasmagi, L.; Salumets, A.; Rumm, A.P.; Jurgenson, M.; Krassohhina, E.; Remm, A.; Sein, H.; Kareinen, L.; Vapalahti, O.; Sironen, T.; et al. Longitudinal Proteomic Profiling Reveals Increased Early Inflammation and Sustained Apoptosis Proteins in Severe Covid-19. Sci. Rep. 2020, 10, 20533. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Bjorkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-Plex Pea Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [Green Version]
- Sims, J.T.; Krishnan, V.; Chang, C.Y.; Engle, S.M.; Casalini, G.; Rodgers, G.H.; Bivi, N.; Nickoloff, B.J.; Konrad, R.J.; de Bono, S.; et al. Characterization of the Cytokine Storm Reflects Hyperinflammatory Endothelial Dysfunction in Covid-19. J. Allergy Clin. Immunol. 2021, 147, 107–111. [Google Scholar] [CrossRef]

| Non-COVID-19 Group | COVID-19 Group | ||||||
|---|---|---|---|---|---|---|---|
| (N = 44) | (N = 97) | ||||||
| Parameter | Valid Values | Median Value | Interquartile Ranges | Valid Values | Median Value | Interquartile Ranges | p-Value |
| ADM | 44 | 8.05 | 6.96, 8.74 | 95 | 7.64 | 7.21, 8.54 | 0.5725400 |
| CCL23 | 41 | 10.43 | 9.99, 11.58 | 96 | 10.41 | 9.65, 10.99 | 0.0787430 |
| CTSL1 | 44 | 8.73 | 8.52, 8.99 | 95 | 8.62 | 8.43, 8.91 | 0.6467800 |
| CXCL10 | 41 | 9.88 | 8.96, 11.07 | 96 | 12.69 | 11.90, 13.34 | 0.0000004 |
| CXCL11 | 41 | 10.30 | 9.90, 11.15 | 96 | 12.05 | 10.75, 12.53 | 0.0000039 |
| CXCL5 | 41 | 12.18 | 11.29, 12.50 | 96 | 11.52 | 10.83, 12.04 | 0.0006775 |
| DCN | 44 | 3.99 | 3.70, 4.39 | 95 | 4.12 | 3.70, 4.57 | 0.4066400 |
| Gal9 | 44 | 4.76 | 4.13, 5.11 | 95 | 5.70 | 5.10, 6.58 | 0.0000001 |
| HGF | 41 | 7.72 | 7.16, 8.19 | 96 | 7.45 | 6.76, 8.15 | 0.4525200 |
| IFNgamma | 41 | 2.33 | 2.33, 3.01 | 96 | 4.67 | 2.96, 6.01 | 0.0000160 |
| IL18 | 44 | 7.64 | 7.22, 8.07 | 95 | 8.33 | 7.80, 8.75 | 0.0007774 |
| IL18R1 | 41 | 7.35 | 7.08, 7.56 | 96 | 7.92 | 7.53, 8.45 | 0.0000445 |
| IL27 | 44 | 3.20 | 2.87, 3.69 | 95 | 3.36 | 3.01, 3.90 | 0.5745600 |
| IL6 | 44 | 4.18 | 2.42, 6.56 | 95 | 4.72 | 3.50, 5.84 | 0.0639610 |
| KIM1 | 44 | 8.21 | 7.47, 8.81 | 95 | 7.86 | 7.08, 8.79 | 0.7888000 |
| LIFR | 41 | 4.33 | 4.04, 4.54 | 96 | 4.63 | 4.35, 4.90 | 0.0005687 |
| LPL | 44 | 7.49 | 6.99, 8.16 | 95 | 7.52 | 6.94, 8.03 | 0.8819400 |
| MCP1 | 41 | 11.72 | 11.33, 12.14 | 96 | 12.33 | 11.65, 12.82 | 0.0240940 |
| MCP2 | 41 | 8.81 | 8.37, 9.16 | 96 | 10.69 | 9.81, 11.58 | 0.00000001 |
| MCP3 | 41 | 3.98 | 3.43, 4.95 | 96 | 5.43 | 4.55, 6.77 | 0.0003295 |
| MERTK | 44 | 6.55 | 6.23, 6.83 | 95 | 6.96 | 6.56, 7.26 | 0.0007555 |
| MMP1 | 41 | 13.25 | 12.28, 14.66 | 96 | 13.00 | 11.86, 13.89 | 0.0007128 |
| MMP12 | 44 | 6.41 | 5.64, 7.37 | 95 | 5.80 | 4.97, 6.93 | 0.1359200 |
| OPG | 41 | 9.35 | 8.83, 9.99 | 96 | 9.11 | 8.65, 9.78 | 0.1229900 |
| PDL1 | 41 | 4.39 | 4.06, 4.64 | 96 | 5.14 | 4.74, 5.61 | 0.0000002 |
| TNF | 41 | 1.59 | 1.21, 2.03 | 96 | 2.15 | 1.80, 2.63 | 0.0002014 |
| TNFRSF10A | 44 | 2.52 | 1.97, 2.96 | 95 | 2.49 | 2.09, 3.23 | 0.8851000 |
| TRAIL-R2 | 44 | 6.33 | 5.76, 7.17 | 95 | 6.05 | 5.48, 7.01 | 0.2625300 |
| Median and Interquartile Ranges for Non-Outcomes | Median and Interquartile Ranges for Outcomes | p-Values Non-Outcome vs. Outcome | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICU | MV | TE | Death | ICU | MV | TE | Death | ICU | MV | TE | Death | |
| 29 (54.72%) | 37 (69.81%) | 47 (88.68%) | 46 (86.79%) | 24 (45.28%) | 16 (30.19%) | 6 (11.32%) | 7 (13.21%) | |||||
| SoC analytes | ||||||||||||
| CRP | 36.5 (14.6–62.6) | 35.3 (15.8–66.1) | 44.4 (16.5–89.7) | 45.3 (16.4–106) | 110 (25.8–155) | 140 (59.8–216) | 187 (39.4–265) | 53.7 (22.8–129) | 0.020 | 0.011 | 0.175 | 0.637 |
| PCT | 0.10 (0.08–0.24) | 0.10 (0.08–0.24) | 0.10 (0.08–0.25) | 0.11 (0.08–0.26) | 0.17 (0.10–0.30) | 0.20 (0.10–0.62) | 0.47 (0.20–0.78) | 0.11 (0.10–0.25) | 0.241 | 0.160 | 0.082 | 0.897 |
| WBC | 6.35 (5.33–8.51) | 6.62 (5.27–8.63) | 6.35 (5.26–8.62) | 7.12 (5.29–10.3) | 7.49 (5.13–10.9) | 7.64 (5.33–12.0) | 9.35 (9.12–10.5) | 5.65 (5.05–7.35) | 0.372 | 0.416 | 0.106 | 0.325 |
| LDH | 300 (266–399) | 295 (263–438) | 322 (271–504) | 340 (269–563) | 509 (439–684) | 509 (453–718) | 461 (447–589) | 432 (300–460) | 0.014 | 0.006 | 0.196 | 0.978 |
| PEA analytes | ||||||||||||
| ADM | 8.37 (7.40–8.78) | 8.30 (7.38–8.66) | 8.37 (7.49–8.80) | 8.28 (7.44–8.65) | 8.41 (7.54–8.82) | 8.64 (8.16–8.90) | 8.50 (7.77–8.75) | 8.98 (8.76–9.12) | 0.1207000 | 0.0472670 | 0.5869700 | 0.0000932 |
| CCL23 | 10.5 (9.92–10.9) | 10.5 (9.92–11.1) | 10.8 (10.2–11.2) | 10.6 (10.1–11.1) | 11.1 (10.6–11.6) | 11.2 (10.7–11.6) | 10.7 (9.81–11.1) | 11.3 (10.8–11.6) | 0.0006146 | 0.0018489 | 0.8367700 | 0.0551560 |
| CTSL1 | 8.72 (8.52–9.03) | 8.75 (8.52–9.03) | 8.75 (8.53–9.04) | 8.80 (8.52–9.10) | 8.86 (8.68–9.18) | 8.86 (8.70–9.26) | 9.17 (8.88–9.18) | 8.78 (8.67–8.99) | 0.0085455 | 0.0472670 | 0.0173740 | 0.3078300 |
| CXCL10 | 12.6 (12.0–13.3) | 12.7 (12.0–13.3) | 12.9 (12.0–13.3) | 12.7 (11.2–13.3) | 13.1 (11.9–13.5) | 13.1 (12.4–13.6) | 13.6 (10.7–13.8) | 13.7 (13.2–13.8) | 0.0669700 | 0.1257600 | 0.0160730 | 0.0024786 |
| CXCL11 | 11.8 (10.9–12.4) | 11.9 (10.8–12.5) | 12.2 (10.9–12.5) | 12.0 (10.7–12.5) | 12.5 (11.3–12.7) | 12.4 (12.1–12.6) | 12.6 (11.2–12.6) | 12.6 (12.6–12.6) | 0.1027300 | 0.1931000 | 0.3580900 | 0.0109360 |
| CXCL5 | 11.6 (11.2–12.2) | 11.5 (11.1–12.0) | 11.5 (11.1–12.0) | 11.4 (11.1–12.0) | 11.3 (10.8–12.0) | 11.3 (11.1–12.2) | 11.2 (11.1–12.0) | 11.7 (10.6–12.3) | 0.6541900 | 0.2407300 | 0.1392900 | 0.0403690 |
| DCN | 4.35 (3.84–4.77) | 4.22 (3.83–4.71) | 4.35 (3.84–4.82) | 4.30 (3.84–4.70) | 4.41 (4.01–4.75) | 4.64 (4.37–5.10) | 4.60 (4.42–4.72) | 5.31 (4.56–5.62) | 0.0475360 | 0.0009899 | 0.0154320 | 0.0074615 |
| Gal9 | 5.90 (5.10–6.77) | 5.90 (5.08–6.77) | 5.93 (5.14–6.72) | 5.78 (5.08–6.72) | 5.99 (5.21–6.82) | 6.05 (5.59–6.82) | 6.27 (4.59–7.00) | 6.60 (6.05–6.86) | 0.7322500 | 0.5908400 | 0.1927500 | 0.0442300 |
| HGF | 7.54 (7.25–8.20) | 7.54 (7.31–8.20) | 7.64 (7.32–8.23) | 7.56 (7.32–8.36) | 8.00 (7.44–8.59) | 8.16 (7.59–8.59) | 8.25 (7.71–8.41) | 8.16 (7.83–8.27) | 0.0354240 | 0.0452030 | 0.3725600 | 0.1236400 |
| IFNgamma | 4.59 (2.60–6.00) | 4.63 (2.79–6.04) | 4.80 (3.19–6.03) | 4.67 (3.15–6.03) | 4.95 (3.64–5.63) | 4.89 (3.20–5.21) | 3.50 (2.40–4.86) | 4.53 (3.09–5.31) | 0.1746500 | 0.6409200 | 0.8324600 | 0.8324600 |
| IL18 | 8.42 (8.09–8.71) | 8.36 (8.05–8.67) | 8.44 (8.14–8.86) | 8.43 (8.09–8.90) | 8.51 (8.16–9.41) | 8.64 (8.27–9.56) | 8.42 (7.60–9.17) | 8.66 (8.26–9.03) | 0.3469200 | 0.0557820 | 0.3616600 | 0.3725600 |
| IL18R1 | 8.05 (7.60–8.42) | 8.05 (7.59–8.53) | 8.09 (7.61–8.49) | 7.98 (7.61–8.47) | 8.35 (7.70–8.62) | 8.23 (7.84–8.55) | 8.12 (7.65–8.59) | 8.55 (8.30–8.64) | 0.4282800 | 0.2964400 | 0.8367700 | 0.1236400 |
| IL27 | 3.70 (3.27–4.15) | 3.62 (3.07–3.98) | 3.68 (3.09–4.11) | 3.62 (3.04–3.96) | 3.67 (3.06–4.04) | 3.90 (3.45–4.35) | 3.68 (3.43–4.13) | 4.42 (4.05–4.49) | 0.3287600 | 0.0472670 | 0.4544500 | 0.0074615 |
| IL6 | 4.84 (4.33–5.55) | 4.86 (4.33–6.01) | 5.27 (4.66–6.61) | 5.05 (4.40–6.26) | 6.43 (5.37–6.96) | 6.74 (5.60–7.31) | 5.84 (4.36–6.54) | 6.62 (5.55–6.80) | 0.0000044 | 0.0000002 | 0.3580900 | 0.0319650 |
| KIM1 | 8.51 (8.13–9.53) | 8.38 (7.75–9.07) | 8.41 (7.71–9.08) | 8.37 (7.69–9.09) | 8.06 (7.62–8.85) | 8.68 (7.78–9.32) | 8.72 (7.96–9.39) | 9.01 (8.68–9.35) | 0.1353200 | 0.1374300 | 0.8455500 | 0.0442300 |
| LIFR | 4.66 (4.42–4.94) | 4.73 (4.49–4.92) | 4.80 (4.50–4.93) | 4.76 (4.50–4.92) | 4.84 (4.66–4.93) | 4.84 (4.67–5.10) | 4.93 (4.66–4.99) | 5.07 (4.86–5.38) | 0.0197430 | 0.0853480 | 0.3725600 | 0.0551560 |
| LPL | 7.72 (6.81–8.44) | 7.72 (7.06–8.29) | 7.66 (7.09–8.35) | 7.62 (7.07–8.10) | 7.65 (7.41–8.14) | 7.65 (7.51–8.67) | 7.78 (7.54–8.37) | 8.73 (8.10–9.06) | 0.4779700 | 0.1412000 | 0.5225300 | 0.0001652 |
| MCP1 | 12.4 (11.8–12.8) | 12.3 (11.8–12.8) | 12.5 (11.8–13.0) | 12.5 (11.7–13.0) | 13.0 (12.1–13.5) | 13.3 (12.6–13.7) | 13.4 (12.0–13.8) | 13.5 (13.0–13.7) | 0.0000581 | 0.0000419 | 0.0545020 | 0.0024786 |
| MCP2 | 10.5 (9.34–11.2) | 10.8 (9.24–11.7) | 10.8 (9.48–11.5) | 10.8 (9.26–11.5) | 11.1 (9.60–12.0) | 11.0 (9.95–12.0) | 12.0 (9.91–12.0) | 12.2 (11.3–12.4) | 0.0934230 | 0.4824300 | 0.0545020 | 0.0024786 |
| MCP3 | 5.42 (4.70–6.70) | 5.59 (4.72–6.74) | 6.11 (4.76–7.09) | 6.11 (4.76–7.16) | 6.93 (5.98–7.87) | 7.43 (6.39–8.22) | 7.66 (5.99–8.11) | 7.29 (5.63–8.11) | 0.0001310 | 0.0000231 | 0.0024786 | 0.0571620 |
| MERTK | 6.96 (6.79–7.39) | 6.96 (6.79–7.40) | 7.02 (6.82–7.37) | 6.97 (6.80–7.37) | 7.10 (6.89–7.43) | 7.16 (6.95–7.43) | 7.32 (6.72–7.79) | 7.25 (7.04–7.52) | 0.1706600 | 0.1503800 | 0.0580550 | 0.2731700 |
| MMP1 | 13.6 (12.8–14.1) | 13.5 (12.5–14.0) | 13.5 (12.6–14.2) | 13.4 (12.5–14.0) | 13.5 (12.5–14.3) | 14.1 (12.6–14.5) | 14.0 (12.9–14.3) | 14.3 (14.1–14.5) | 0.1102800 | 0.0158300 | 0.5116700 | 0.0551560 |
| MMP12 | 6.34 (5.38–7.30) | 5.98 (5.27–7.14) | 6.10 (5.28–7.30) | 5.84 (5.23–6.98) | 5.87 (5.22–7.21) | 6.36 (5.24–7.54) | 5.75 (5.34–6.70) | 7.32 (6.92–7.99) | 0.2720700 | 0.2977100 | 0.8455500 | 0.1061200 |
| OPG | 9.54 (8.97–10.1) | 9.50 (8.97–9.92) | 9.52 (8.97–10.1) | 9.55 (8.97–9.93) | 9.59 (9.02–10.1) | 9.82 (9.40–10.2) | 9.72 (9.59–10.0) | 10.1 (9.69–10.2) | 0.0499440 | 0.0242520 | 0.0109360 | 0.0551560 |
| PDL1 | 5.04 (4.81–5.46) | 5.19 (4.81–5.61) | 5.34 (4.90–5.73) | 5.29 (4.82–5.72) | 5.49 (5.17–5.82) | 5.49 (5.29–5.80) | 5.25 (4.59–5.68) | 5.49 (5.33–5.77) | 0.0006113 | 0.0528220 | 0.8324600 | 0.1714200 |
| TNF | 2.49 (2.02–2.87) | 2.33 (1.89–2.79) | 2.32 (1.92–2.77) | 2.18 (1.83–2.65) | 2.05 (1.85–2.41) | 2.23 (1.92–2.63) | 2.31 (1.76–2.46) | 2.72 (2.63–3.10) | 0.0669310 | 0.4650900 | 0.9491500 | 0.1859300 |
| TNFRSF10A | 2.74 (2.40–3.25) | 2.65 (2.40–3.27) | 2.87 (2.44–3.43) | 2.73 (2.41–3.41) | 3.16 (2.50–3.61) | 3.30 (2.68–3.72) | 3.02 (2.46–3.53) | 3.25 (2.91–3.68) | 0.0168280 | 0.0074506 | 0.5225300 | 0.1002400 |
| TRAILR2 | 6.40 (5.83–7.09) | 6.40 (5.77–7.08) | 6.75 (5.91–7.22) | 6.52 (5.85–7.09) | 7.01 (6.46–7.35) | 7.16 (6.71–7.63) | 7.06 (6.41–7.16) | 7.18 (7.07–7.85) | 0.0004393 | 0.0000823 | 0.5869700 | 0.0125550 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, W.; Weber, M.; Diehl-Wiesenecker, E.; Galtung, N.; Prpic, M.; Somasundaram, R.; Tauber, R.; Schwenk, J.M.; Micke, P.; Kappert, K. Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection. Viruses 2021, 13, 2456. https://doi.org/10.3390/v13122456
Bauer W, Weber M, Diehl-Wiesenecker E, Galtung N, Prpic M, Somasundaram R, Tauber R, Schwenk JM, Micke P, Kappert K. Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection. Viruses. 2021; 13(12):2456. https://doi.org/10.3390/v13122456
Chicago/Turabian StyleBauer, Wolfgang, Marcus Weber, Eva Diehl-Wiesenecker, Noa Galtung, Monika Prpic, Rajan Somasundaram, Rudolf Tauber, Jochen M. Schwenk, Patrick Micke, and Kai Kappert. 2021. "Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection" Viruses 13, no. 12: 2456. https://doi.org/10.3390/v13122456
APA StyleBauer, W., Weber, M., Diehl-Wiesenecker, E., Galtung, N., Prpic, M., Somasundaram, R., Tauber, R., Schwenk, J. M., Micke, P., & Kappert, K. (2021). Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection. Viruses, 13(12), 2456. https://doi.org/10.3390/v13122456








