INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. Virus Stock Preparation
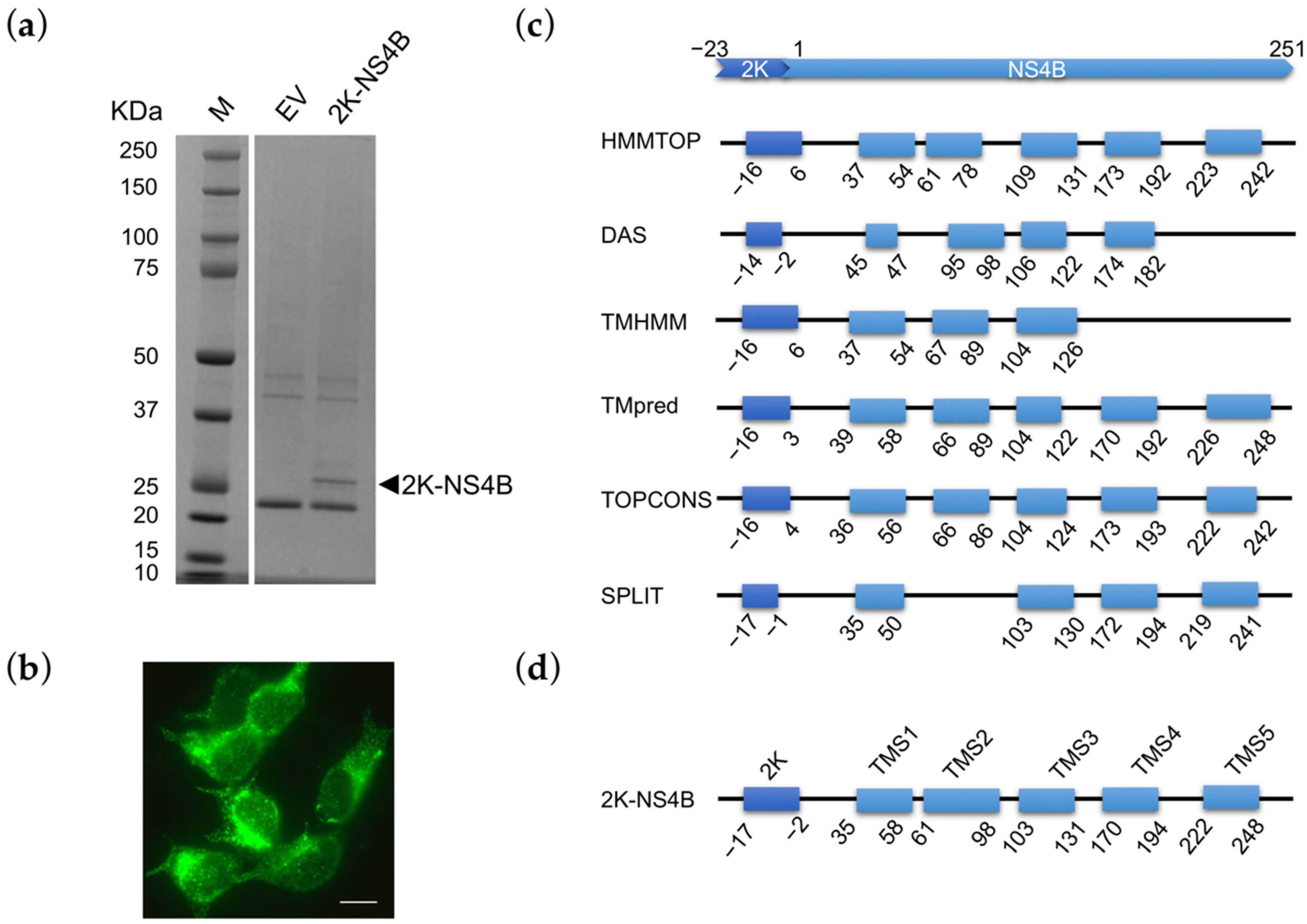
2.3. Construction of 2K-NS4B Expression Plasmid
2.4. Luciferase Reporter Gene Assays
2.5. EC50 and FIC50 Calculations
2.6. Immunoprecipitation
2.7. Immunoblot Analysis
2.8. Immunofluorescence
2.9. RNA Extraction and Quantitative Real-Time PCR
2.10. Bioinformatics
2.10.1. NS4B Topology Prediction
2.10.2. Alignments
2.10.3. Phylogenetic Analysis
2.11. Graphics
3. Results
3.1. NS4B Genomic Characterization and Prediction of Membrane Topology
3.2. Sequence and Phylogenic Analysis of NS4B
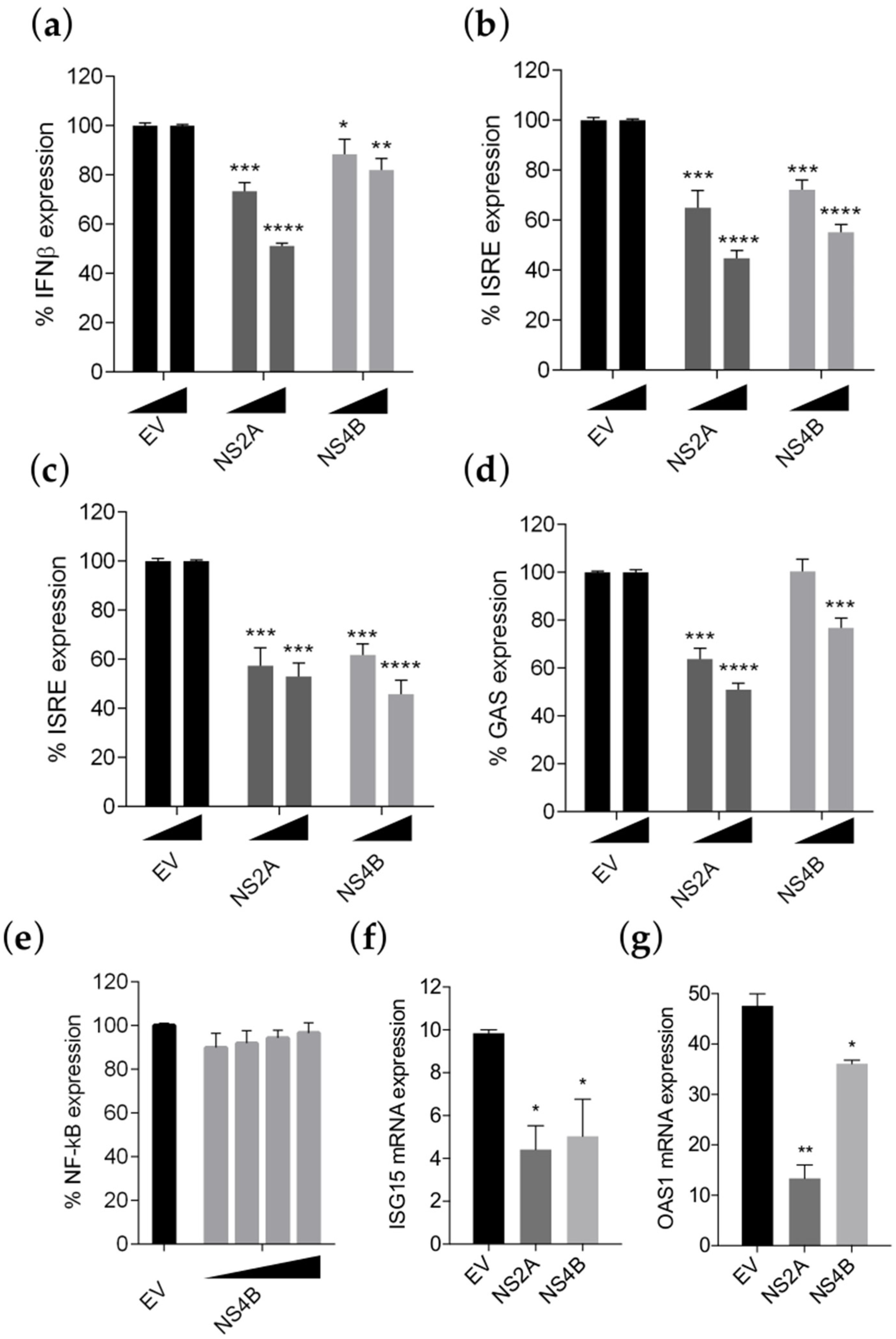
3.3. NS4B Inhibits IFN-β Production and Type I and II IFN Signaling
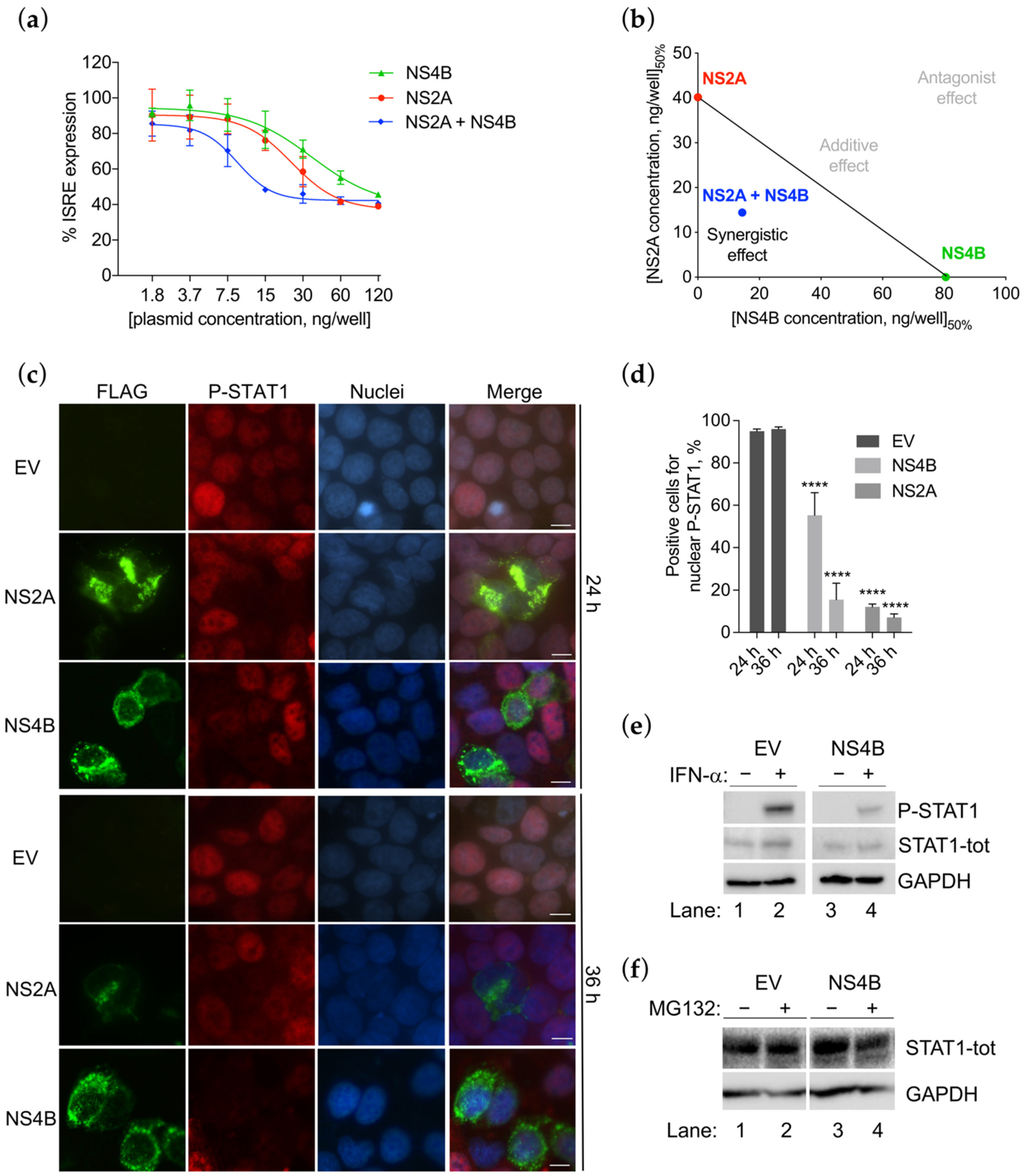
3.4. NS4B Inhibits the STAT1 Phosphorylation
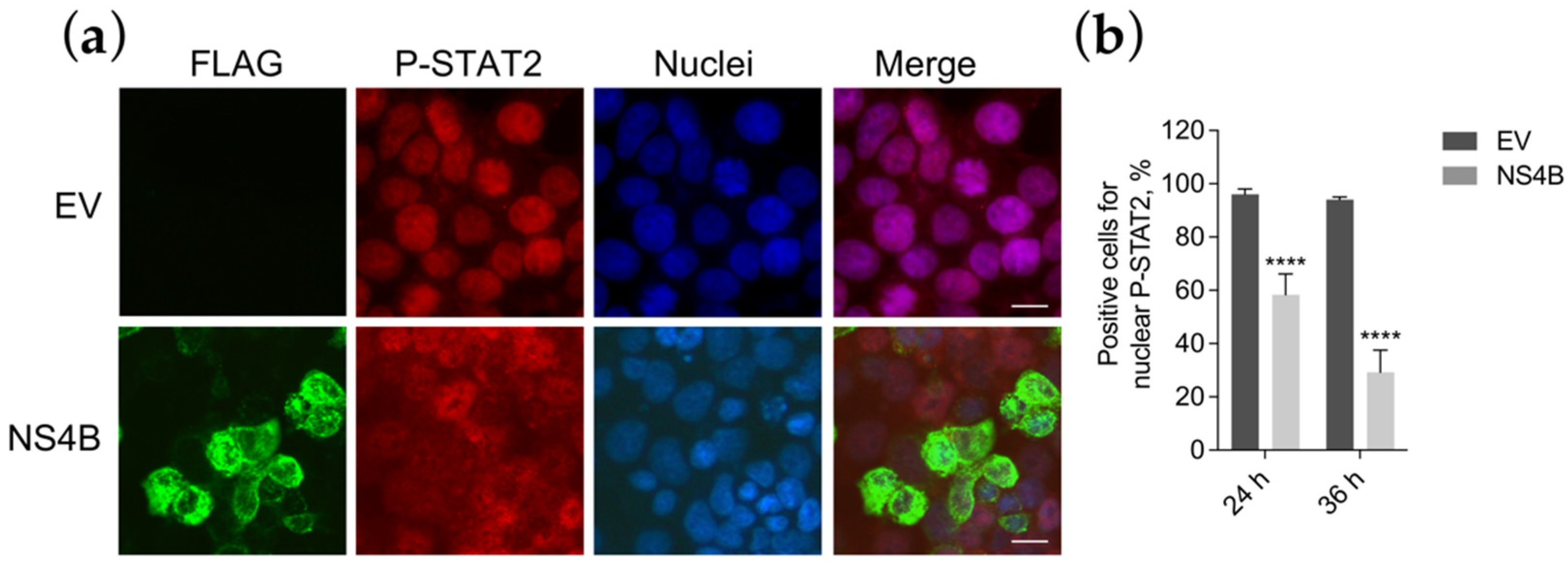
3.5. NS4B Blocks P-STAT2 Nuclear Transport
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zanluca, C.; De Melo, V.C.A.; Mosimann, A.L.P.; Dos Santos, G.I.V.; dos Santos, C.N.D.; Luz, K. First report of autochthonous transmission of Zika virus in Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 569–572. [Google Scholar] [CrossRef]
- Gasco, S.; Muñoz-Fernández, M.Á. A review on the current knowledge on zikv infection and the interest of organoids and nanotechnology on development of effective therapies against zika infection. Int. J. Mol. Sci. 2020, 22, 35. [Google Scholar] [CrossRef]
- Barzon, L.; Trevisan, M.; Sinigaglia, A.; Lavezzo, E.; Palù, G. Zika virus: From pathogenesis to disease control. FEMS Microbiol. Lett. 2016, 363, fnw202. [Google Scholar] [CrossRef]
- Westaway, E.G.; Khromykh, A.A.; Kenney, M.T.; MacKenzie, J.M.; Jones, M.K. Proteins C and NS4B of the flavivirus kunjin translocate independently into the nucleus. Virology 1997, 234, 31–41. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Lee, L.T.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.-Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of Flavivirus NS4B Protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef]
- Kakumani, P.K.; Ponia, S.S.; Rajgokul, K.S.; Sood, V.; Chinnappan, M.; Banerjea, A.C.; Medigeshi, G.R.; Malhotra, P.; Mukherjee, S.K.; Bhatnagar, R.K. Role of RNA Interference (RNAi) in Dengue Virus Replication and Identification of NS4B as an RNAi Suppressor. J. Virol. 2013, 87, 8870–8883. [Google Scholar] [CrossRef]
- Cumberworth, S.L.; Clark, J.J.; Kohl, A.; Donald, C.L. Inhibition of type I interferon induction and signalling by mosquito-borne flaviviruses. Cell. Microbiol. 2017, 19, e12737. [Google Scholar] [CrossRef]
- Coldbeck-Shackley, R.C.; Eyre, N.S.; Beard, M.R. The molecular interactions of zikv and denv with the type-i IFN response. Vaccines 2020, 8, 530. [Google Scholar] [CrossRef]
- Gorman, M.J.; Caine, E.A.; Zaitsev, K.; Begley, M.C.; Weger-Lucarelli, J.; Uccellini, M.B.; Tripathi, S.; Morrison, J.; Yount, B.L.; Dinnon, K.H.; et al. An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe 2018, 23, 672–685. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type i interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.J.; Kato, N.; Shu, H.B.; Zhong, J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef]
- Yi, G.; Wen, Y.; Shu, C.; Han, Q.; Konan, K.V.; Li, P.; Kao, C.C. Hepatitis C Virus NS4B Can Suppress STING Accumulation To Evade Innate Immune Responses. J. Virol. 2016, 90, 254–265. [Google Scholar] [CrossRef]
- Zhang, W.; Zeng, M.; Jiang, B.; Lu, T.; Guo, J.; Hu, T.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; et al. Amelioration of Beta Interferon Inhibition by NS4B Contributes to Attenuating Tembusu Virus Virulence in Ducks. Front. Immunol. 2021, 12, 1864. [Google Scholar] [CrossRef]
- Shan, Y.; Tong, Z.; Jinzhu, M.; Yu, L.; Zecai, Z.; Chenhua, W.; Wenjing, H.; Siyu, L.; Nannan, C.; Siyu, S.; et al. Bovine viral diarrhea virus NS4B protein interacts with 2CARD of MDA5 domain and negatively regulates the RLR-mediated IFN-β production. Virus Res. 2021, 302, 198471. [Google Scholar] [CrossRef]
- Munoz-Jordan, J.L.; Sanchez-Burgos, G.G.; Laurent-Rolle, M.; Garcia-Sastre, A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 2003, 100, 14333–14338. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Laurent-rolle, M.; Martínez-sobrido, L.; Ashok, M.; Ian, W.; Mun, J.L. Inhibition of Alpha/Beta Interferon Signaling by the NS4B Protein of Flaviviruses Inhibition of Alpha / Beta Interferon Signaling by the NS4B Protein of Flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Fredericksen, B.L. How flaviviruses activate and suppress the interferon response. Viruses 2010, 2, 676–691. [Google Scholar] [CrossRef]
- Liu, W.J.; Wang, X.J.; Mokhonov, V.V.; Shi, P.-Y.; Randall, R.; Khromykh, A.A. Inhibition of Interferon Signaling by the New York 99 Strain and Kunjin Subtype of West Nile Virus Involves Blockage of STAT1 and STAT2 Activation by Nonstructural Proteins. J. Virol. 2005, 79, 1934–1942. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Q.; Zhou, J.; Xie, W.; Chen, C.; Wang, Z.; Yang, H.; Cui, J. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017, 3, 17006. [Google Scholar] [CrossRef]
- Xia, H.; Luo, H.; Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Medeiros, D.B.A.; Zou, J.; Xie, X.; Giraldo, M.I.; Vasconcelos, P.F.C.; et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat. Commun. 2018, 9, 414. [Google Scholar] [CrossRef]
- Kumar, A.; Hou, S.; Airo, A.M.; Limonta, D.; Mancinelli, V.; Branton, W.; Power, C.; Hobman, T.C. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016, 17, 1766–1775. [Google Scholar] [CrossRef]
- Colavita, F.; Bordoni, V.; Caglioti, C.; Biava, M.; Castilletti, C.; Bordi, L.; Quartu, S.; Iannetta, M.; Ippolito, G.; Agrati, C.; et al. ZIKV infection induces an inflammatory response but fails to activate types I, II, and III IFN response in human PBMC. Mediators Inflamm. 2018, 2450540. [Google Scholar] [CrossRef]
- Fanunza, E.; Carletti, F.; Quartu, M.; Grandi, N.; Ermellino, L.; Milia, J.; Corona, A.; Capobianchi, M.R.; Ippolito, G.; Tramontano, E. Zika virus NS2A inhibits interferon signaling by degradation of STAT1 and STAT2. Virulence 2021, 12, 1580–1596. [Google Scholar] [CrossRef] [PubMed]
- Cannas, V.; Daino, G.L.; Corona, A.; Esposito, F.; Tramontano, E. A Luciferase Reporter Gene Assay to Measure Ebola Virus Viral Protein 35-Associated Inhibition of Double-Stranded RNA-Stimulated, Retinoic Acid-Inducible Gene 1-Mediated Induction of Interferon β. J. Infect. Dis. 2015, 212, 277–281. [Google Scholar] [CrossRef][Green Version]
- Fanunza, E.; Frau, A.; Sgarbanti, M.; Orsatti, R.; Corona, A.; Tramontano, E. Development and validation of a novel dual luciferase reporter gene assay to quantify ebola virus VP24 inhibition of IFN signaling. Viruses 2018, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Akoachere, M.; Buchholz, K.; Fischer, E.; Burhenne, J.; Haefeli, W.E.; Schirmer, R.H.; Becker, K. In vitro assessment of methylene blue on chloroquine-sensitive and -resistant Plasmodium falciparum strains reveals synergistic action with artemisinins. Antimicrob. Agents Chemother. 2005, 49, 4592–4597. [Google Scholar] [CrossRef]
- Fivelman, Q.L.; Adagu, I.S.; Warhurst, D.C. Modified fixed-ratio isobologram method for studying in vitro interactions between atovaquone and proguanil or dihydroartemisinin against drug-resistant strains of Plasmodium falciparum. Antimicrob. Agents Chemother. 2004, 48, 4097–4102. [Google Scholar] [CrossRef] [PubMed]
- Fanunza, E.; Iampietro, M.; Distinto, S.; Corona, A.; Quartu, M.; Maccioni, E.; Horvat, B.; Tramontano, E. Quercetin blocks Ebola Virus infection by counteracting the VP24 Interferon inhibitory function. Antimicrob. Agents Chemother. 2020, 64, e00530-20. [Google Scholar] [CrossRef]
- Abner, E.; Stoszko, M.; Zeng, L.; Chen, H.-C.; Izquierdo-Bouldstridge, A.; Konuma, T.; Zorita, E.; Fanunza, E.; Zhang, Q.; Mahmoudi, T.; et al. A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for re-activation from other ‘shock’ drugs. J. Virol. 2018, 92, e02056-17. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Wang, L.; Valderramos, S.G.; Wu, A.; Ouyang, S.; Li, C.; Brasil, P.; Bonaldo, M.; Coates, T.; Nielsen-Saines, K.; Jiang, T.; et al. From Mosquitos to Humans: Genetic Evolution of Zika Virus. Cell Host Microbe 2016, 19, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Amberg, S.M.; Chambers, T.J.; Rice, C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 1993, 67, 2327–2335. [Google Scholar] [CrossRef]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef] [PubMed]
- Welsch, C.; Albrecht, M.; Maydt, J.; Herrmann, E.; Welker, M.W.; Sarrazin, C.; Scheidig, A.; Lengauer, T.; Zeuzem, S. Structural and functional comparison of the non-structural protein 4B in flaviviridae. J. Mol. Graph. Model. 2007, 26, 546–557. [Google Scholar] [CrossRef]
- Welker, M.W.; Welsch, C.; Meyer, A.; Antes, I.; Albrecht, M.; Forestier, N.; Kronenberger, B.; Lengauer, T.; Piiper, A.; Zeuzem, S.; et al. Dimerization of the hepatitis C virus nonstructural protein 4B depends on the integrity of an aminoterminal basic leucine zipper. Protein Sci. 2010, 19, 1327–1336. [Google Scholar] [CrossRef]
- Guo, F.; Wu, S.; Julander, J.; Ma, J.; Zhang, X.; Kulp, J.; Cuconati, A.; Block, T.M.; Du, Y.; Guo, J.-T.; et al. A Novel Benzodiazepine Compound Inhibits Yellow Fever Virus Infection by Specifically Targeting NS4B Protein. J. Virol. 2016, 90, 10774–10788. [Google Scholar] [CrossRef]
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.A.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.Y.; Vasilakis, N. Zika virus: History, emergence, biology, and prospects for control. Antivir. Res. 2016, 130, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Seeger, C. Differential Effects of Mutations in NS4B on West Nile Virus Replication and Inhibition of Interferon Signaling. J. Virol. 2007, 81, 11809–11816. [Google Scholar] [CrossRef]
- Welte, T.; Xie, G.; Wicker, J.A.; Whitemanb, M.C.; Li, L.; Rachamallu, A.; Barrett, A.; Wang, T. Immune responses to an attenuated West Nile virus NS4B-P38G mutant strain. Vaccine 2011, 29, 4853–4861. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Adam, A.; Luo, H.; Shan, C.; Cao, Z.; Fontes-Garfias, C.R.; Sarathy, V.V.; Teleki, C.; Winkelmann, E.R.; Liang, Y.; et al. An attenuated Zika virus NS4B protein mutant is a potent inducer of antiviral immune responses. NPJ Vaccines 2019, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.-H.; Tsai, M.-H.; Tsai, K.-N.; Tian, J.N.; Wu, J.-S.; Wu, S.-Y.; Chern, J.-H.; Chen, C.-H.; Yueh, A. Mutagenesis of Dengue Virus Protein NS2A Revealed a Novel Domain Responsible for Virus-Induced Cytopathic Effect and Interactions between NS2A and NS2B Transmembrane Segments. J. Virol. 2017, 91, e01836-16. [Google Scholar] [CrossRef]
- Banningeri, G.; Reich, N.C. STAT2 nuclear trafficking. J. Biol. Chem. 2004, 279, 39199–39206. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Serman, T.M.; Gack, M.U. Evasion of innate and intrinsic antiviral pathways by the Zika Virus. Viruses 2019, 11, 970. [Google Scholar] [CrossRef]
- Shu, J.; Ma, X.; Zhang, Y.; Zou, J.; Yuan, Z.; Yi, Z. NS5-independent ablation of STAT2 by Zika virus to antagonize interferon signaling. Emerg. Microbes Infect. 2021, 10, 1609–1625. [Google Scholar] [CrossRef]
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sánchez-Seco, M.P.; Evans, M.J.; Best, S.M.; et al. Zika Virus Targets Human STAT2 to Inhibit Type i Interferon Signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef]
- Zhao, Z.; Tao, M.; Han, W.; Fan, Z.; Imran, M.; Cao, S.; Ye, J. Nuclear localization of Zika virus NS5 contributes to suppression of type I interferon production and response. J. Gen. Virol. 2019, 102, 001376. [Google Scholar] [CrossRef]
- Esteves, E.; Rosa, N.; Correia, M.J.; Arrais, J.P.; Barros, M. New Targets for Zika Virus Determined by Human-Viral Interactomic: A Bioinformatics Approach. Biomed. Res. Int. 2017, 2017, 1734151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Miner, J.J.; Gorman, M.J.; Rausch, K.; Ramage, H.; White, J.P.; Zuiani, A.; Zhang, P.; Fernandez, E.; Zhang, Q.; et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 2016, 535, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.Z.; Moraes, S.N.; Shaban, N.M.; Fanunza, E.; Bierle, C.J.; Southern, P.J.; Bresnahan, W.A.; Rice, S.A.; Harris, R.S. APOBECs and Herpesviruses. Viruses 2021, 13, 390. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 402–424. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef]
- Fanunza, E.; Frau, A.; Corona, A.; Tramontano, E. Insights into Ebola Virus VP35 and VP24 Interferon Inhibitory Functions and their Initial Exploitation as Drug Targets. Infect. Disord. Drug Targets 2019, 19, 362–374. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, X.; Du, L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin. Ther. Targets 2021, 25, 415–421. [Google Scholar] [CrossRef]
- Kuo, Y.T.; Liu, C.H.; Corona, A.; Fanunza, E.; Tramontano, E.; Lin, L.T. The methanolic extract of perilla frutescens robustly restricts ebola virus glycoprotein-mediated entry. Viruses 2021, 13, 1793. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Shi, P.Y. Flavivirus Entry Inhibitors. ACS Infect. Dis. 2016, 9, 428–434. [Google Scholar] [CrossRef]
- Flego, M.; Frau, A.; Accardi, L.; Mallano, A.; Ascione, A.; Gellini, M.; Fanunza, E.; Vella, S.; Di Bonito, P.; Tramontano, E. Intracellular human antibody fragments recognizing the VP35 protein of Zaire Ebola filovirus inhibit the protein activity. BMC Biotechnol. 2019, 19, 64. [Google Scholar] [CrossRef]
- Daino, G.L.; Frau, A.; Sanna, C.; Rigano, D.; Distinto, S.; Madau, V.; Esposito, F.; Fanunza, E.; Bianco, G.; Taglialatela-Scafati, O.; et al. Identification of Myricetin as an Ebola Virus VP35-Double-Stranded RNA Interaction Inhibitor through a Novel Fluorescence-Based Assay. Biochemistry 2018, 57, 6367–6378. [Google Scholar] [CrossRef] [PubMed]
- Fanunza, E.; Frau, A.; Corona, A.; Tramontano, E. Antiviral Agents Against Ebola Virus Infection: Repositioning Old Drugs and Finding Novel Small Molecules. Annu. Rep. Med. Chem. 2018, 51, 135–173. [Google Scholar]
- Jasenosky, L.D.; Cadena, C.; Mire, C.E.; Borisevich, V.; Haridas, V.; Ranjbar, S.; Nambu, A.; Bavari, S.; Soloveva, V.; Sadukhan, S.; et al. The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus. iScience 2019, 19, 1279–1290. [Google Scholar] [CrossRef]
- Di Palma, F.; Daino, G.L.; Ramaswamy, V.K.; Corona, A.; Frau, A.; Fanunza, E.; Vargiu, A.V.; Tramontano, E.; Ruggerone, P. Relevance of Ebola virus VP35 homo-dimerization on the type I interferon cascade inhibition. Antivir. Chem. Chemother. 2019, 27, 2040206619889220. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zou, J.; Wang, Q.Y.; Shi, P.Y. Targeting dengue virus NS4B protein for drug discovery. Antivir. Res. 2015, 118, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.; Wu, Y.H.; Tseng, C.K.; Lin, C.K.; Hsu, Y.C.; Chen, Y.H.; Lee, J.C. Schisandrin A inhibits dengue viral replication via upregulating antiviral interferon responses through STAT signaling pathway. Sci. Rep. 2017, 7, 45171. [Google Scholar] [CrossRef]
- Panayiotou, C.; Lindqvist, R.; Kurhade, C.; Vonderstein, K.; Pasto, J.; Edlund, K.; Upadhyay, A.S.; Överby, A.K. Viperin Restricts Zika Virus and Tick-Borne Encephalitis Virus Replication by Targeting NS3 for Proteasomal Degradation. J. Virol. 2018, 92, e02054-17. [Google Scholar] [CrossRef]
- Li, L.; Zhao, H.; Liu, P.; Li, C.; Quanquin, N.; Ji, X.; Sun, N.; Du, P.; Qin, C.F.; Lu, N.; et al. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci. Signal. 2018, 11, eaas9332. [Google Scholar] [CrossRef] [PubMed]
- Morchang, A.; Malakar, S.; Poonudom, K.; Noisakran, S.; Yenchitsomanus, P.T.; Limjindaporn, T. Melatonin inhibits dengue virus infection via the sirtuin 1-mediated interferon pathway. Viruses 2021, 13, 659. [Google Scholar] [CrossRef]
- Zmurko, J.; Neyts, J.; Dallmeier, K. Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev. Med. Virol. 2015, 25, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Nitta, S.; Sakamoto, N.; Nakagawa, M.; Kakinuma, S.; Mishima, K.; Kusano-Kitazume, A.; Kiyohashi, K.; Murakawa, M.; Nishimura-Sakurai, Y.; Azuma, S.; et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 2013, 57, 46–58. [Google Scholar] [CrossRef] [PubMed]

| Primer Name 1 | Sequence (5′ to 3′) |
|---|---|
| 2K-NS4B_F | TCTCCCCAGGACAACCAAATG |
| 2K-NS4B_R | ACGTCTCTTGACCAAGCCAGC |
| Primer Name 1 | Sequence (5′ to 3′) |
|---|---|
| GAPDH_F | GAGTCAACGGATTTTGGTCGT |
| GAPDH_R | TTGATTTTGGAGGGATCTCG |
| ISG15_F | TCCTGGTGAGGAATAACAAGGG |
| ISG15_R | CTCAGCCAGAACAGGTCGTC |
| OAS1_F | AGCTTCATGGAGAGGGGCA |
| OAS1_R | AGGCCTGGCTGAATTACCCAT |
| Protein | EC50 (ng/well) | FIC50 A/B 1 | FIC50 1 | Combination Effect |
|---|---|---|---|---|
| NS2A (A) | 40.15 | 0.35 | 0.5 | Synergism |
| NS4B (B) | 93 | 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanunza, E.; Grandi, N.; Quartu, M.; Carletti, F.; Ermellino, L.; Milia, J.; Corona, A.; Capobianchi, M.R.; Ippolito, G.; Tramontano, E. INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation. Viruses 2021, 13, 2448. https://doi.org/10.3390/v13122448
Fanunza E, Grandi N, Quartu M, Carletti F, Ermellino L, Milia J, Corona A, Capobianchi MR, Ippolito G, Tramontano E. INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation. Viruses. 2021; 13(12):2448. https://doi.org/10.3390/v13122448
Chicago/Turabian StyleFanunza, Elisa, Nicole Grandi, Marina Quartu, Fabrizio Carletti, Laura Ermellino, Jessica Milia, Angela Corona, Maria Rosaria Capobianchi, Giuseppe Ippolito, and Enzo Tramontano. 2021. "INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation" Viruses 13, no. 12: 2448. https://doi.org/10.3390/v13122448
APA StyleFanunza, E., Grandi, N., Quartu, M., Carletti, F., Ermellino, L., Milia, J., Corona, A., Capobianchi, M. R., Ippolito, G., & Tramontano, E. (2021). INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation. Viruses, 13(12), 2448. https://doi.org/10.3390/v13122448











